SakA Regulates Morphological Development, Ochratoxin A Biosynthesis and Pathogenicity of Aspergillus westerdijkiae and the Response to Different Environmental Stresses
Abstract
1. Introduction
2. Results
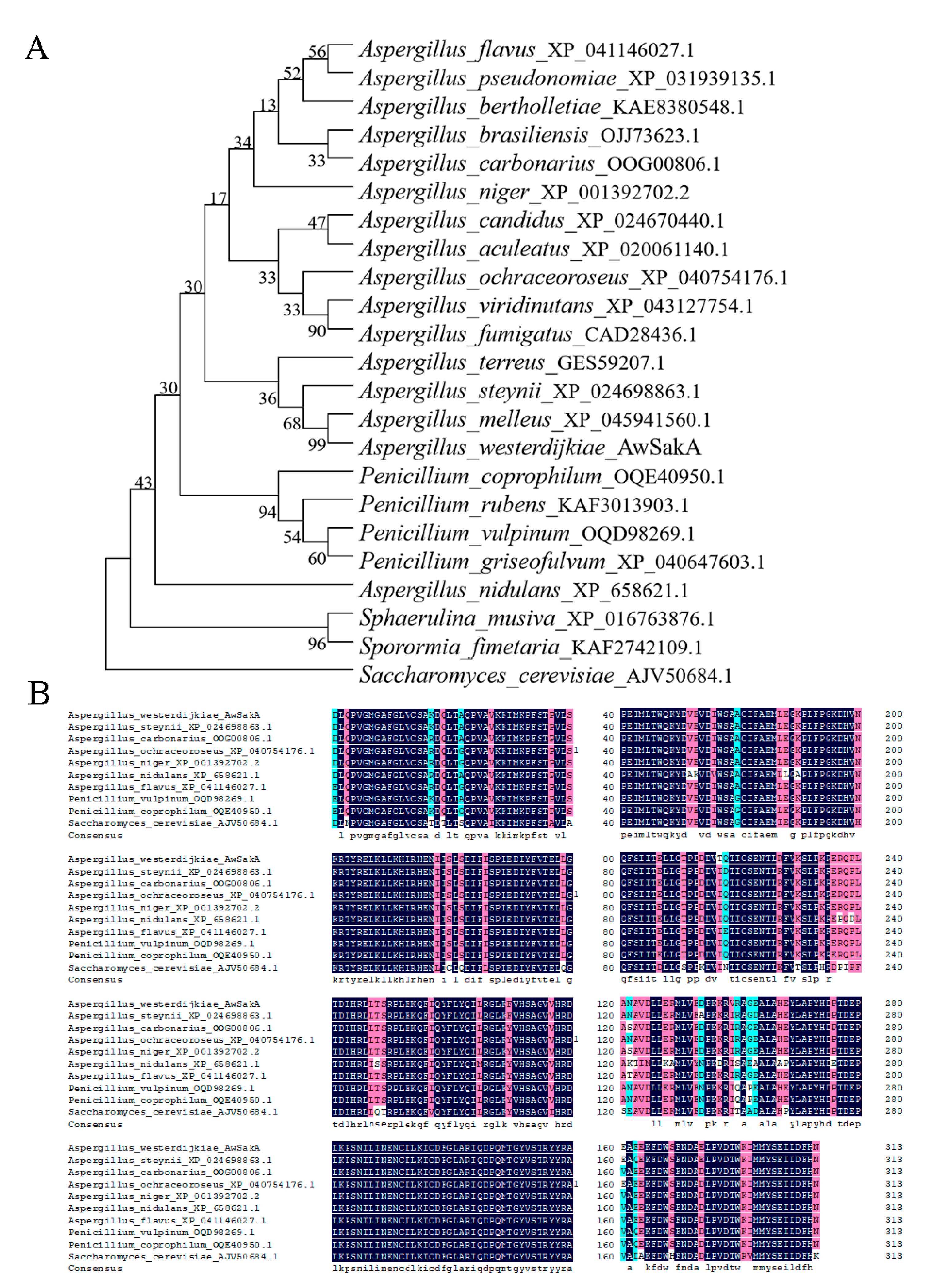
2.1. Identification of SakA in A. westerdijkiae
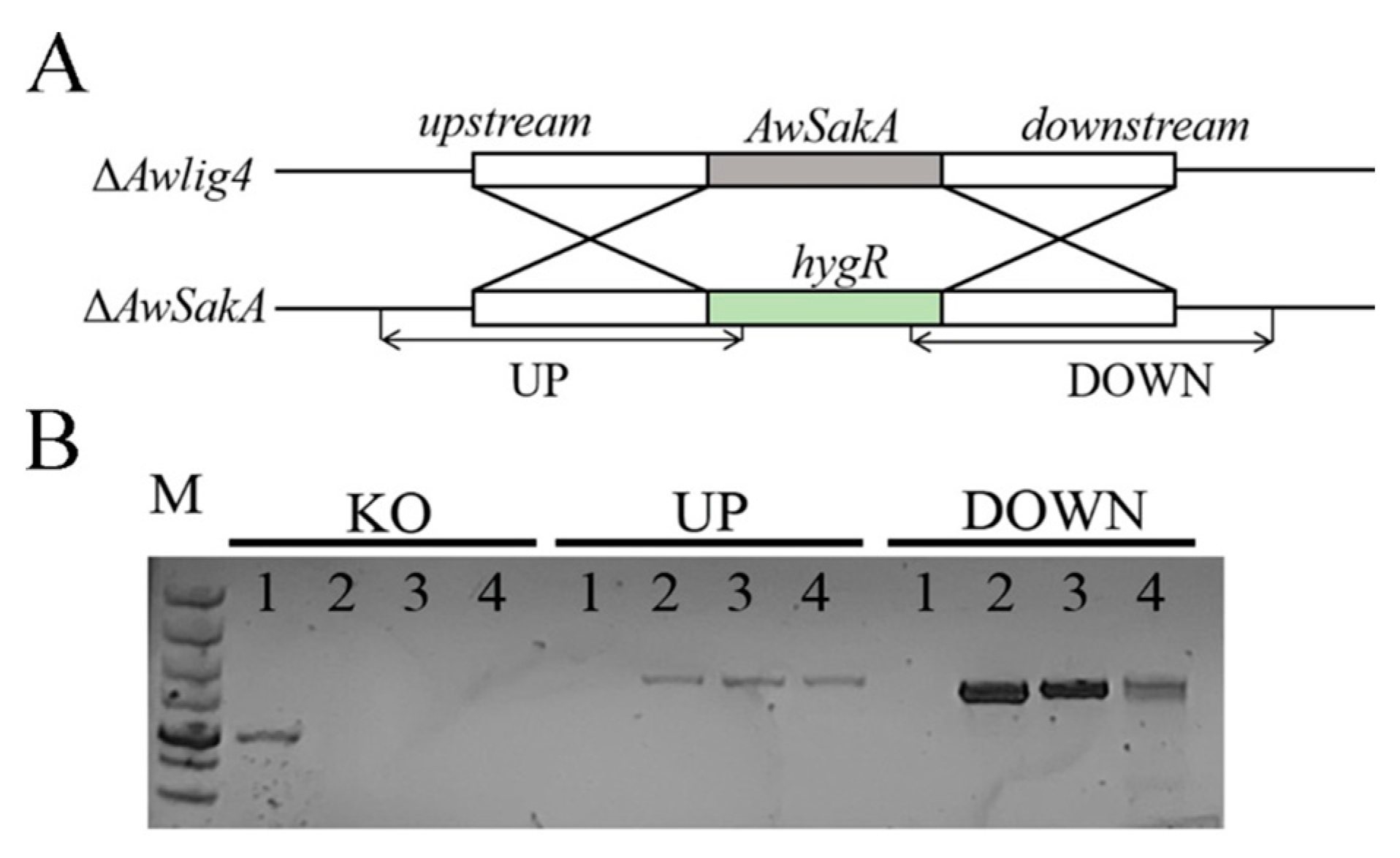
2.2. Generation of AwSakA Deletion Mutants
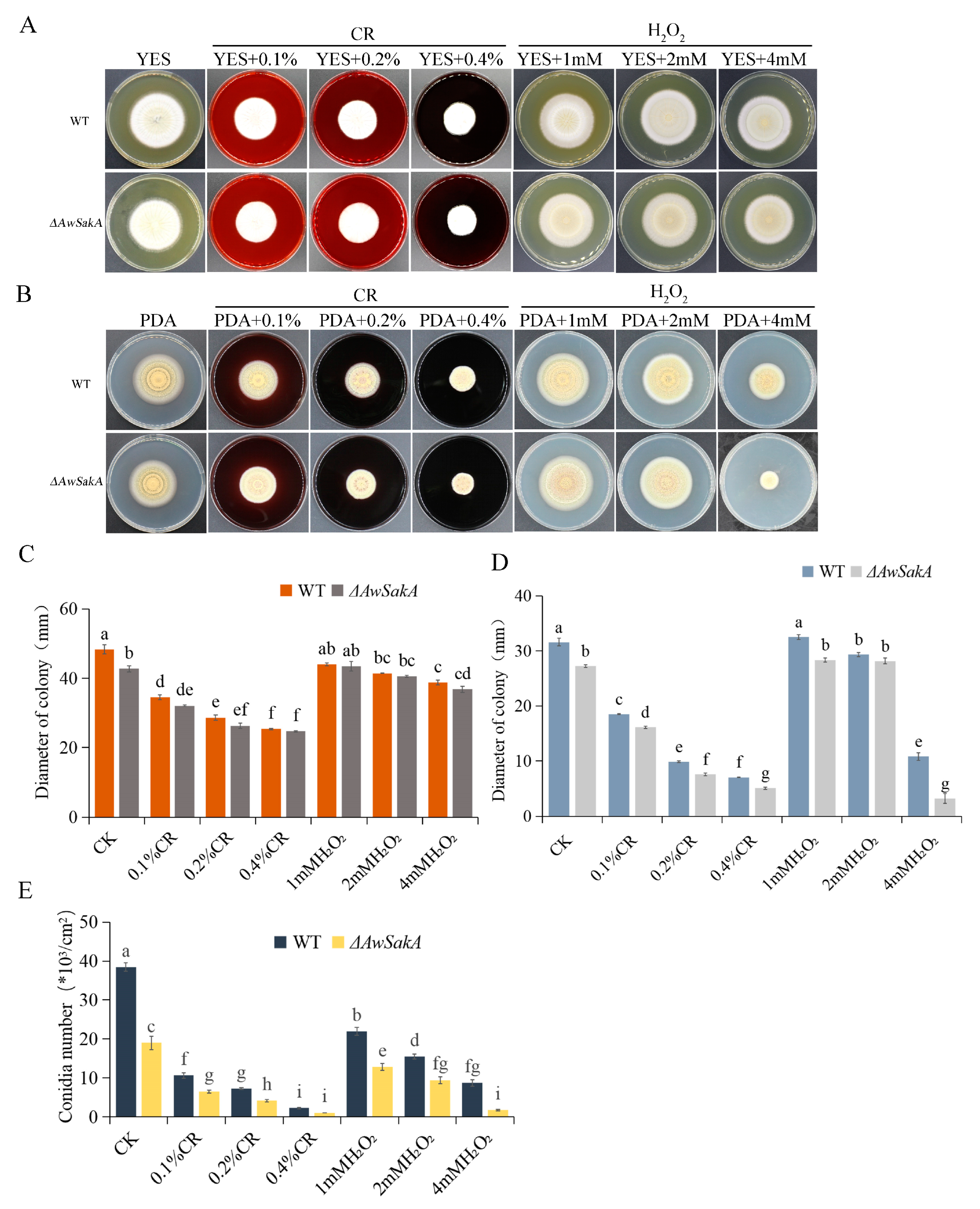
2.3. Growth, Conidiation and Sclerotia Formation of A. westerdijkiae Were Modulated by AwSakA
2.4. The Effect of Environmental Stress Agents on Growth and Conidial Production of A. westerdijkiae Are Regulated by AwSakA
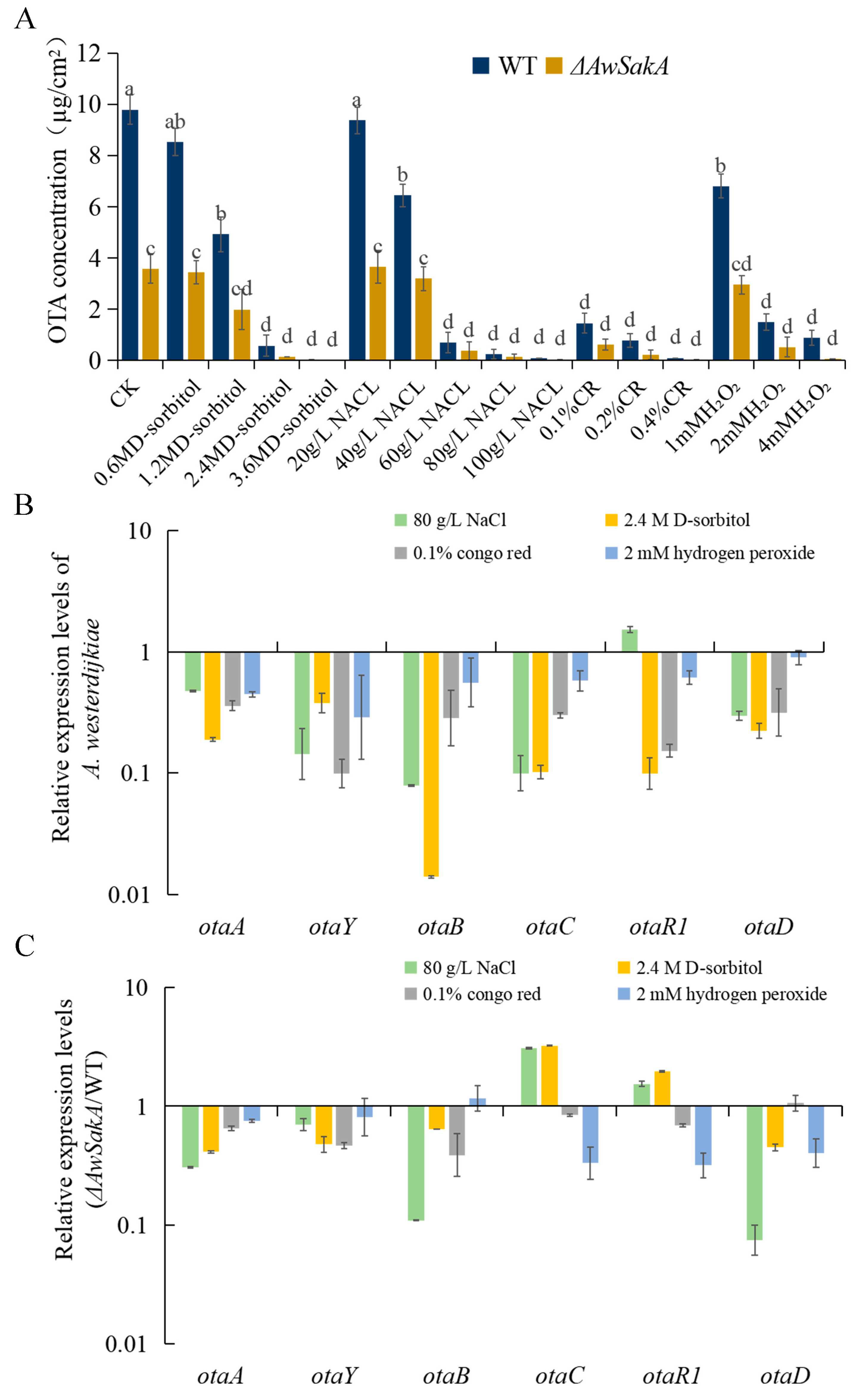
2.5. AwSakA Was Involved in the Biosynthesis of OTA
2.6. The Role of AwSakA in the Pathogenicity of A. westerdijkiae
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Strains and Culture Conditions
5.2. AwSakA Identification and Phylogenetic Analysis
5.3. Acquisition of SakA Nucleic Acid Sequences and Primer Design
5.4. Construction of Gene Deletion Mutant Strains
5.5. Growth Phenotype Analysis
5.6. OTA Production Analysis
5.7. Extraction of RNA and preparation of cDNA
5.8. Quantitative Real-Time Polymerase Chain Reaction Analysis
5.9. Pathogenicity Assay
5.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Malir, F.; Ostry, V.; Pfohl-Leszkowicz, A.; Malir, J.; Toman, J. Ochratoxin A: 50 Years of Research. Toxins 2016, 8, 191. [Google Scholar] [CrossRef] [PubMed]
- Malir, F.; Ostry, V.; Novotna, E. Toxicity of the mycotoxin ochratoxin A in the light of recent data. Toxin Rev. 2013, 32, 19–33. [Google Scholar] [CrossRef]
- Pfohl-Leszkowicz, A. Ochratoxin A and aristolochic acid involvement in nephropathies and associated urothelial tract tumours. Arh. Hig. Rada Toksikol. 2009, 60, 465–482. [Google Scholar] [CrossRef]
- IARC (International Agency for Research on Cancer). Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; IARC: Lyon, France, 1993; Volume 56, pp. 489–521.6. [Google Scholar]
- Wang, Y.; Wang, L.; Wu, F.; Liu, F.; Wang, Q.; Zhang, X.; Selvaraj, J.N.; Zhao, Y.; Xing, F.; Yin, W.-B.; et al. A Consensus Ochratoxin A Biosynthetic Pathway: Insights from the Genome Sequence of Aspergillus ochraceus and a Comparative Genomic Analysis. Appl. Environ. Microbiol. 2018, 84, e01009–e01018. [Google Scholar] [CrossRef] [PubMed]
- Gil-Serna, J.; Vazquez, C.; Sardinas, N.; Gonzalez-Jaen, M.T.; Patino, B. Discrimination of the main Ochratoxin A-producing species in Aspergillus section Circumdati by specific PCR assays. Int. J. Food Microbiol. 2009, 136, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Visagie, C.M.; Varga, J.; Houbraken, J.; Meijer, M.; Kocsube, S.; Yilmaz, N.; Fotedar, R.; Seifert, K.A.; Frisvad, J.C.; Samson, R.A. Ochratoxin production and taxonomy of the yellow aspergilli (Aspergillus section Circumdati). Stud. Mycol. 2014, 78, 1–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Liu, F.; Wang, Q.; Selvaraj, J.N.; Xing, F.; Zhao, Y.; Liu, Y. Ochratoxin A Producing Fungi, Biosynthetic Pathway and Regulatory Mechanisms. Toxins 2016, 8, 83. [Google Scholar] [CrossRef]
- Gil-Serna, J.; Patino, B.; Cortes, L.; Gonzalez-Jaen, M.T.; Vazquez, C. Aspergillus steynii and Aspergillus westerdijkiae as potential risk of OTA contamination in food products in warm climates. Int. J. Food Microbiol. 2015, 46, 168–175. [Google Scholar] [CrossRef]
- Vipotnik, Z.; Rodriguez, A.; Rodrigues, P. Aspergillus westerdijkiae as a major ochratoxin A risk in dry-cured ham based-media. Int. J. Food Microbiol. 2017, 241, 244–251. [Google Scholar] [CrossRef]
- Tumukunde, E.; Li, D.; Qin, L.; Li, Y.; Shen, J.; Wang, S.; Yuan, J. Osmotic-Adaptation Response of sakA/hogA Gene to Aflatoxin Biosynthesis, Morphology Development and Pathogenicity in Aspergillus flavus . Toxins 2019, 11, 41. [Google Scholar] [CrossRef]
- Lewis, T.S.; Shapiro, P.S.; Ahn, N.G. Signal Transduction through MAP Kinase Cascades. Adv. Cancer Res. 1998, 74, 49–139. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gibson, T.B.; Robinson, F.; Silvestro, L.; Pearson, G.; Xu, B.E.; Wright, A.; Vanderbilt, C.; Cobb, M.H. MAP Kinases. Chem. Rev. 2001, 101, 2449–2476. [Google Scholar] [CrossRef] [PubMed]
- Takayama, T.; Yamamoto, K.; Saito, H.; Tatebayashi, K. Interaction between the transmembrane domains of Sho1 and Opy2 enhances the signaling efficiency of the Hog1 MAP kinase cascade in Saccharomyces cerevisiae . PLoS ONE 2019, 14, e0211380. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.L.; Xia, L.M.; Wang, P.; Zhu, L.H.; Ye, J.R.; Huang, L. The MAPKKK CgMck1 Is Required for Cell Wall Integrity, Appressorium Development, and Pathogenicity in Colletotrichum gloeosporioides . Genes 2018, 9, 543. [Google Scholar] [CrossRef]
- Liao, X.; Long, X.; He, Q.; Song, M.; Li, X.; Liu, W.; Zhang, Y.; Lin, C.; Miao, W. Screening of binding proteins that interact with two components of the HOG MAPK pathway by the yeast two-hybrid method in Colletotrichum siamense . Eur. J. Plant Pathol. 2021, 159, 949–958. [Google Scholar] [CrossRef]
- Stoll, D.; Schmidt-Heydt, M.; Geisen, R. Differences in the regulation of ochratoxin A by the HOG pathway in Penicillium and Aspergillus in response to high osmolar environments. Toxins 2013, 5, 1282–1298. [Google Scholar] [CrossRef]
- Kawasaki, L.; Sánchez, O.; Shiozaki, K.; Aguirre, J. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans . Mol. Microbiol. 2002, 45, 1153–1163. [Google Scholar] [CrossRef]
- Jaimes-Arroyo, R.; Lara-Rojas, F.; Bayram, O.; Valerius, O.; Braus, G.H.; Aguirre, J. The SrkA Kinase Is Part of the SakA Mitogen-Activated Protein Kinase Interactome and Regulates Stress Responses and Development in Aspergillus nidulans . Eukaryot. Cell 2015, 14, 495–510. [Google Scholar] [CrossRef]
- Zhang, Y.; Choi, Y.E.; Zou, X.; Xu, J.R. The FvMK1 mitogen-activated protein kinase gene regulates conidiation, pathogenesis, and fumonisin production in Fusarium verticillioides . Fungal Genet. Biol. 2011, 48, 71–79. [Google Scholar] [CrossRef]
- Nimmanee, P.; Woo, P.C.; Kummasook, A.; Vanittanakom, N. Characterization of sakA gene from pathogenic dimorphic fungus Penicillium marneffei . Int. J. Med. Microbiol. 2015, 305, 65–74. [Google Scholar] [CrossRef]
- Furukawa, K.; Hoshi, Y.; Maeda, T.; Nakajima, T.; Abe, K. Aspergillus nidulans HOG pathway is activated only by two-component signalling pathway in response to osmotic stress. Mol. Microbiol. 2005, 56, 1246–1261. [Google Scholar] [CrossRef] [PubMed]
- Graf, E.; Schmidt-Heydt, M.; Geisen, R. HOG MAP kinase regulation of alternariol biosynthesis in Alternaria alternata is important for substrate colonization. Int. J. Food Microbiol. 2012, 157, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Igbalajobi, O.; Gao, J.; Fischer, R. The HOG Pathway Plays Different Roles in Conidia and Hyphae During Virulence of Alternaria alternata . Mol. Plant-Microbe Interact. 2020, 33, 1405–1410. [Google Scholar] [CrossRef] [PubMed]
- Manfiolli, A.O.; Mattos, E.C.; De Assis, L.J.; Silva, L.P.; Ulaş, M.; Brown, N.A.; Silva-Rocha, R.; Bayram, Ö.; Goldman, G.H. Aspergillus fumigatus high osmolarity glycerol mitogen activated protein kinases SakA and MpkC physically interact during osmotic and cell wall stresses. Front. Microbiol. 2019, 10, 918. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Bazan, V.; Jaimes-Arroyo, R.; Sanchez, O.; Lara-Rojas, F.; Aguirre, J. SakA and MpkC Stress MAPKs Show Opposite and Common Functions During Stress Responses and Development in Aspergillus nidulans . Front. Microbiol. 2018, 9, 2518. [Google Scholar] [CrossRef]
- Schmidt-Heydt, M.; Stoll, D.A.; Mrohs, J.; Geisen, R. Intraspecific variability of HOG1 phosphorylation in Penicillium verrucosum reflects different adaptation levels to salt rich habitats. Int. J. Food Microbiol. 2013, 165, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Duran, R.; Cary, J.W.; Calvo, A.M. Role of the osmotic stress regulatory pathway in morphogenesis and secondary metabolism in filamentous fungi. Toxins 2010, 2, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, Y.; Yang, B.; Li, E.; Wu, W.; Si, P.; Xing, F. AwAreA Regulates Morphological Development, Ochratoxin A Production, and Fungal Pathogenicity of Food Spoilage Fungus Aspergillus westerdijkiae Revealed by an Efficient Gene Targeting System. Front. Microbiol. 2022, 13, 857726. [Google Scholar] [CrossRef]
- Nguyen, A.N.; Lee, A.; Place, W.; Shiozaki, K. Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol. Biol. Cell 2000, 11, 1169–1181. [Google Scholar] [CrossRef]
- Buck, V.; Quinn, J.; Soto Pino, T.; Martin, H.; Saldanha, J.; Makino, K.; Morgan, B.A.; Millar, J.B. Peroxide sensors for the fission yeast stress-activated mitogen-activated protein kinase pathway. Mol. Biol. Cell 2001, 12, 407–419. [Google Scholar] [CrossRef]
- Ma, L.; Li, X.; Xing, F.; Ma, J.; Ma, X.; Jiang, Y. Fus3, as a Critical Kinase in MAPK Cascade, Regulates Aflatoxin Biosynthesis by Controlling the Substrate Supply in Aspergillus flavus, Rather than the Cluster Genes Modulation. Microbiol. Spectr. 2022, 10, e01221–e01269. [Google Scholar] [CrossRef] [PubMed]
- Rubenstein, E.M.; McCartney, R.R.; Zhang, C.; Shokat, K.M.; Shirra, M.K.; Arndt, K.M.; Schmidt, M.C. Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase. J. Biol. Chem. 2008, 283, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Román, E.; Arana, D.M.; Nombela, C.; Alonso-Monge, R.; Pla, J. MAP kinase pathways as regulators of fungal virulence. Trends Microbiol. 2007, 15, 181–190. [Google Scholar] [CrossRef]
- Mattos, E.C.; Silva, L.P.; Valero, C.; Castro, P.A.d.; Reis, T.F.d.; Ribeiro, L.F.C.; Marten, M.R.; Silva-Rocha, R.; Westmann, C.; Silva, C.H.T.d.P.d.; et al. The Aspergillus fumigatus Phosphoproteome Reveals Roles of High-Osmolarity Glycerol Mitogen-Activated Protein Kinases in Promoting Cell Wall Damage and Caspofungin Tolerance. mBio 2020, 11, e02919–e02962. [Google Scholar] [CrossRef] [PubMed]
- Bourret, R.B.; Kennedy, E.N.; Foster, C.A.; Sepúlveda, V.E.; Goldman, W.E. A radical reimagining of fungal two-component regulatory systems. Trends Microbiol. 2021, 29, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Roman, E.; Alonso-Monge, R.; Gong, Q.; Li, D.; Calderone, R.; Pla, J. The Cek1 MAPK is a short-lived protein regulated by quorum sensing in the fungal pathogen Candida albicans. FEMS Yeast Res. 2009, 9, 942–955. [Google Scholar] [CrossRef]
- Frawley, D.; Bayram, Ö. The pheromone response module, a mitogen-activated protein kinase pathway implicated in the regulation of fungal development, secondary metabolism and pathogenicity. Fungal Genet. Biol. 2020, 144, 103469. [Google Scholar] [CrossRef]
- Lara-Rojas, F.; Sanchez, O.; Kawasaki, L.; Aguirre, J. Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol. Microbiol. 2011, 80, 436–454. [Google Scholar] [CrossRef]
- Du, C.; Sarfati, J.; Latge, J.P.; Calderone, R. The role of the sakA (Hog1) and tcsB (sln1) genes in the oxidant adaptation of Aspergillus fumigatus . Med. Mycol. 2006, 44, 211–218. [Google Scholar] [CrossRef]
- Ochiai, N.; Tokai, T.; Nishiuchi, T.; Takahashi-Ando, N.; Fujimura, M.; Kimura, M. Involvement of the osmosensor histidine kinase and osmotic stress-activated protein kinases in the regulation of secondary metabolism in Fusarium graminearum . Biochem. Biophys. Res. Commun. 2007, 363, 639–644. [Google Scholar] [CrossRef]
- Alves de Castro, P.; Dos Reis, T.F.; Dolan, S.K.; Oliveira Manfiolli, A.; Brown, N.A.; Jones, G.W.; Doyle, S.; Riano-Pachon, D.M.; Squina, F.M.; Caldana, C.; et al. The Aspergillus fumigatus SchA(SCH9) kinase modulates SakA(HOG1) MAP kinase activity and it is essential for virulence. Mol. Microbiol. 2016, 102, 642–671. [Google Scholar] [CrossRef] [PubMed]
- Bruder Nascimento, A.C.M.d.O.; Dos Reis, T.F.; de Castro, P.A.; Hori, J.I.; Bom, V.L.P.; de Assis, L.J.; Ramalho, L.N.Z.; Rocha, M.C.; Malavazi, I.; Brown, N.A. Mitogen activated protein kinases SakAHOG1 and MpkC collaborate for Aspergillus fumigatus virulence. Mol. Microbiol. 2016, 100, 841–859. [Google Scholar] [CrossRef] [PubMed]
- Fabri, J.; Godoy, N.L.; Rocha, M.C.; Munshi, M.; Cocio, T.A.; von Zeska Kress, M.R.; Fill, T.P.; da Cunha, A.F.; Del Poeta, M.; Malavazi, I. The AGC Kinase YpkA Regulates Sphingolipids Biosynthesis and Physically Interacts with SakA MAP Kinase in Aspergillus fumigatus . Front. Microbiol. 2018, 9, 3347. [Google Scholar] [CrossRef] [PubMed]
- Román, E.; Correia, I.; Prieto, D.; Alonso, R.; Pla, J. The HOG MAPK pathway in Candida albicans: More than an osmosensing pathway. Int. Microbiol. 2020, 23, 23–29. [Google Scholar] [CrossRef]
- Liang, J.; Slingerland, J.M. Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle 2003, 2, 336–342. [Google Scholar] [CrossRef]
- Hemmings, B.A.; Restuccia, D.F. The PI3K-PKB/Akt Pathway. Cold Spring Harb. Perspect. Biol. 2015, 7, a011189. [Google Scholar] [CrossRef] [PubMed]
- Ikner, A.; Shiozaki, K. Yeast signaling pathways in the oxidative stress response. Mutat. Res. 2005, 569, 13–27. [Google Scholar] [CrossRef]
- Wang, G.; Wang, Y.; Yang, B.; Zhang, C.; Zhang, H.; Xing, F.; Liu, Y. Carbon Catabolite Repression Gene AoCreA Regulates Morphological Development and Ochratoxin A Biosynthesis Responding to Carbon Sources in Aspergillus ochraceus . Toxins 2020, 12, 697. [Google Scholar] [CrossRef]
- Han, X.; Chakrabortti, A.; Zhu, J.; Liang, Z.X.; Li, J. Sequencing and functional annotation of the whole genome of the filamentous fungus Aspergillus westerdijkiae . BMC Genom. 2016, 17, 1–14. [Google Scholar] [CrossRef]
- Yu, J.H.; Hamari, Z.; Han, K.H.; Seo, J.A.; Reyes-Dominguez, Y.; Scazzocchio, C. Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, H.; Wang, Y.; Liu, F.; Li, E.; Ma, J.; Yang, B.; Zhang, C.; Li, L.; Liu, Y. Requirement of LaeA, VeA, and VelB on Asexual Development, Ochratoxin A Biosynthesis, and Fungal Virulence in Aspergillus ochraceus . Front. Microbiol. 2019, 10, 2759. [Google Scholar] [CrossRef] [PubMed]




| Primers | Sequence (5′ to 3′) |
|---|---|
| SakA-up-F | CAGCCCAGATTGTATCTGAC |
| SakA-up-R | GCATTGATGTGTTGACCTCCCAGCCATACCAGACCAGTCC |
| SakA-down-F | CGAGGGCAAAGGAATAGAGTAGACGATGCTGAGCTACCAGTG |
| SakA-down-R | AGATCGCGTCAATAACTCCG |
| SakA-knock-F | TTAGGCATCGTGGGCTGACAC |
| SakA-knock-R | AACTTCTCTTCCGCCTCAGG |
| hygR-F | GGAGGTCAACACATCAATGCCTATTTTGGT |
| hygR-R | CTACTCTATTCCTTTGCCCTCGGACGAGTG |
| N-SakA-F | AGGAACATCTTGCATGGGCA |
| N-SakA-R | TCCTGGTGATACGGAGGGAG |
| C-SakA-up-R | GGCTGATCTGACCAGTTGCC |
| C-SakA-down-F | GGCTGTGTAGAAGTACTCGCC |
| Primers | Sequence (5′ to 3′) |
|---|---|
| AwGADPH-RT-F | CGGCAAGAAGGTTCAGTT |
| AwGADPH-RT R | CTCGTTGGTGGTGAAGAC |
| AwotaA-RT-F | CGCCACGTCAATAAGTCTCGG |
| AwotaA-RT-R | GTATGGAGCGTGCAGATCTG |
| AwotaY-RT-F | ACACACAATCGTGAACAAGCG |
| AwotaY-RT-R | CATCCAGAAGTCCTCCACCG |
| AwotaB-RT-F | CTCGGCTACCTGCCTTCATG |
| AwotaB-RT-R | CAATGCCAACGCAATCAACG |
| AwotaC-RT-F | CGTGCGTTCAACTACCTAGACG |
| AwotaC-RT-R | CCTAGCCTCGGTGACATCAG |
| AwotaR1-RT-F | CCAGGACTCGTTCAGTCTCC |
| AwotaR1-RT-R | GAGCACCCTGCGACATCATG |
| AwotaD-RT-F | GCTGAACAACCAGAAGGAAGC |
| AwotaD-RT-R | GCCATCTCCTGGTTCATGACC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Si, P.; Wang, G.; Wu, W.; Hussain, S.; Guo, L.; Wu, W.; Yang, Q.; Xing, F. SakA Regulates Morphological Development, Ochratoxin A Biosynthesis and Pathogenicity of Aspergillus westerdijkiae and the Response to Different Environmental Stresses. Toxins 2023, 15, 292. https://doi.org/10.3390/toxins15040292
Si P, Wang G, Wu W, Hussain S, Guo L, Wu W, Yang Q, Xing F. SakA Regulates Morphological Development, Ochratoxin A Biosynthesis and Pathogenicity of Aspergillus westerdijkiae and the Response to Different Environmental Stresses. Toxins. 2023; 15(4):292. https://doi.org/10.3390/toxins15040292
Chicago/Turabian StyleSi, Peidong, Gang Wang, Wenqing Wu, Sarfaraz Hussain, Ling Guo, Wei Wu, Qingli Yang, and Fuguo Xing. 2023. "SakA Regulates Morphological Development, Ochratoxin A Biosynthesis and Pathogenicity of Aspergillus westerdijkiae and the Response to Different Environmental Stresses" Toxins 15, no. 4: 292. https://doi.org/10.3390/toxins15040292
APA StyleSi, P., Wang, G., Wu, W., Hussain, S., Guo, L., Wu, W., Yang, Q., & Xing, F. (2023). SakA Regulates Morphological Development, Ochratoxin A Biosynthesis and Pathogenicity of Aspergillus westerdijkiae and the Response to Different Environmental Stresses. Toxins, 15(4), 292. https://doi.org/10.3390/toxins15040292






