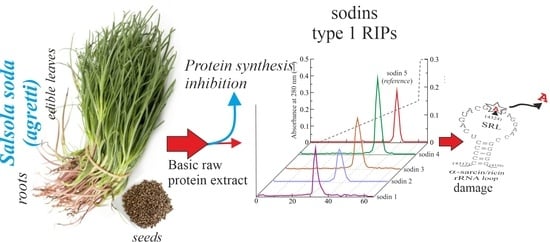
Isolation, Characterization and Biological Action of Type-1 Ribosome-Inactivating Proteins from Tissues of Salsola soda L.
Abstract
:1. Introduction
2. Results and Discussion
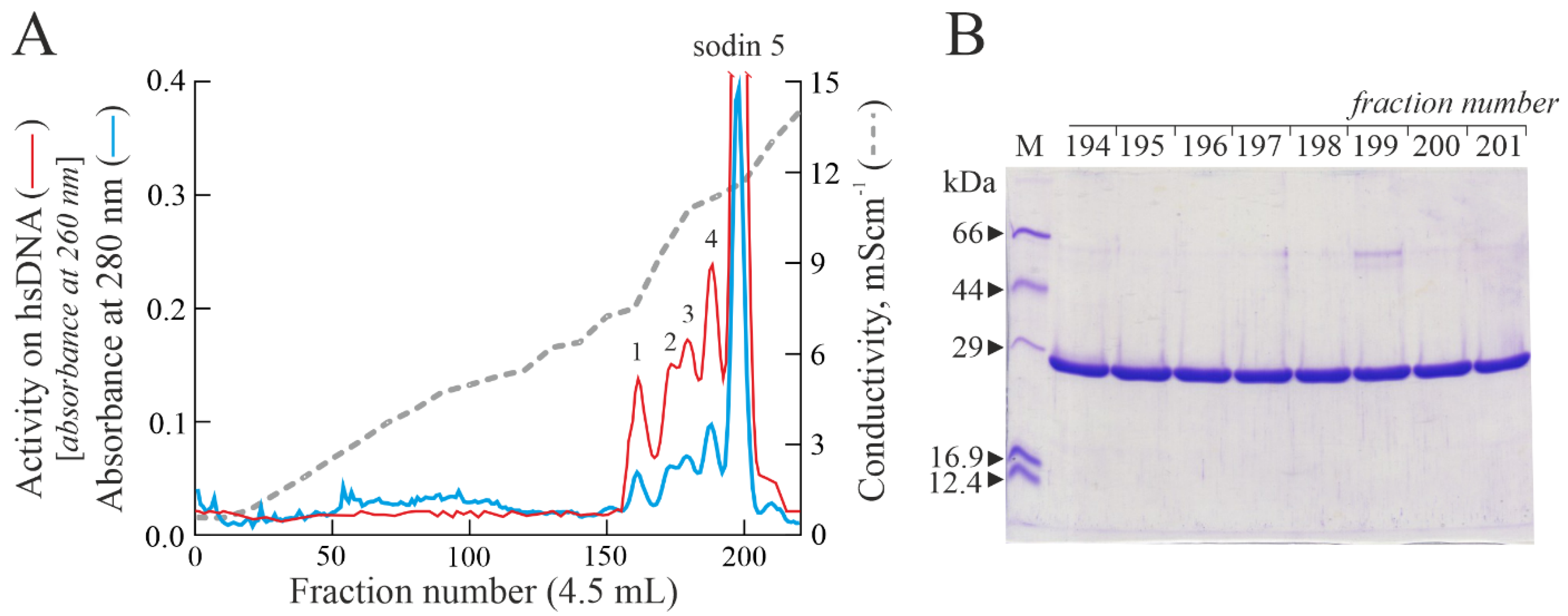
2.1. Purification of Type-1 RIPs from Salsola soda Seeds
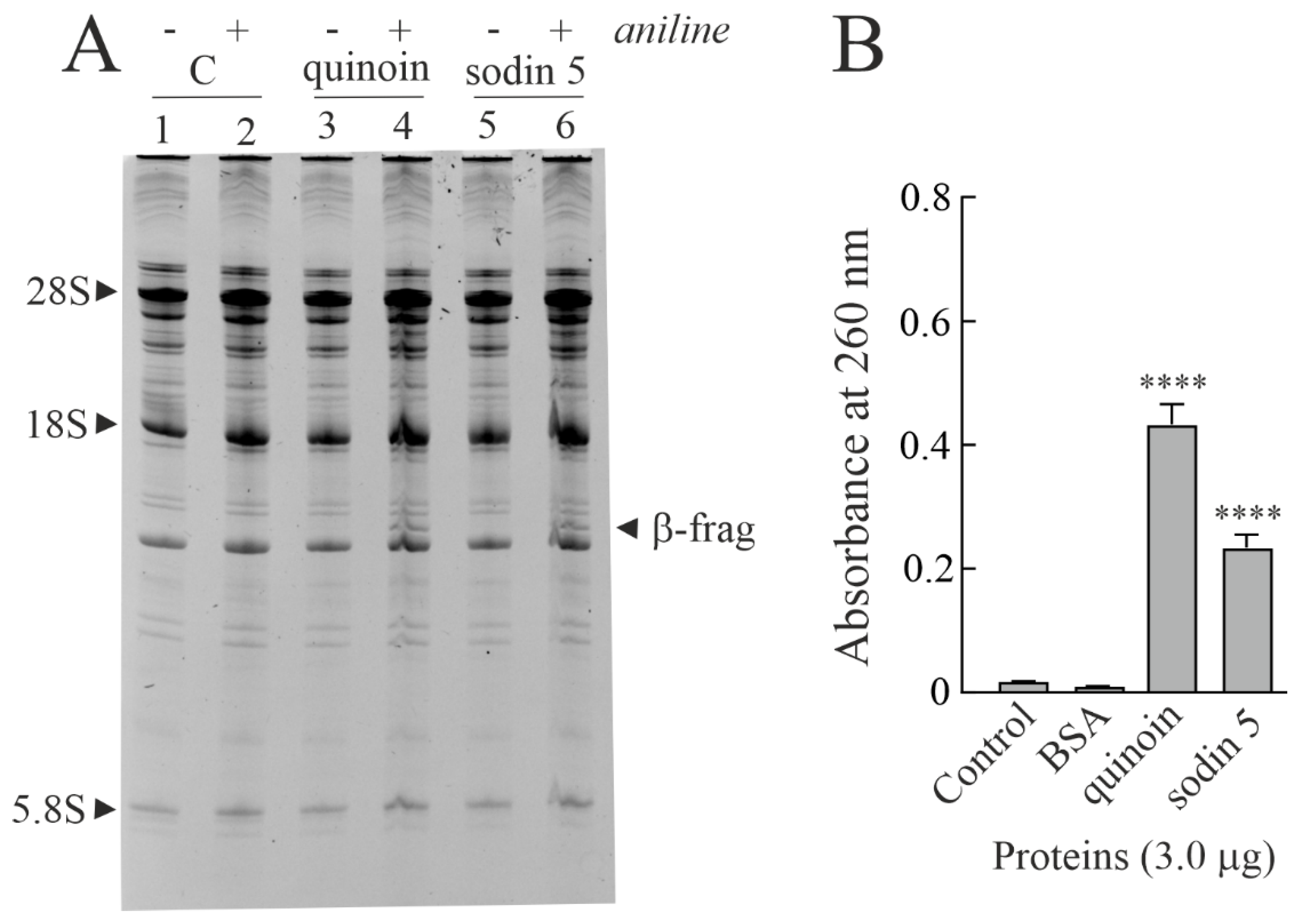
2.2. Enzymatic and Structural Features of Sodin 5
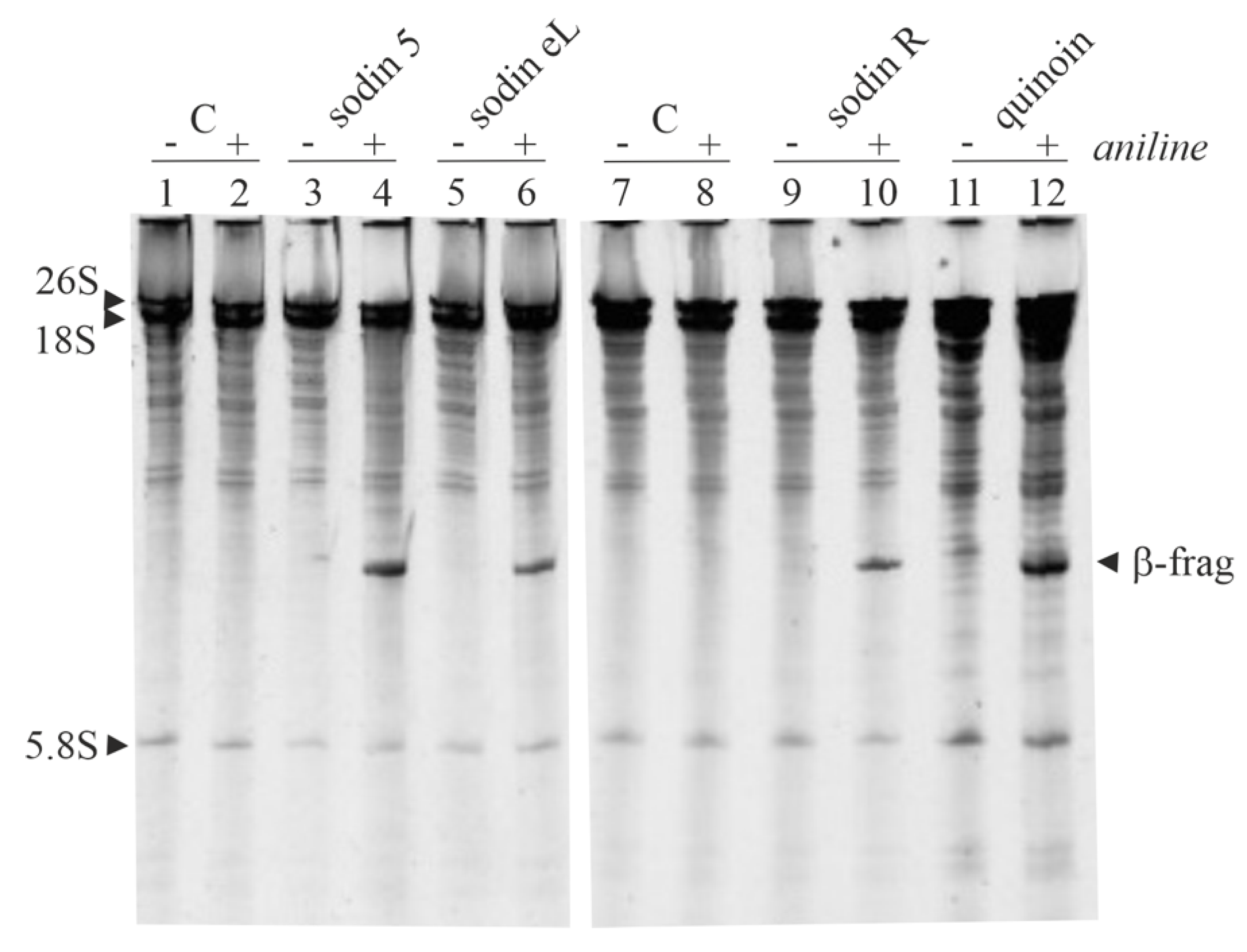
2.3. Minor Forms of Type-1 RIPs from Salsola soda Seeds
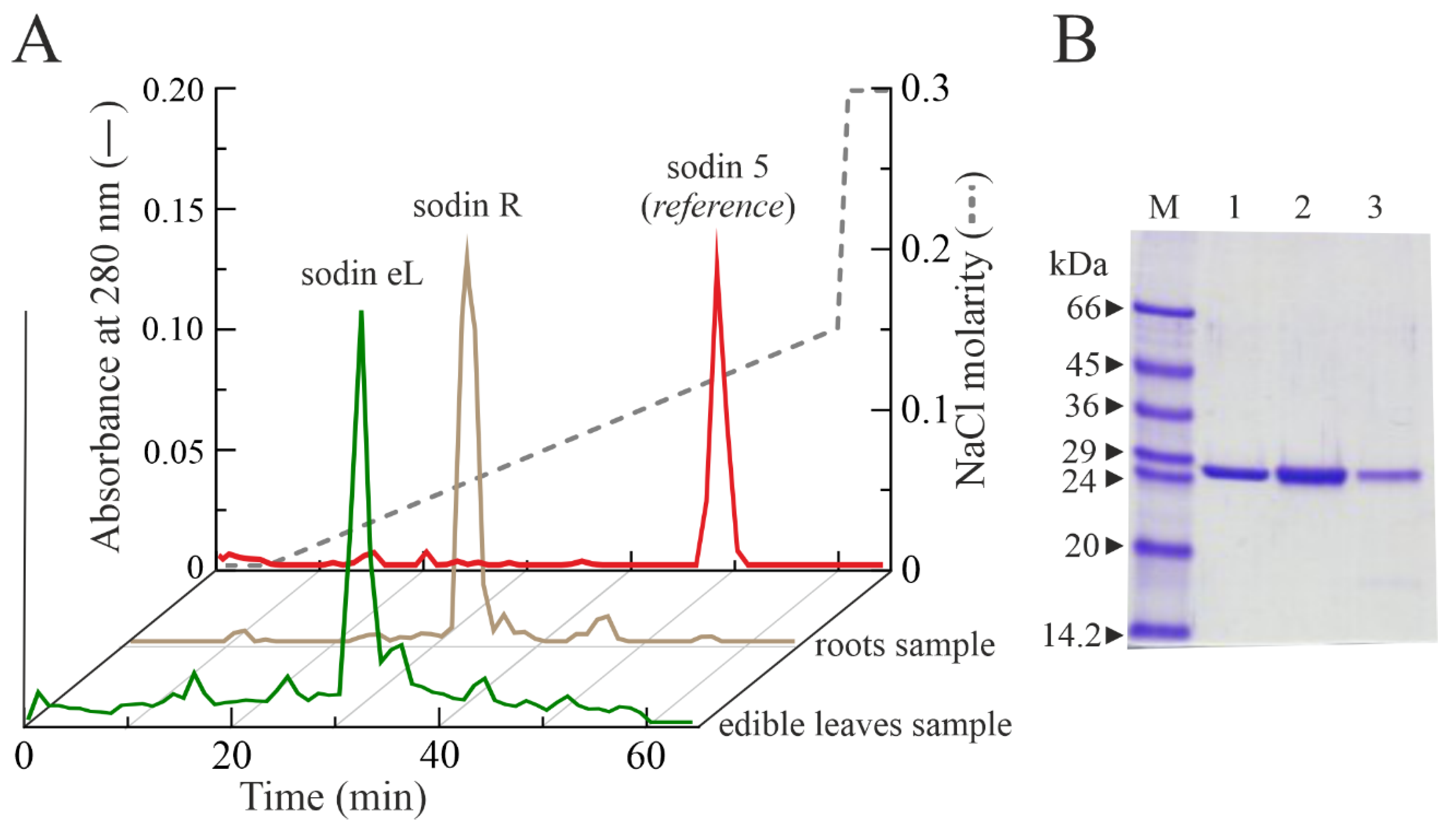
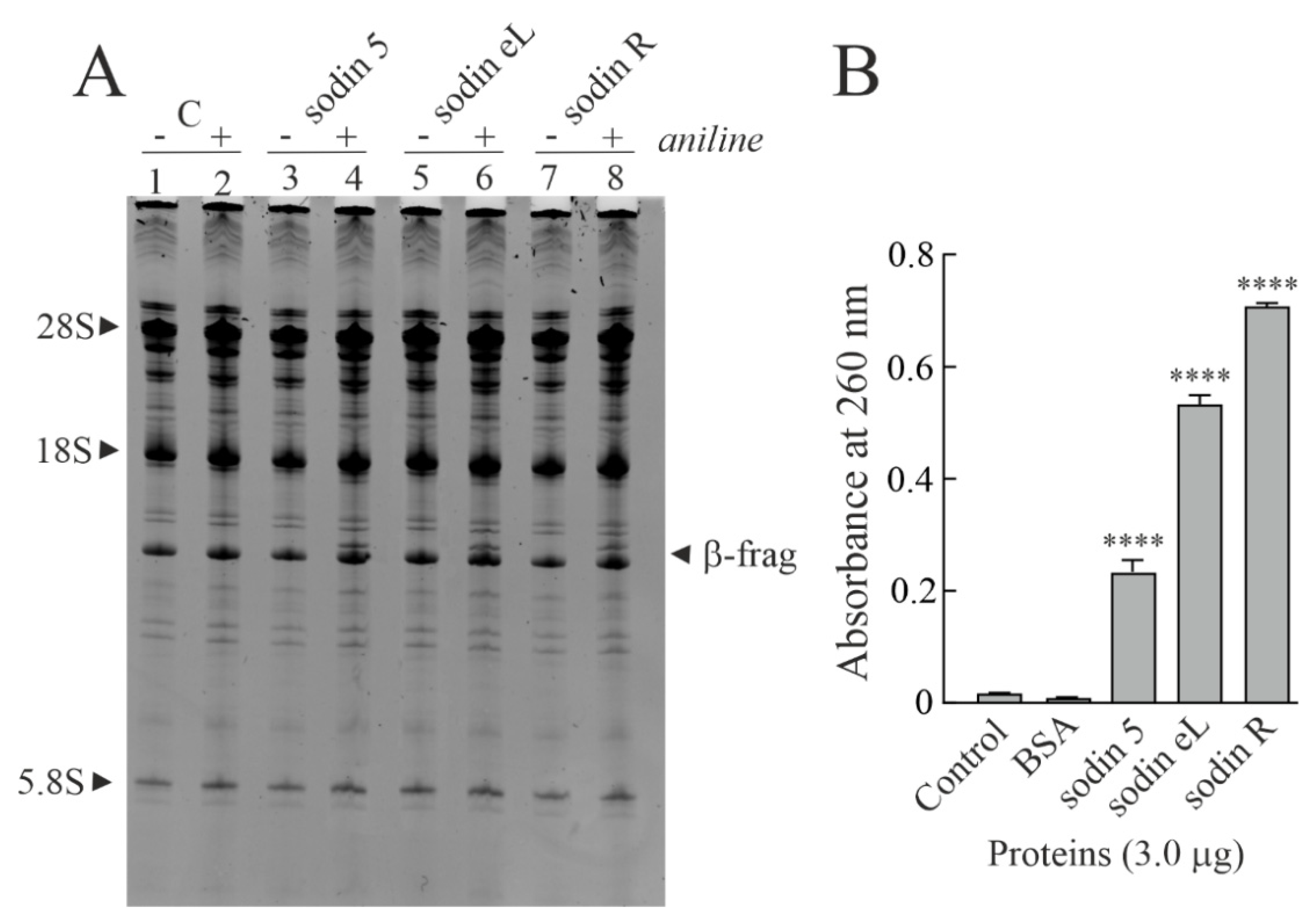
2.4. Type-1 RIPs from Edible Leaves and Roots of Salsola soda
2.5. Cytotoxic Effects of Sodins from S. soda Tissues in Cell Cultures
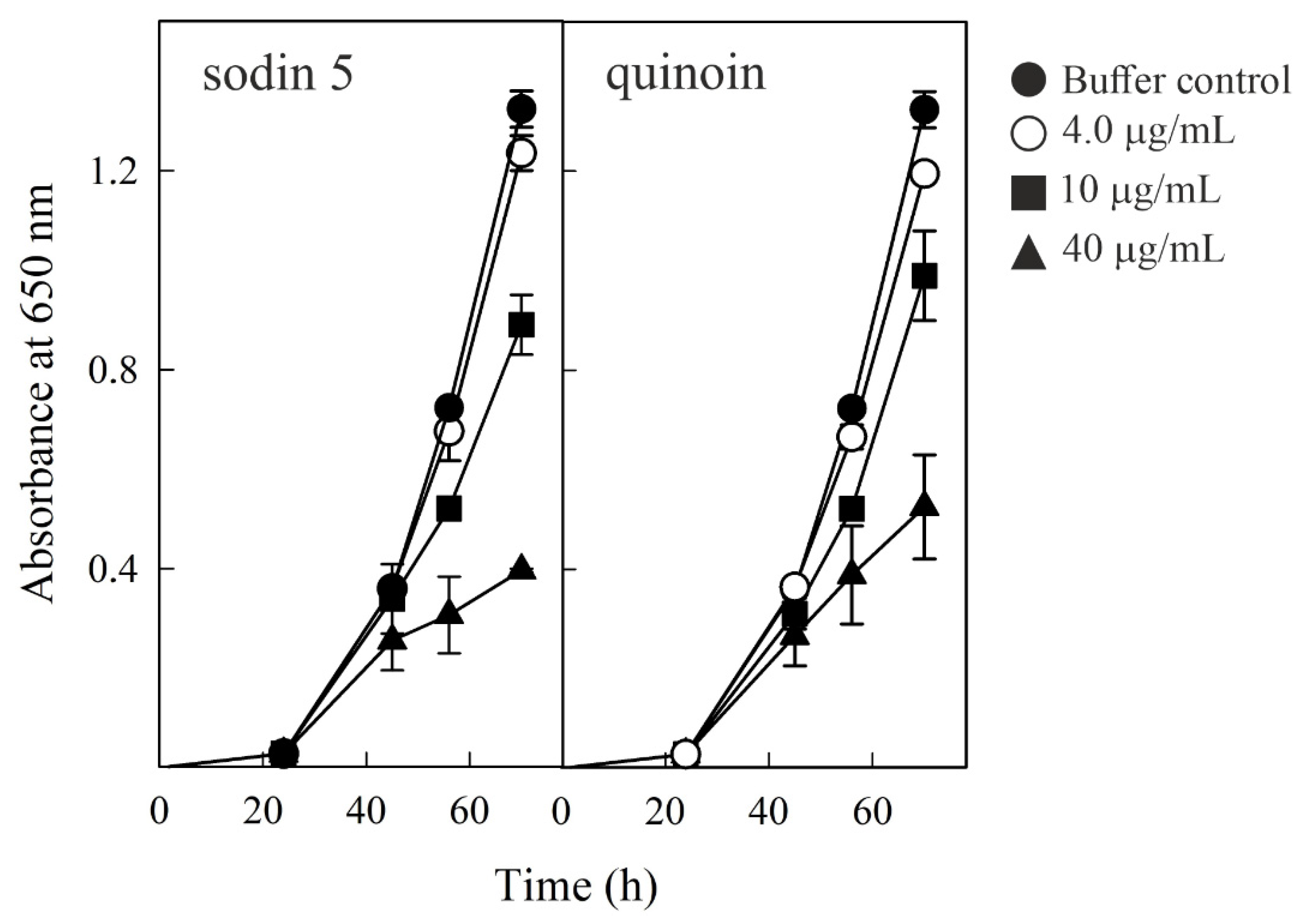
2.6. Effect of Sodin 5 and Quinoin on the Growth of P. digitatum
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Purification of Type-1 RIPs from Seeds, Roots and Edible Leaves of S. soda
4.3. Enzymatic Assays
4.3.1. rRNA N-Glycosylase Activity of RIPs on Rabbit Ribosomes
4.3.2. rRNA N-Glycosylase Activity of RIPs on Yeast Ribosomes
4.3.3. Polynucleotide: Adenosine Glycosylase Activity on Salmon Sperm DNA
4.3.4. Cell-Free Protein Synthesis Inhibition
4.4. Analytical Procedures
4.5. Circular Dichroism and Thermal Stability Determination
4.6. Cell Viability Assays
4.7. DNA Fragmentation Analysis
4.8. Antifungal Activity Measurements
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Endo, Y.; Huber, P.W.; Wool, I.G. The ribonuclease activity of the cytotoxin alpha-sarcin. The characteristics of the enzymatic activity of alpha-sarcin with ribosomes and ribonucleic acids as substrates. J. Biol. Chem. 1983, 258, 2662–2667. [Google Scholar] [CrossRef]
- Shi, X.; Khade, P.K.; Sanbonmatsu, K.Y.; Joseph, S. Functional role of the sarcin-ricin loop of the 23S rRNA in the elongation cycle of protein synthesis. J. Mol. Biol. 2012, 419, 125–138. [Google Scholar] [CrossRef]
- Barbieri, L.; Valbonesi, P.; Bonora, E.; Gorini, P.; Bolognesi, A.; Stirpe, F. Polynucleotide:adenosine glycosidase activity of ribosome-inactivating proteins: Effect on DNA, RNA and poly(A). Nucleic Acids Res. 1997, 25, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Valbonesi, P.; Righi, F.; Zuccheri, G.; Monti, F.; Gorini, P.; Samorí, B.; Stirpe, F. Polynucleotide:Adenosine glycosidase is the sole activity of ribosome-inactivating proteins on DNA. J. Biochem. 2000, 128, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Gorini, P.; Valbonesi, P.; Castiglioni, P.; Stirpe, F. Unexpected activity of saporins. Nature 1994, 372, 624. [Google Scholar] [CrossRef]
- Ruggiero, A.; Chambery, A.; Di Maro, A.; Mastroianni, A.; Parente, A.; Berisio, R. Crystallization and preliminary X-ray diffraction analysis of PD-L1, a highly glycosylated ribosome inactivating protein with DNase activity. Protein Pept. Lett. 2007, 14, 407–409. [Google Scholar] [CrossRef]
- Aceto, S.; Di Maro, A.; Conforto, B.; Siniscalco, G.G.; Parente, A.; Delli Bovi, P.; Gaudio, L. Nicking activity on pBR322 DNA of ribosome inactivating proteins from Phytolacca dioica L. leaves. Biol. Chem. 2005, 386, 307–317. [Google Scholar] [CrossRef]
- Mock, J.W.; Ng, T.B.; Wong, R.N.; Yao, Q.Z.; Yeung, H.W.; Fong, W.P. Demonstration of ribonuclease activity in the plant ribosome-inactivating proteins alpha- and beta-momorcharins. Life Sci. 1996, 59, 1853–1859. [Google Scholar] [CrossRef]
- Shih, N.R.; McDonald, K.A.; Jackman, A.P.; Girbés, T.; Iglesias, R. Bifunctional plant defence enzymes with chitinase and ribosome inactivating activities from Trichosanthes kirilowii cell cultures. Plant Sci. 1997, 130, 145–150. [Google Scholar] [CrossRef]
- Lombard, S.; Helmy, M.E.; Piéroni, G. Lipolytic activity of ricin from Ricinus sanguineus and Ricinus communis on neutral lipids. Biochem. J. 2001, 358, 773–781. [Google Scholar] [CrossRef]
- Li, X.-D.; Chen, W.-F.; Liu, W.-Y.; Wang, G.-H. Large-scale preparation of two new ribosome-inactivating proteins--cinnamomin and camphorin from the seeds of Cinnamomum camphora. Protein Expr. Purif. 1997, 10, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Stirpe, F. Ribosome-inactivating proteins. Toxicon 2004, 44, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Hao, Q.; Van Damme, E.J. Ribosome-inactivating proteins from plants: More than RNA N-glycosidases? FASEB J. 2001, 15, 1493–1506. [Google Scholar] [CrossRef] [PubMed]
- Day, P.J.; Lord, J.M.; Roberts, L.M. The deoxyribonuclease activity attributed to ribosome-inactivating proteins is due to contamination. Eur. J. Biochem. 1998, 258, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, A.; Bortolotti, M.; Maiello, S.; Battelli, M.G.; Polito, L. Ribosome-Inactivating Proteins from Plants: A Historical Overview. Molecules 2016, 21, 1627. [Google Scholar] [CrossRef] [PubMed]
- Di Maro, A.; Citores, L.; Russo, R.; Iglesias, R.; Ferreras, J.M. Sequence comparison and phylogenetic analysis by the Maximum Likelihood method of ribosome-inactivating proteins from angiosperms. Plant Mol. Biol 2014, 85, 575–588. [Google Scholar] [CrossRef]
- Landi, N.; Hussain, H.Z.F.; Pedone, P.V.; Ragucci, S.; Di Maro, A. Ribotoxic Proteins, Known as Inhibitors of Protein Synthesis, from Mushrooms and Other Fungi According to Endo’s Fragment Detection. Toxins 2022, 14, 403. [Google Scholar] [CrossRef]
- O’Loughlin, E.V.; Robins-Browne, R.M. Effect of Shiga toxin and Shiga-like toxins on eukaryotic cells. Microbes Infect. 2001, 3, 493–507. [Google Scholar] [CrossRef]
- Liu, R.S.; Yang, J.H.; Liu, W.Y. Isolation and enzymatic characterization of lamjapin, the first ribosome-inactivating protein from cryptogamic algal plant (Laminaria japonica A). Eur. J. Biochem. 2002, 269, 4746–4752. [Google Scholar] [CrossRef]
- Becker, W.; Apel, K. Isolation and characterization of a cDNA clone encoding a novel jasmonate-induced protein of barley (Hordeum vulgare L.). Plant Mol. Biol. 1992, 19, 1065–1067. [Google Scholar] [CrossRef]
- Chaudhry, B.; Müller-Uri, F.; Cameron-Mills, V.; Gough, S.; Simpson, D.; Skriver, K.; Mundy, J. The barley 60 kDa jasmonate-induced protein (JIP60) is a novel ribosome-inactivating protein. Plant J. 1994, 6, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.A.; Morgan, A.E.; Hey, T.D. Characterization and molecular cloning of a proenzyme form of a ribosome-inactivating protein from maize. Novel mechanism of proenzyme activation by proteolytic removal of a 2.8-kilodalton internal peptide segment. J. Biol. Chem. 1991, 266, 23422–23427. [Google Scholar] [CrossRef]
- Lapadula, W.J.; Sánchez Puerta, M.V.; Juri Ayub, M. Revising the taxonomic distribution, origin and evolution of ribosome inactivating protein genes. PLoS ONE 2013, 8, e72825. [Google Scholar] [CrossRef] [PubMed]
- Akkouh, O.; Ng, T.B.; Cheung, R.C.; Wong, J.H.; Pan, W.; Ng, C.C.; Sha, O.; Shaw, P.C.; Chan, W.Y. Biological activities of ribosome-inactivating proteins and their possible applications as antimicrobial, anticancer, and anti-pest agents and in neuroscience research. Appl. Microbiol. Biotechnol. 2015, 99, 9847–9863. [Google Scholar] [CrossRef]
- Rotondo, R.; Ragucci, S.; Castaldo, S.; Oliva, M.A.; Landi, N.; Pedone, P.V.; Arcella, A.; Di Maro, A. Cytotoxicity Effect of Quinoin, Type 1 Ribosome-Inactivating Protein from Quinoa Seeds, on Glioblastoma Cells. Toxins 2021, 13, 684. [Google Scholar] [CrossRef]
- Citores, L.; Iglesias, R.; Ferreras, J.M. Antiviral Activity of Ribosome-Inactivating Proteins. Toxins 2021, 13, 80. [Google Scholar] [CrossRef]
- Zhu, F.; Zhou, Y.-K.; Ji, Z.-L.; Chen, X.-R. The Plant Ribosome-Inactivating Proteins Play Important Roles in Defense against Pathogens and Insect Pest Attacks. Front. Plant Sci. 2018, 9, 146. [Google Scholar] [CrossRef]
- Pizzo, E.; Di Maro, A. A new age for biomedical applications of Ribosome Inactivating Proteins (RIPs): From bioconjugate to nanoconstructs. J. Biomed. Sci. 2016, 23, 54. [Google Scholar] [CrossRef] [PubMed]
- Hammer, K.; Pignone, D.; Cifarelli, S.; Perrino, P. Barilla (Salsola soda, Chenopodiaceae). Econ. Bot. 1990, 44, 410–412. [Google Scholar] [CrossRef]
- Iannuzzi, A.M.; Moschini, R.; De Leo, M.; Pineschi, C.; Balestri, F.; Cappiello, M.; Braca, A.; Del-Corso, A. Chemical profile and nutraceutical features of Salsola soda (agretti): Anti-inflammatory and antidiabetic potential of its flavonoids. Food Biosci. 2020, 37, 100713. [Google Scholar] [CrossRef]
- Murshid, S.S.A.; Atoum, D.; Abou-Hussein, D.R.; Abdallah, H.M.; Hareeri, R.H.; Almukadi, H.; Edrada-Ebel, R. Genus Salsola: Chemistry, Biological Activities and Future Prospective-A Review. Plants 2022, 11, 714. [Google Scholar] [CrossRef] [PubMed]
- Ragucci, S.; Bulgari, D.; Landi, N.; Russo, R.; Clemente, A.; Valletta, M.; Chambery, A.; Gobbi, E.; Faoro, F.; Di Maro, A. The Structural Characterization and Antipathogenic Activities of Quinoin, a Type 1 Ribosome-Inactivating Protein from Quinoa Seeds. Int. J. Mol. Sci. 2021, 22, 8964. [Google Scholar] [CrossRef] [PubMed]
- Chow, T.P.; Feldman, R.A.; Lovett, M.; Piatak, M. Isolation and DNA sequence of a gene encoding alpha-trichosanthin, a type I ribosome-inactivating protein. J. Biol. Chem. 1990, 265, 8670–8674. [Google Scholar] [CrossRef]
- Di Maro, A.; Valbonesi, P.; Bolognesi, A.; Stirpe, F.; De Luca, P.; Siniscalco Gigliano, G.; Gaudio, L.; Delli Bovi, P.; Ferranti, P.; Malorni, A.; et al. Isolation and characterization of four type-1 ribosome-inactivating proteins, with polynucleotide:adenosine glycosidase activity, from leaves of Phytolacca dioica L. Planta 1999, 208, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, A.; Polito, L.; Lubelli, C.; Barbieri, L.; Parente, A.; Stirpe, F. Ribosome-inactivating and adenine polynucleotide glycosylase activities in Mirabilis jalapa L. tissues. J. Biol. Chem. 2002, 277, 13709–13716. [Google Scholar] [CrossRef]
- Parente, A.; De Luca, P.; Bolognesi, A.; Barbieri, L.; Battelli, M.G.; Abbondanza, A.; Sande, M.J.; Gigliano, G.S.; Tazzari, P.L.; Stirpe, F. Purification and partial characterization of single-chain ribosome-inactivating proteins from the seeds of Phytolacca dioica L. Biochim. Biophys. Acta-Gene Struct. Expr. 1993, 1216, 43–49. [Google Scholar] [CrossRef]
- Barbieri, L.; Polito, L.; Bolognesi, A.; Ciani, M.; Pelosi, E.; Farini, V.; Jha, A.K.; Sharma, N.; Vivanco, J.M.; Chambery, A.; et al. Ribosome-inactivating proteins in edible plants and purification and characterization of a new ribosome-inactivating protein from Cucurbita moschata. Biochim. Biophys. Acta-Gen. Subj. 2006, 1760, 783–792. [Google Scholar] [CrossRef]
- Di Maro, A.; Chambery, A.; Daniele, A.; Casoria, P.; Parente, A. Isolation and characterization of heterotepalins, type 1 ribosome-inactivating proteins from Phytolacca heterotepala leaves. Phytochemistry 2007, 68, 767–776. [Google Scholar] [CrossRef]
- Landi, N.; Ruocco, M.R.; Ragucci, S.; Aliotta, F.; Nasso, R.; Pedone, P.V.; Di Maro, A. Quinoa as source of type 1 ribosome inactivating proteins: A novel knowledge for a revision of its consumption. Food Chem. 2021, 342, 128337. [Google Scholar] [CrossRef]
- Stirpe, F.; Gasperi-Campani, A.; Barbieri, L.; Falasca, A.; Abbondanza, A.; Stevens, W.A. Ribosome-inactivating proteins from the seeds of Saponaria officinalis L. (soapwort), of Agrostemma githago L. (corn cockle) and of Asparagus officinalis L. (asparagus), and from the latex of Hura crepitans L. (sandbox tree). Biochem. J. 1983, 216, 617–625. [Google Scholar] [CrossRef]
- Stirpe, F.; Gilabert-Oriol, R. Ribosome-Inactivating Proteins: An Overview. In Plant Toxins; Carlini, C.R., Ligabue-Braun, R., Gopalakrishnakone, P., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 153–182. [Google Scholar] [CrossRef]
- Monzingo, A.F.; Collins, E.J.; Ernst, S.R.; Irvin, J.D.; Robertus, J.D. The 2.5 A structure of pokeweed antiviral protein. J. Mol. Biol. 1993, 233, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Savino, C.; Federici, L.; Ippoliti, R.; Lendaro, E.; Tsernoglou, D. The crystal structure of saporin SO6 from Saponaria officinalis and its interaction with the ribosome. FEBS Lett. 2000, 470, 239–243. [Google Scholar] [CrossRef]
- Sánchez, M.; Scirè, A.; Tanfani, F.; Ausili, A. The thermal unfolding of the ribosome-inactivating protein saporin-S6 characterized by infrared spectroscopy. Biochim. Biophys. Acta-Proteins Proteom. 2015, 1854, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, R.; Citores, L.; Ragucci, S.; Russo, R.; Di Maro, A.; Ferreras, J.M. Biological and antipathogenic activities of ribosome-inactivating proteins from Phytolacca dioica L. Biochim. Biophys. Acta-Gen. Subj. 2016, 1860, 1256–1264. [Google Scholar] [CrossRef]
- Iglesias, R.; Citores, L.; Di Maro, A.; Ferreras, J.M. Biological activities of the antiviral protein BE27 from sugar beet (Beta vulgaris L.). Planta 2015, 241, 421–433. [Google Scholar] [CrossRef]
- Citores, L.; Iglesias, R.; Gay, C.; Ferreras, J.M. Antifungal activity of the ribosome-inactivating protein BE27 from sugar beet (Beta vulgaris L.) against the green mould Penicillium digitatum. Mol. Plant Pathol. 2016, 17, 261–271. [Google Scholar] [CrossRef]
- Landi, N.; Pacifico, S.; Ragucci, S.; Iglesias, R.; Piccolella, S.; Amici, A.; Di Giuseppe, A.M.A.; Di Maro, A. Purification, characterization and cytotoxicity assessment of Ageritin: The first ribotoxin from the basidiomycete mushroom Agrocybe aegerita. Biochim. Biophys. Acta-Gen. Subj. 2017, 1861, 1113–1121. [Google Scholar] [CrossRef]
- Di Maro, A.; Terracciano, I.; Sticco, L.; Fiandra, L.; Ruocco, M.; Corrado, G.; Parente, A.; Rao, R. Purification and characterization of a viral chitinase active against plant pathogens and herbivores from transgenic tobacco. J. Biotechnol. 2010, 147, 1–6. [Google Scholar] [CrossRef]
- Iglesias, R.; Citores, L.; Ferreras, J.M. Ribosomal RNA N-glycosylase Activity Assay of Ribosome-inactivating Proteins. Bio-Protocol 2017, 7, e2180. [Google Scholar] [CrossRef]
- Ferreras, J.M.; Citores, L.; Iglesias, R.; Jiménez, P.; Souza, A.M.; Gayoso, M.J.; Girbés, T. Occurrence and new procedure of preparation of nigrin, an antiribosomal lectin present in elderberry bark. Food Res. Int. 2011, 44, 2798–2805. [Google Scholar] [CrossRef]
- Miles, A.J.; Ramalli, S.G.; Wallace, B.A. DichroWeb, a website for calculating protein secondary structure from circular dichroism spectroscopic data. Protein Sci. 2022, 31, 37–46. [Google Scholar] [CrossRef] [PubMed]


| Type-1 RIP | HeLa 48 h | COLO 48 h | COLO 72 h | |
|---|---|---|---|---|
| -- | +Z-VAD Pretreatment | -- | ||
| quinoin | 1.9 × 10−9 | 1.0 × 10−7 | 1.0 × 10−6 | 3.9 × 10−7 |
| sodin 5 | 2.0 × 10−9 | 2.5 × 10−7 | 1.2 × 10−6 | 3.3 × 10−7 |
| sodin eL | 1.3 × 10−9 | -- | -- | >1.2 × 10−7 |
| sodin R | 4.1 × 10−10 | -- | -- | 1.6 × 10−7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landi, N.; Ragucci, S.; Citores, L.; Clemente, A.; Hussain, H.Z.F.; Iglesias, R.; Ferreras, J.M.; Di Maro, A. Isolation, Characterization and Biological Action of Type-1 Ribosome-Inactivating Proteins from Tissues of Salsola soda L. Toxins 2022, 14, 566. https://doi.org/10.3390/toxins14080566
Landi N, Ragucci S, Citores L, Clemente A, Hussain HZF, Iglesias R, Ferreras JM, Di Maro A. Isolation, Characterization and Biological Action of Type-1 Ribosome-Inactivating Proteins from Tissues of Salsola soda L. Toxins. 2022; 14(8):566. https://doi.org/10.3390/toxins14080566
Chicago/Turabian StyleLandi, Nicola, Sara Ragucci, Lucía Citores, Angela Clemente, Hafiza Z. F. Hussain, Rosario Iglesias, José M. Ferreras, and Antimo Di Maro. 2022. "Isolation, Characterization and Biological Action of Type-1 Ribosome-Inactivating Proteins from Tissues of Salsola soda L." Toxins 14, no. 8: 566. https://doi.org/10.3390/toxins14080566
APA StyleLandi, N., Ragucci, S., Citores, L., Clemente, A., Hussain, H. Z. F., Iglesias, R., Ferreras, J. M., & Di Maro, A. (2022). Isolation, Characterization and Biological Action of Type-1 Ribosome-Inactivating Proteins from Tissues of Salsola soda L. Toxins, 14(8), 566. https://doi.org/10.3390/toxins14080566