Growth and Toxigenicity of A. flavus on Resistant and Susceptible Peanut Genotypes
Abstract
:1. Introduction
2. Results
2.1. Characterisation of A. Flavus into Aflatoxigenic and Non-Aflatoxigenic Isolates
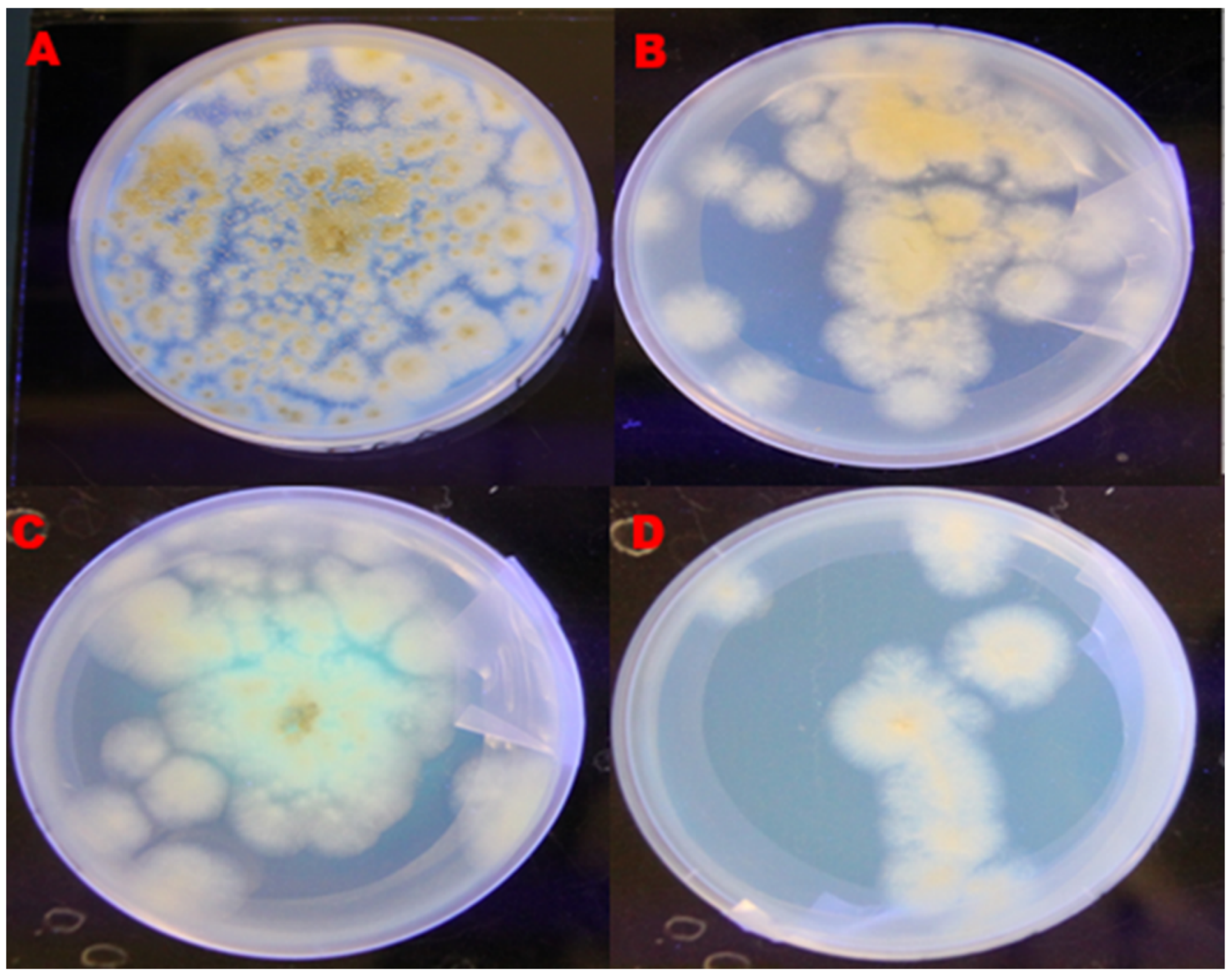
2.2. Morphological Variation in the Growth of Aflatoxigenic and Non-Aflatoxigenic A. flavus
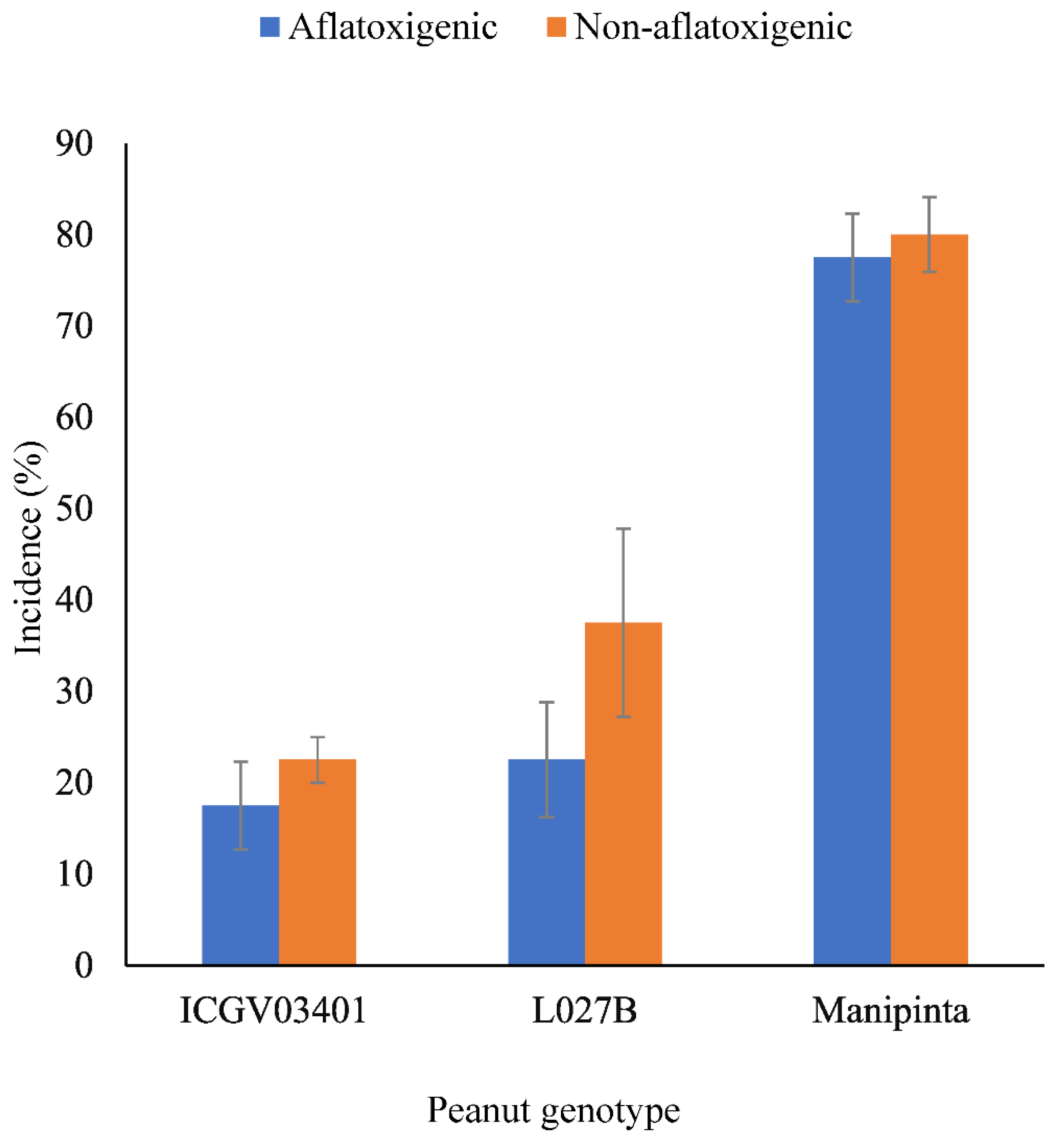
2.3. Incidence of Aflatoxigenic and Non-Aflatoxigenic A. flavus
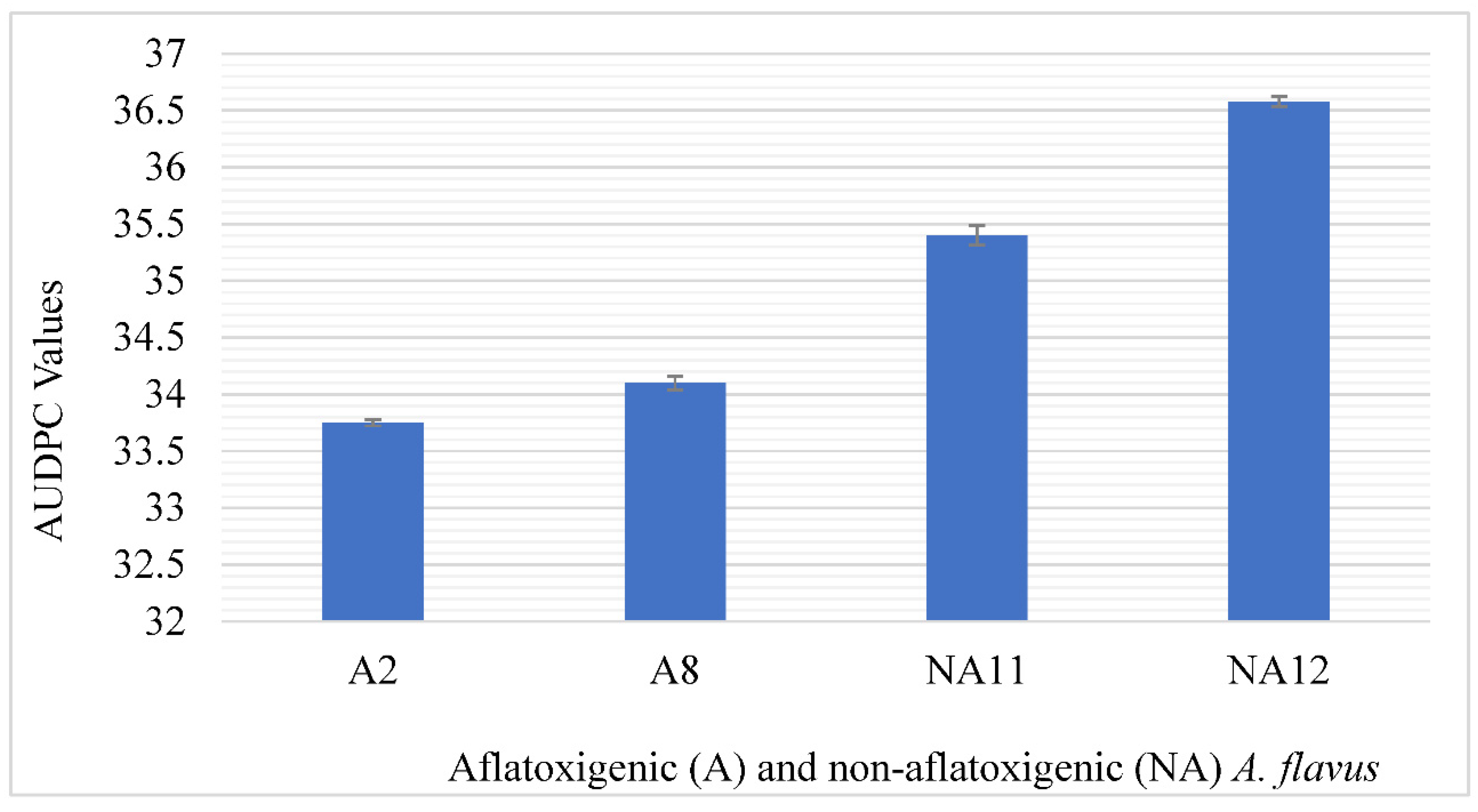
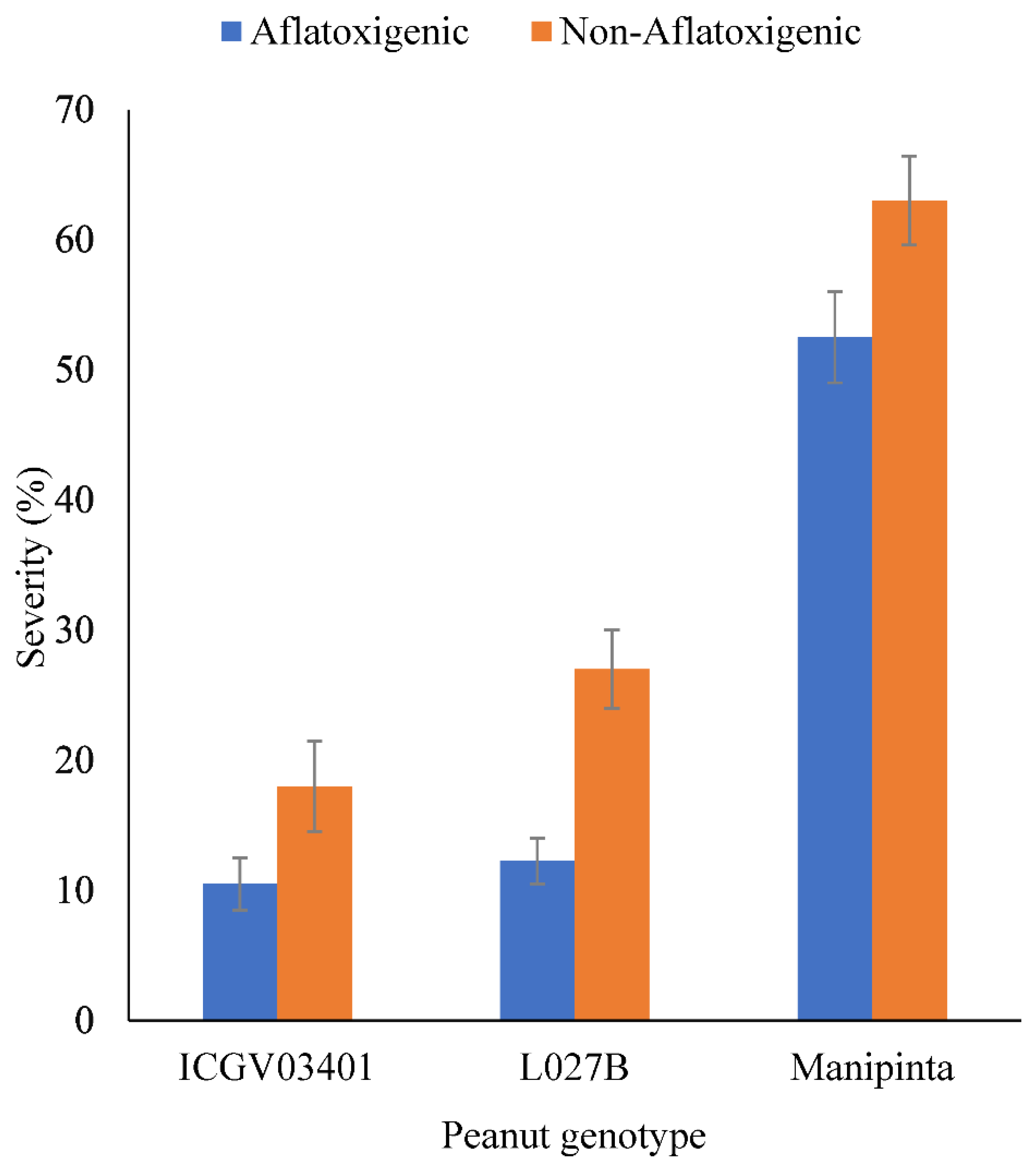
2.4. Severity of Aflatoxigenic and Non-Aflatoxigenic A. flavus
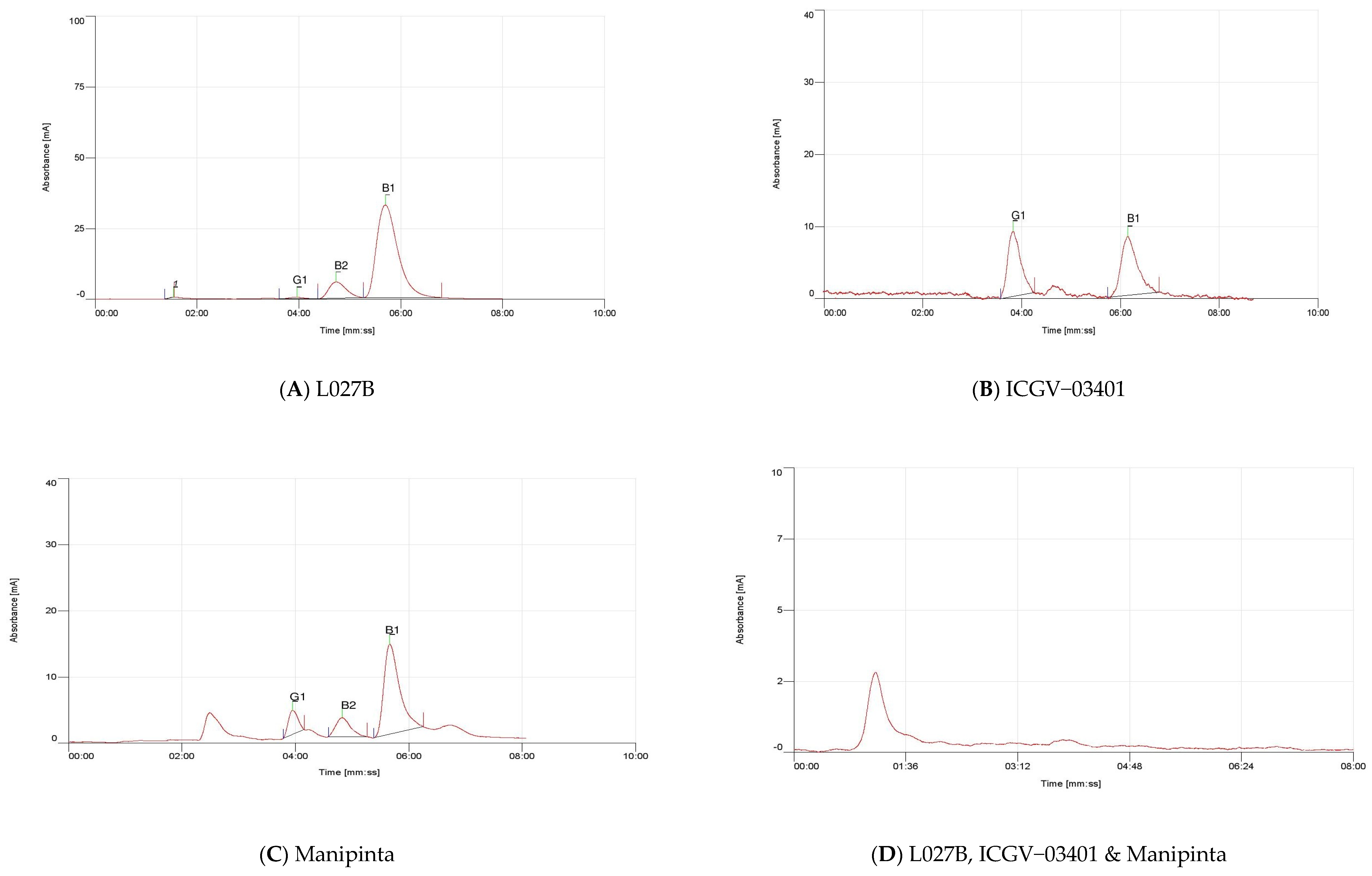
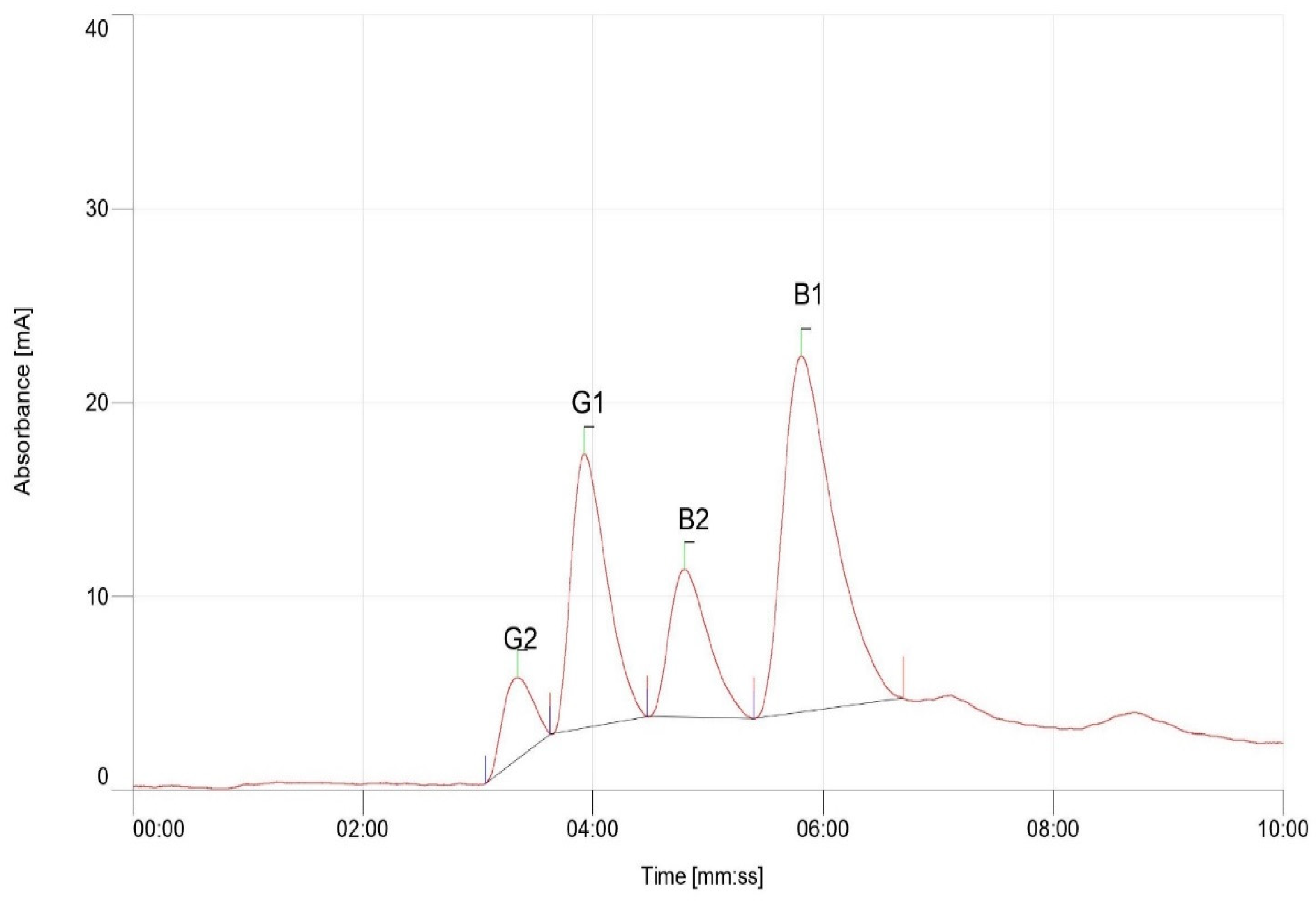
2.5. Quantification of Aflatoxin
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Study Area
5.2. Media Preparation for Fungal Isolation
5.2.1. Preparation of Potato Dextrose Agar (PDA)
5.2.2. Preparation of Yeast Extract Sucrose Agar (YESA)
5.2.3. Source of Infected Peanuts for A. flavus Isolation
5.2.4. Isolation of A. flavus Isolates
5.2.5. Obtaining Pure Cultures of A. flavus
5.2.6. Identification of A. flavus
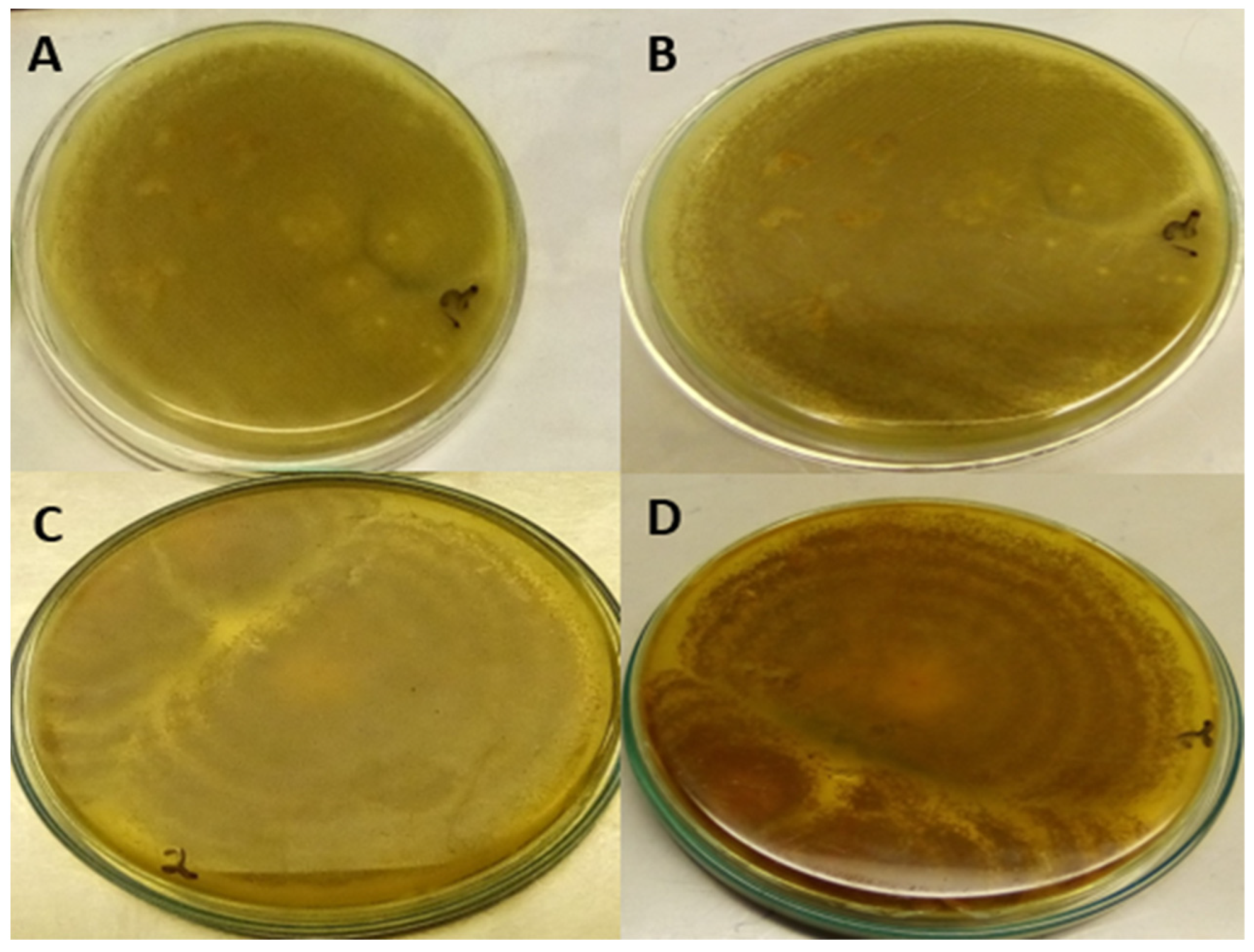
5.2.7. Detection of Aflatoxigenic and Non-Aflatoxigenic A. flavus
YESA Medium under UV Light
Exposure of Isolates on PDA to Ammonia Vapour
5.2.8. Radial Growth of Aflatoxigenic and Non-Aflatoxigenic Isolate on PDA
5.2.9. Peanut Genotypes
5.3. In Vitro Seed Colonisation
5.3.1. Experimental Design
5.3.2. Seed Sterilisation and Inoculation with Aflatoxigenic and Non-Aflatoxigenic Isolate
5.3.3. Data Collection
5.3.4. Detection of Aflatoxins Using High Performance Liquid Chromatography
Sample Extraction
HPLC Analysis Technique
5.3.5. Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raza, A.; Khan, Z.H.; Khan, K.; Anjum, M.M.; Ali, N.; Owais, M. Evaluation of Groundnut Varieties for the Agro-Ecological Zone of Malak and Division. Int. J. Environ. Sci. Nat. Resour. 2017, 5, 1–4. [Google Scholar] [CrossRef]
- Onat, B.; Bakal, H.; Gulluoglu, L.; Arioglu, H. The effects of row spacing and plant density on yield and yield components of peanut grown as a double crop in Mediterranean environment in Turkey. Turkish J. F. Crop. 2017, 22, 71–80. [Google Scholar] [CrossRef]
- Bakal, H.; Kenetli, A.; Arioglu, H. The effect of plant density on pod yield and some agronomic characteristics of different growthtype peanut varieties (Arachis hypogaea L.) grown as a main crop. Turkish J. F. Crop. 2020, 25, 92–99. [Google Scholar] [CrossRef]
- Sylvanus, U.J. Effect of Defoliation on The Growth and Seed Yield of Four Groundnut (Arachis Hypogaea L.) Cultivars. Master’s Thesis, Ahmadu Bello University, Faculty of Science, Department of Biological Sciences, Zaria, Nigeria, 2014. [Google Scholar]
- Mwatawala, H.W.; Kyaruzi, P.P. An Exploration of Factors Affecting Groundnut Production in Central Tanzania: Empirical Evidence from Kongwa District, Dodoma Region. Int. J. Progress. Sci. Technol. 2019, 14, 122–130. [Google Scholar]
- Danso-abbeam, G.; Dahamani, A.M.; Bawa, G.A.-S. Resource-use-efficiency among smallholder groundnut farmers in Northern Region, Ghana. Am. J. Exp. Agric. 2015, 6, 290–304. [Google Scholar] [CrossRef]
- Tsigbey, F.; Brandenburg, R.L.; Clottey, V. Peanut Production Methods in Northern Ghana and Some Disease Perspectives. In World Geography of the Peanut Knowledge Base Website; 2003; Volume 9, pp. 33–38. [Google Scholar]
- Jayaprakash, A.; Thanmalagan, R.R.; Roy, A.; Arunachalam, A.; Lakshmi, P. Strategies to understand Aspergillus flavus resistance mechanism in Arachis hypogaea L. Curr. Plant Biol. 2019, 20, 100123. [Google Scholar] [CrossRef]
- Ajeigbe, H.A.; Waliyar, F.; Echekwu, C.A.; Kunihya, A.; Motagi, B.N.; Eniaiyeju, D.; Inuwa, A. A Farmer’s Guide to Profitable Groundnut Production in Nigeria; International Crops Research Institute for the Semi-Arid Tropics (ICRISAT): Kano, Nigeria, 2015. [Google Scholar]
- Guo, B.; Fedorova, N.D.; Chen, X.; Wan, C.-H.; Wang, W.; Nierman, W.C.; Bhatnagar, D.; Yu, J. Gene expression profiling and identification of resistance genes to Aspergillus flavus infection in peanut through EST and microarray strategies. Toxins 2011, 3, 737–753. [Google Scholar] [CrossRef] [PubMed]
- Dube, M.; Maphosa, M. Prevalence of Aflatoxigenic Aspergillus spp and Groundnut Resistance in Zimbabwe. J. Agric. Vet. Sci. 2014, 7, 8–12. [Google Scholar] [CrossRef]
- Korani, W.A.; Chu, Y.; Holbrook, C.; Clevenger, J.; Ozias-Akins, P. Genotypic regulation of aflatoxin accumulation but not Aspergillus fungal growth upon post-Harvest infection of peanut (Arachis hypogaea L.) seeds. Toxins 2017, 9, 218. [Google Scholar] [CrossRef]
- Navya, H.M.; Hariprasad, P.; Naveen, J.; Chandranayaka, S.; Niranjana, S.R. Natural occurrence of aflatoxin, aflatoxigenic and non-aflatoxigenic Aspergillus flavus in groundnut seeds across India. Afr. J. Biotechnol. 2013, 12, 2587–2597. [Google Scholar] [CrossRef]
- Bediako, K.A.; Ofori, K.; Offei, S.K.; Dzidzienyo, D.; Asibuo, J.Y.; Amoah, R.A. A flatoxin contamination of groundnut (Arachis hypogaea L.): Predisposing factors and management interventions. Food Control 2019, 98, 61–67. [Google Scholar] [CrossRef]
- Bediako, K.A.; Dzidzienyo, D.; Ofori, K.; Offei, S.K.; Asibuo, J.Y.; Amoah, R.A.; Obeng, J. Prevalence of fungi and aflatoxin contamination in stored groundnut in Ghana. Food Control 2019, 104, 152–156. [Google Scholar] [CrossRef]
- Pandey, M.K.; Kumar, R.; Pandey, A.K.; Soni, P.; Gangurde, S.S.; Sudini, H.K.; Fountain, J.C.; Liao, B.; Desmae, H.; Okori, P.; et al. Mitigating aflatoxin contamination in groundnut through a combination of genetic resistance and post-harvest management practices. Toxins 2019, 11, 315. [Google Scholar] [CrossRef] [Green Version]
- Guchi, E.; Ayalew, A.; Dejene, M.; Ketema, M.; Asalf, B.; Fininsa, C. Occurrence of Aspergillus species in groundnut (Arachis hypogaea L.) along the value chain in different agro-ecological zones of Eastern Ethiopia. J. Appl. Environ. Microbiol. 2014, 2, 309–317. [Google Scholar] [CrossRef]
- Mohammed, A.; Chala, A.; Dejene, M.; Fininsa, C.; Hoisington, D.A.; Sobolev, V.S.; Arias, R.S. Aspergillus and aflatoxin in groundnut (Arachis hypogaea L.) and groundnut cake in Eastern Ethiopia. Food Addit. Contam. 2016, 9, 290–298. [Google Scholar] [CrossRef] [Green Version]
- Bharose, A.A.; Gajera, H.P.; Hirpara, D.G.; Kachhadia, V.H.; Golakiya, B.A. Morphological Credentials of Afla-Toxigenic and Non-Toxigenic Aspergillus Using Polyphasic Taxonomy. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 2450–2465. [Google Scholar] [CrossRef] [Green Version]
- Dieme, R.M.A.; Faye, I.; Zoclanclounon, Y.A.B.; Fonceka, D.; Ndoye, O.; Diedhiou, P.M. Identification of sources of resistance for peanut Aspergillus flavus colonization and aflatoxin contamination. Int. J. Agron. 2018, 2018, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Sudini, H.; Srilakshmi, P.; Kumar KV, K.; Njoroge, S.M.; Osiru, M.; Seetha, A.; Waliyar, F. Detection of aflatoxigenic Aspergillus strains by cultural and molecular methods: A critical review. Afr. J. Agric. Res. 2015, 9, 484–491. [Google Scholar]
- El-Aziz, A.R.M.A.; Shehata, S.M.; Hisham, S.M.; Alobathani, A.A. Molecular profile of aflatoxigenic and non-aflatoxigenic isolates of Aspergillus flavus isolated from stored maize. Saudi J. Biol. Sci. 2021, 28, 1383–1391. [Google Scholar] [CrossRef]
- Mahuku, G.; Warburton, M.L.; Makumbi, D.; Vicente, F.S. Managing aflatoxin contamination of maize: Developing host resistance. Aflatoxins Finding Solutions to Improve Food Safety 2020 Vision. 2013, p. 62. Available online: http://cdm15738.contentdm.oclc.org/utils/getfile/collection/p15738coll2/id/127887/filename/128098.pdf (accessed on 2 May 2022).
- Widstrom, N.; Butron, A.; Guo, B.; Wilson, D.; Snook, M.; Cleveland, T.; Lynch, R. Control of preharvest aflatoxin contamination in maize by pyramiding QTL involved in resistance to ear-feeding insects and invasion by Aspergillus spp. Eur. J. Agron. 2003, 19, 563–572. [Google Scholar] [CrossRef]
- Warburton, M.L.; Williams, W.P. Aflatoxin resistance in maize: What have we learned lately? Adv. Bot. 2014, 2014, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Guo, B.; Chen, Z.Y.; Lee, R.D.; Scully, B.T. Drought stress and preharvest aflatoxin contamination in agricultural commodity: Genetics, genomics and proteomics. J. Integr. Plant Biol. 2008, 50, 1281–1291. [Google Scholar] [CrossRef]
- Brown, R.L.; Chen, Z.Y.; Cleveland, T.E.; Russin, J.S. Advances in the development of host resistance in corn to aflatoxin contamination by Aspergillus flavus. Phytopathology 1999, 89, 113–117. [Google Scholar] [CrossRef] [Green Version]
- Williams, W.; Krakowsky, M.; Scully, B.; Brown, R.; Menkir, A.; Warburton, M.; Windham, G. Identifying and developing maize germplasm with resistance to accumulation of aflatoxins. World Mycotoxin J. 2015, 8, 193–209. [Google Scholar] [CrossRef]
- Soni, P.; Gangurde, S.S.; Ortega-Beltran, A.; Kumar, R.; Parmar, S.; Sudini, H.K.; Lei, Y.; Ni, X.; Huai, D.; Fountain, J.C.; et al. Functional biology and molecular mechanisms of host-pathogen interactions for aflatoxin contamination in groundnut (Arachis hypogaea L.) and maize (Zea mays L.). Front. Microbiol. 2020, 11, 227. [Google Scholar] [CrossRef]
- Garrido-Bazan, V.; Mahuku, G.; Bibbins-Martinez, M.; Arroyo-Bacerra, A.; Villalobos-López, M.Á. Dissection of mechanisms of resistance to Aspergillus flavus and aflatoxin using tropical maize germplasm. World Mycotoxin J. 2018, 11, 215–224. [Google Scholar] [CrossRef]
- Khan, R.; Ghazali, F.M.; Mahyudin, N.A.; Samsudin, N.I.P. Morphological Characterization and Determination of Aflatoxigenic and Non-Aflatoxigenic Aspergillus flavus Isolated from Sweet Corn Kernels and Soil in Malaysia. Agriculture 2020, 10, 450. [Google Scholar] [CrossRef]
- Mahmoud, M.A.; Ali, H.M.; El-Aziz, A.R.M.; Al-Othman, M.R.; Al-Wadai, A.S. Molecular characterization of aflatoxigenic and non-aflatoxigenic Aspergillus flavus isolates collected from corn grains. Genet. Mol. Res. 2014, 13, 9352–9370. [Google Scholar] [CrossRef]
- Guezlane-Tebibel, N.; Bouras, N.; Mokrane, S.; Benayad, T.; Mathieu, F. Aflatoxigenic strains of Aspergillus section Flavi isolated from marketed peanuts (Arachis hypogaea) in Algiers (Algeria). Ann. Microbiol. 2013, 63, 295–305. [Google Scholar] [CrossRef]
- Okayo, R.O.; Andika, D.O.; Dida, M.M.; K’Otuto, G.O.; Gichimu, B.M. Morphological and Molecular Characterization of Toxigenic Aspergillus flavus from Groundnut Kernels in Kenya. Int. J. Microbiol. 2020, 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C.; Wei, Q.; Brown, R.L.; Bhatnagar, D. Inverse correlation of ability to produce aflatoxin and Aspergillus colonization of maize seed. Food Nutr. Sci. 2011, 2, 486–489. [Google Scholar] [CrossRef] [Green Version]
- Kasno, A.; Trustinah, T.; Purnomo, J.; Sumartini, S. Seed coat resistance of groundnut to Aspergillus flavus and their stability performance in the field. AGRIVITA J. Agric. Sci. 2011, 33, 53–62. [Google Scholar] [CrossRef]
- Commey, L.; Tengey, T.K.; Cobos, C.J.; Dampanaboina, L.; Dhillon, K.K.; Pandey, M.K.; Sudini, H.K.; Falalou, H.; Varshney, R.K.; Burow, M.D.; et al. Peanut seed coat acts as a physical and biochemical barrier against Aspergillus flavus infection. J. Fungi 2021, 7, 1000. [Google Scholar] [CrossRef]
- Yeboah, A.; Ahiakpa, K.; Adjei-Nsiah, S. Aflatoxin levels in seeds of commonly grown groundnut varieties (Arachis hypogaea L.) in Ghana as influenced by storage method. Afr. J. Food Agric. Nutr. Dev. 2020, 20, 15402–15414. [Google Scholar] [CrossRef]
- Astoreca, A.; Vaamonde, G.; Dalcero, A.; Marin, S.; Ramos, A. Abiotic factors and their interactions influence on the co-production of aflatoxin B1 and cyclopiazonic acid by Aspergillus flavus isolated from corn. Food Microbiol. 2014, 38, 276–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahouar, A.; Marin, S.; Crespo-Sempere, A.; Saïd, S.; Sanchis, V. Effects of temperature, water activity and incubation time on fungal growth and aflatoxin B1 production by toxinogenic Aspergillus flavus isolates on sorghum seeds. Rev. Argent. Microbiol. 2016, 48, 78–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agbetiameh, D.; Ortega-Beltran, A.; Awuah, R.T.; Atehnkeng, J.; Islam, M.S.; Callicott, K.A.; Bandyopadhyay, R. Potential of Atoxigenic Aspergillus flavus Vegetative Compatibility Groups Associated With Maize and Groundnut in Ghana as Biocontrol Agents for Aflatoxin Management. Front. Microbiol. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Xu, J.; Wang, P.; Zhou, Z.; Cotty, P.J.; Kong, Q. Selection of Atoxigenic Aspergillus flavus for Potential Use in Aflatoxin Prevention in Shandong Province, China. J. Fungi 2021, 7, 773. [Google Scholar] [CrossRef] [PubMed]
- Senghor, L.A.; Ortega-Beltran, A.; Atehnkeng, J.; Callicott, K.A.; Cotty, P.J.; Bandyopadhyay, R. The atoxigenic biocontrol product Aflasafe SN01 is a valuable tool to mitigate aflatoxin contamination of both maize and groundnut cultivated in Senegal. Plant Dis. 2020, 104, 510–520. [Google Scholar] [CrossRef]
- Bandyopadhyay, R.; Ortega-Beltran, A.; Akande, A.; Mutegi, C.; Atehnkeng, J.; Kaptoge, L.; Senghor, A.; Adhikari, B.; Cotty, P. Biological control of aflatoxins in Africa: Current status and potential challenges in the face of climate change. World Mycotoxin J. 2016, 9, 771–789. [Google Scholar] [CrossRef] [Green Version]
- Raed, N.K.A. Cultural and Molecular Detection of Aflatoxigenic Activity in Aspergillus flavus Isolated from Poultry Feed. Master’s Thesis, University of Basrah-College of Veterinary Medicine, Basra, Iraq, 2016. [Google Scholar]
- Abdi, M.; Alemayehu, C. Incidence of Aspergillus contamination of groundnut (Arachis hypogaea L.) in Eastern Ethiopia. Afr. J. Microbiol. Res. 2014, 8, 759–765. [Google Scholar] [CrossRef] [Green Version]
- Klich, M.A. Identification of Common Aspergillus Species; Centraalbureau voor Schimmelcultures (CBS) Fungal Biodiversity Centre, Royal Netherlands Academy of Arts and Sciences: Utrecht, The Netherlands, 2002. [Google Scholar]
- Saito, M.; Machida, S. A rapid identification method for aflatoxin-producing strains of Aspergillus flavus and A. parasiticus by ammonia vapor. Mycoscience 1999, 40, 205–208. [Google Scholar] [CrossRef]
- Sirhan, A.Y.; Tan, G.H.; Al-Shunnaq, A.; Abdulra’uf, L.; Wong, R.C.S. QuEChERS-HPLC method for aflatoxin detection of domestic and imported food in Jordan. J. Liq. Chromatogr. Relat. Technol. 2014, 37, 321–342. [Google Scholar] [CrossRef]

| Isolates Code | UV Fluorescence | Concentrated Ammonia Solution | |
|---|---|---|---|
| PDA | YESA+β-CD | PDA | |
| AF01 | + | + | + |
| AF02 | + | + | + |
| AF03 | + | − | + |
| AF04 | + | + | + |
| AF05 | + | + | + |
| AF06 | + | + | + |
| AF07 | + | + | + |
| AF08 | + | + | + |
| AF09 | − | + | + |
| AF10 | − | + | + |
| AF11 | − | + | − |
| AF12 | − | + | − |
| AF13 | − | − | − |
| AF14 | − | − | + |
| AF15 | + | + | + |
| AF16 | + | + | + |
| Genotype | Concentration of Aflatoxin (ppb) | ||
|---|---|---|---|
| AFB1 | AFB2 | G1 | |
| ICGV–03401 | 0.86 | Nil | 0.76 |
| L027B | 2.10 | 0.85 | 0.40 |
| Manipinta | 0.61 | 1.09 | 0.65 |
| Aflatoxins | LOD (ppb) | LOQ (ppb) | R2 | Recovery (%) |
|---|---|---|---|---|
| Aflatoxin B1 | 0.2 | 0.4 | 0.999 | 98 ± 0.71 |
| Aflatoxin B2 | 0.1 | 0.2 | 0.999 | 98 ± 1.05 |
| Aflatoxin G1 | 0.2 | 0.4 | 0.999 | 99 ± 0.14 |
| Aflatoxin G2 | 0.1 | 0.2 | 0.995 | 99 ± 0.62 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tengey, T.K.; Kankam, F.; Ndela, D.N.; Frempong, D.; Appaw, W.O. Growth and Toxigenicity of A. flavus on Resistant and Susceptible Peanut Genotypes. Toxins 2022, 14, 536. https://doi.org/10.3390/toxins14080536
Tengey TK, Kankam F, Ndela DN, Frempong D, Appaw WO. Growth and Toxigenicity of A. flavus on Resistant and Susceptible Peanut Genotypes. Toxins. 2022; 14(8):536. https://doi.org/10.3390/toxins14080536
Chicago/Turabian StyleTengey, Theophilus Kwabla, Frederick Kankam, Dominic Ngagmayan Ndela, Daniel Frempong, and William Ofori Appaw. 2022. "Growth and Toxigenicity of A. flavus on Resistant and Susceptible Peanut Genotypes" Toxins 14, no. 8: 536. https://doi.org/10.3390/toxins14080536
APA StyleTengey, T. K., Kankam, F., Ndela, D. N., Frempong, D., & Appaw, W. O. (2022). Growth and Toxigenicity of A. flavus on Resistant and Susceptible Peanut Genotypes. Toxins, 14(8), 536. https://doi.org/10.3390/toxins14080536





