Cloning of Metalloproteinase 17 Genes from Oriental Giant Jellyfish Nemopilema nomurai (Scyphozoa: Rhizostomeae)
Abstract
:1. Introduction
2. Results
2.1. Metalloproteinases Components of NnV
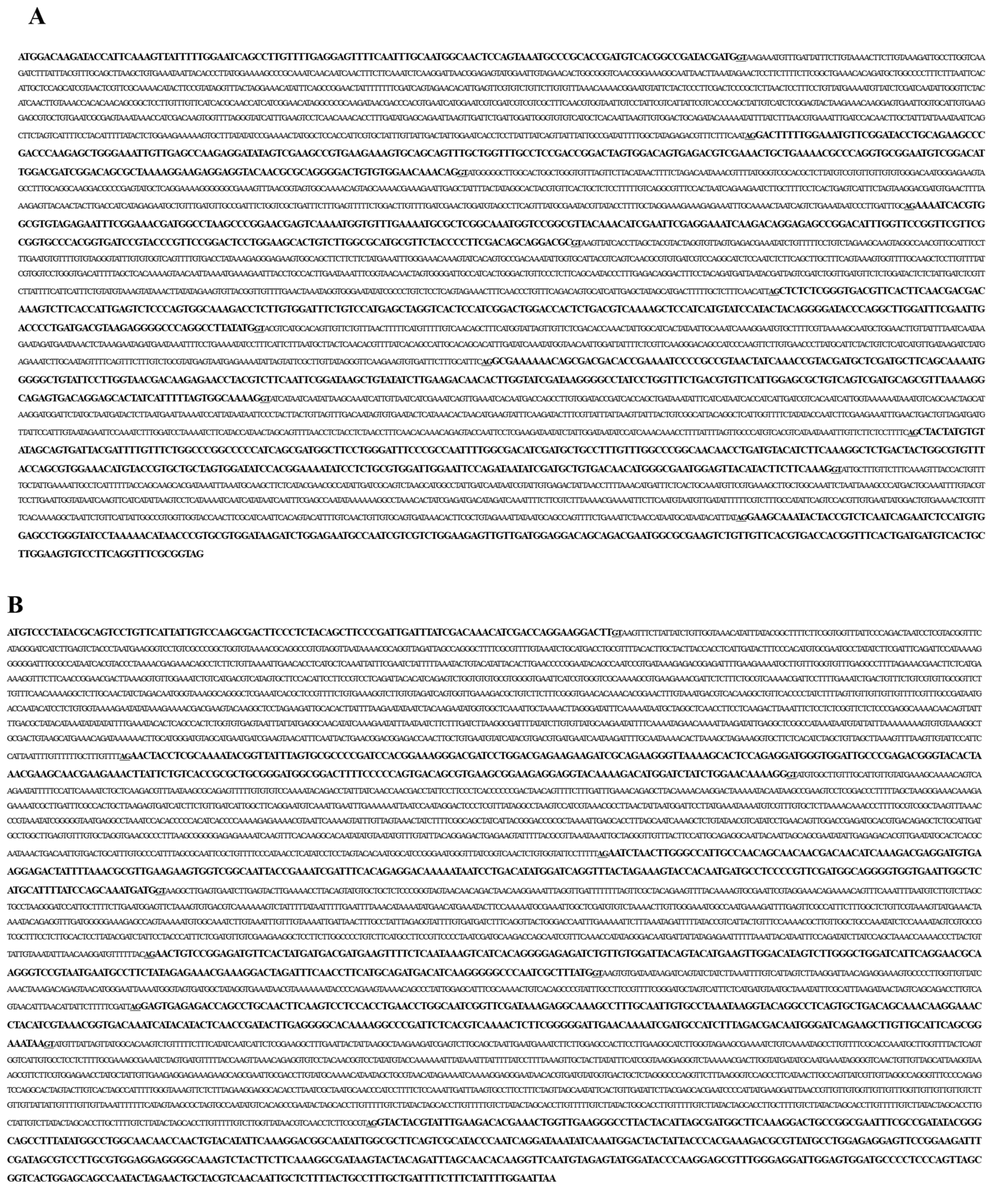
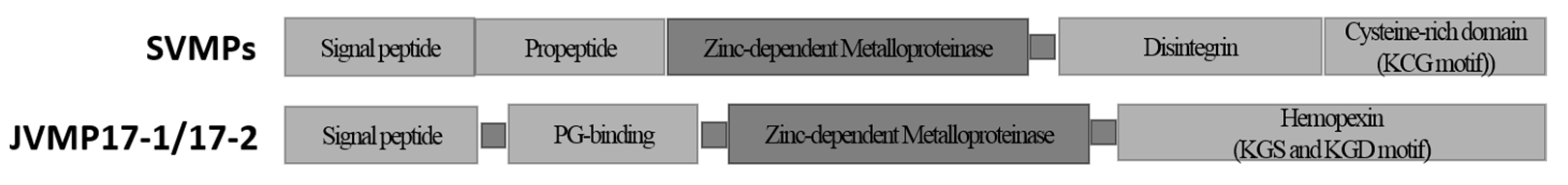
2.2. Full-Length cDNA Sequence Analysis of N. nomurai JVMP17-1 and 17-2
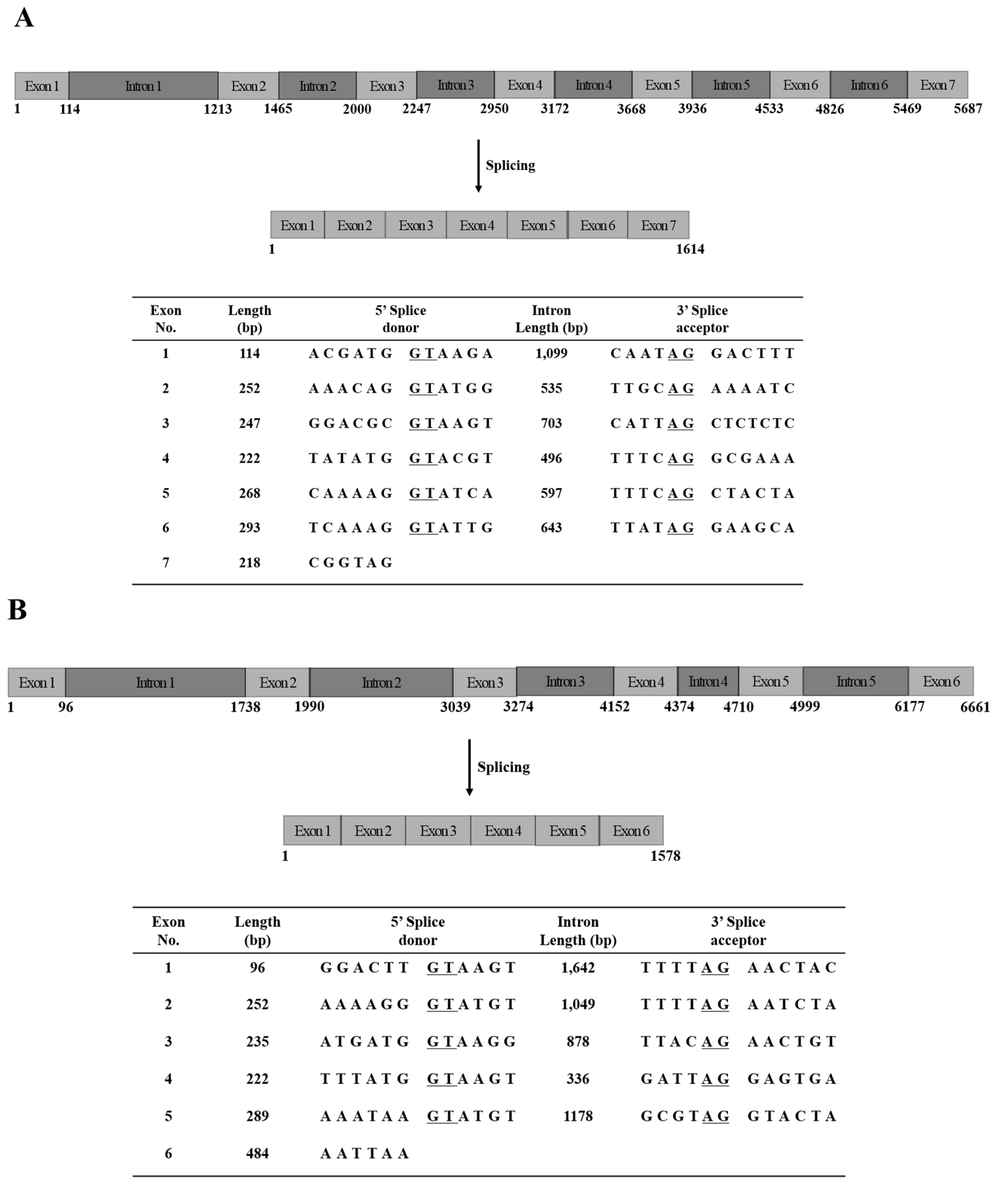
2.3. Genomic DNA Sequences of N. nomurai JVMP17-1 and 17-2
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Jellyfish Collection and Nematocyst Preparation
5.2. Venom Preparation
5.3. Metalloproteinase Analysis of NnV
5.4. Total RNA Extraction
5.5. Rapid Amplification of cDNA Ends (RACE)
5.6. Genomic DNA Sequences of N. nomurai JVMP17-1 and JVMP17-2
5.7. Nucleotide Sequence Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- El-Safory, N.S.; Fazary, A.E.; Lee, C. Hyaluronidases, a Group of Glycosidases: Current and Future Perspectives. Carbohydr. Polym. 2010, 81, 165–181. [Google Scholar] [CrossRef]
- Ramos, O.H.P.; Selistre-de-Araujo, H.S. Snake Venom Metalloproteases—Structure and Function of Catalytic and Disintegrin Domains. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006, 142, 328–346. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.F.; Mellows, B.A.; Mitchell, R.; Sfyri, P.; Layfield, H.J.; Salamah, M.; Vaiyapuri, R.; Collins-Hooper, H.; Bicknell, A.B.; Matsakas, A.; et al. Mechanisms Underpinning the Permanent Muscle Damage Induced by Snake Venom Metalloprotease. PLoS Negl. Trop. Dis. 2019, 13, e0007041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Jung, E.S.; Kang, C.; Yoon, W.D.; Kim, J.S.; Kim, E. Scyphozoan Jellyfish Venom Metalloproteinases and Their Role in the Cytotoxicity. Toxicon 2011, 58, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, I.; Hwang, D.H.; Lee, H.; Yoon, W.D.; Chae, J.; Han, C.H.; Yum, S.; Kang, C.; Kim, E. Proteomic Analysis of Novel Components of Nemopilema Nomurai Jellyfish Venom: Deciphering the Mode of Action. Toxins 2019, 11, 153. [Google Scholar] [CrossRef] [Green Version]
- Moura-da-Silva, A.M.; Baldo, C. Jararhagin, a Hemorrhagic Snake Venom Metalloproteinase from Bothrops jararaca. Toxicon 2012, 60, 280–289. [Google Scholar] [CrossRef]
- Bjarnason, J.B.; Fox, J.W. Hemorrhagic Metalloproteinases from Snake Venoms. Pharmacol. Ther. 1994, 62, 325–372. [Google Scholar] [CrossRef]
- Fox, J.W.; Serrano, S.M. Structural Considerations of the Snake Venom Metalloproteinases, Key Members of the M12 Reprolysin Family of Metalloproteinases. Toxicon 2005, 45, 969–985. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M. Insights into and Speculations About Snake Venom Metalloproteinase (SVMP) Synthesis, Folding and Disulfide Bond Formation and Their Contribution to Venom Complexity. FEBS J. 2008, 275, 3016–3030. [Google Scholar] [CrossRef] [PubMed]
- Clemetson, K.J.; Morita, T.; Kini, R.M. Scientific and Standardization Committee Communications: Classification and Nomenclature of Snake Venom C-Type Lectins and Related Proteins. J. Thromb. Haemost. 2009, 7, 360. [Google Scholar] [CrossRef] [PubMed]
- Uri, S.; Marina, G.; Liubov, G. Severe Delayed Cutaneous Reaction Due to Mediterranean Jellyfish (Rhopilema Nomadica) Envenomation. Contact Dermat. 2005, 52, 282–283. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; He, Q.; Guo, Y.; Zhang, J.; Nie, F.; Li, Y.; Ye, X.; Zhang, L.; Capillata, C. Cyanea Capillata Tentacle-Only Extract as a Potential Alternative of Nematocyst Venom: Its Cardiovascular Toxicity and Tolerance to Isolation and Purification Procedures. Toxicon 2009, 53, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Jin, Y.B.; Kwak, J.; Jung, H.; Yoon, W.D.; Yoon, T.J.; Kim, J.S.; Kim, E. Protective Effect of Tetracycline Against Dermal Toxicity Induced by Jellyfish Venom. PLoS ONE 2013, 8, e57658. [Google Scholar] [CrossRef]
- Hwang, D.H.; Lee, H.; Choudhary, I.; Kang, C.; Chae, J.; Kim, E. Protective Effect of Epigallocatechin-3-Gallate (EGCG) on Toxic Metalloproteinases-Mediated Skin Damage Induced by Scyphozoan Jellyfish Envenomation. Sci. Rep. 2020, 10, 18644. [Google Scholar] [CrossRef] [PubMed]
- Mohan Prakash, R.L.M.; Hwang, D.H.; Hong, I.H.; Chae, J.; Kang, C.; Kim, E. Danio rerio as an Alternative Vertebrate Model for Jellyfish Venom Study: The Toxinological Aspects of Nemopilema Nomurai Venom. Toxicol. Lett. 2020, 335, 91–97. [Google Scholar] [CrossRef]
- Nevalainen, T.J.; Peuravuori, H.J.; Quinn, R.J.; Llewellyn, L.E.; Benzie, J.A.; Fenner, P.J.; Winkel, K.D. Phospholipase A2 in Cnidaria. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2004, 139, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Helmholz, H.; Ruhnau, C.; Schütt, C.; Prange, A. Comparative Study on the Cell Toxicity and Enzymatic Activity of Two Northern Scyphozoan Species Cyanea capillata (L.) and Cyanea lamarckii (Péron & Léslieur). Toxicon 2007, 50, 53–64. [Google Scholar] [CrossRef]
- Radwan, F.F.; Burnett, J.W.; Bloom, D.A.; Coliano, T.; Eldefrawi, M.E.; Erderly, H.; Aurelian, L.; Torres, M.; Heimer-de la Cotera, E.P. A Comparison of the Toxinological Characteristics of Two Cassiopea and Aurelia Species. Toxicon 2001, 39, 245–257. [Google Scholar] [CrossRef]
- Takeda, S.; Takeya, H.; Iwanaga, S. Snake Venom Metalloproteinases: Structure, Function and Relevance to the Mammalian ADAM/ADAMTS Family Proteins. Biochim. Biophys. Acta 2012, 1824, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Krogh, S.; Jørgensen, S.T.; Devine, K.M. Lysis Genes of the Bacillus subtilis Defective Prophage PBSX. J. Bacteriol. 1998, 180, 2110–2117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dideberg, O.; Charlier, P.; Dive, G.; Joris, B.; Frère, J.M.; Ghuysen, J.M. Structure of a Zn 2 -Containing D-Alanyl-D-alanine-Cleaving Carboxypeptidase at 2.5 Å Resolution. Nature 1982, 299, 469–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charlier, P.; Dideberg, O.; Jamoulle, J.; Frère, J.; Ghuysen, J.; Dive, G.; Lamotte-Brasseur, J. Active-Site-Directed Inactivators of the Zn2-Containing D-Alanyl-D-alanine-Cleaving Carboxypeptidase of Streptomyces albus G. Biochim. J. 1984, 219, 763–772. [Google Scholar] [CrossRef] [Green Version]
- Smeets, T.J.; Barg, E.C.; Kraan, M.C.; Smith, M.D.; Breedveld, F.C.; Tak, P.P. Analysis of the Cell Infiltrate and Expression of Proinflammatory Cytokines and Matrix Metalloproteinases in Arthroscopic Synovial Biopsies: Comparison with Synovial Samples from Patients with End Stage, Destructive Rheumatoid Arthritis. Ann. Rheum. Dis. 2003, 62, 635–638. [Google Scholar] [CrossRef] [Green Version]
- Hornebeck, W.; Maquart, F.X. Proteolyzed Matrix as a Template for the Regulation of Tumor Progression. Biomed. Pharmacother. 2003, 57, 223–230. [Google Scholar] [CrossRef]
- Van den Steen, P.E.; Wuyts, A.; Husson, S.J.; Proost, P.; Van Damme, J.; Opdenakker, G. Gelatinase B/MMP-9 and Neutrophil Collagenase/MMP-8 Process the Chemokines Human GCP-2/CXCL6, ENA-78/CXCL5 and Mouse GCP-2/LIX and Modulate Their Physiological Activities. Eur. J. Biochem. 2003, 270, 3739–3749. [Google Scholar] [CrossRef] [PubMed]
- Dufour, A.; Sampson, N.S.; Zucker, S.; Cao, J. Role of the Hemopexin Domain of Matrix Metalloproteinases in Cell Migration. J. Cell. Physiol. 2008, 217, 643–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolosano, E.; Altruda, F. Hemopexin: Structure, Function, and Regulation. DNA Cell Biol. 2002, 21, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Bloom, D.A.; Burnett, J.W.; Alderslade, P. Partial Purification of Box Jellyfish (Chironex Fleckeri) Nematocyst Venom Isolated at the Beachside. Toxicon 1998, 36, 1075–1085. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Heussen, C.; Dowdle, E.B. Electrophoretic Analysis of Plasminogen Activators in Polyacrylamide Gels Containing Sodium Dodecyl Sulfate and Copolymerized Substrates. Anal. Biochem. 1980, 102, 196–202. [Google Scholar] [CrossRef]
- Heo, Y.; Kwon, Y.C.; Bae, S.K.; Hwang, D.; Yang, H.R.; Choudhary, I.; Lee, H.; Yum, S.; Shin, K.; Yoon, W.D.; et al. Cloning a Chymotrypsin-Like 1 (CTRL-1) Protease cDNA from the Jellyfish Nemopilema Nomurai. Toxins 2016, 8, 205. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, D.H.; Heo, Y.; Kwon, Y.C.; Prakash, R.L.M.; Kim, K.; Oh, H.; Seyedian, R.; Munawir, A.; Kang, C.; Kim, E. Cloning of Metalloproteinase 17 Genes from Oriental Giant Jellyfish Nemopilema nomurai (Scyphozoa: Rhizostomeae). Toxins 2022, 14, 519. https://doi.org/10.3390/toxins14080519
Hwang DH, Heo Y, Kwon YC, Prakash RLM, Kim K, Oh H, Seyedian R, Munawir A, Kang C, Kim E. Cloning of Metalloproteinase 17 Genes from Oriental Giant Jellyfish Nemopilema nomurai (Scyphozoa: Rhizostomeae). Toxins. 2022; 14(8):519. https://doi.org/10.3390/toxins14080519
Chicago/Turabian StyleHwang, Du Hyeon, Yunwi Heo, Young Chul Kwon, Ramachandran Loganathan Mohan Prakash, Kyoungyeon Kim, Hyunju Oh, Ramin Seyedian, Al Munawir, Changkeun Kang, and Euikyung Kim. 2022. "Cloning of Metalloproteinase 17 Genes from Oriental Giant Jellyfish Nemopilema nomurai (Scyphozoa: Rhizostomeae)" Toxins 14, no. 8: 519. https://doi.org/10.3390/toxins14080519
APA StyleHwang, D. H., Heo, Y., Kwon, Y. C., Prakash, R. L. M., Kim, K., Oh, H., Seyedian, R., Munawir, A., Kang, C., & Kim, E. (2022). Cloning of Metalloproteinase 17 Genes from Oriental Giant Jellyfish Nemopilema nomurai (Scyphozoa: Rhizostomeae). Toxins, 14(8), 519. https://doi.org/10.3390/toxins14080519





