Vegetative Insecticidal Protein Vip3Aa Is Transported via Membrane Vesicles in Bacillus thuringiensis BMB171
Abstract
:1. Introduction
2. Results
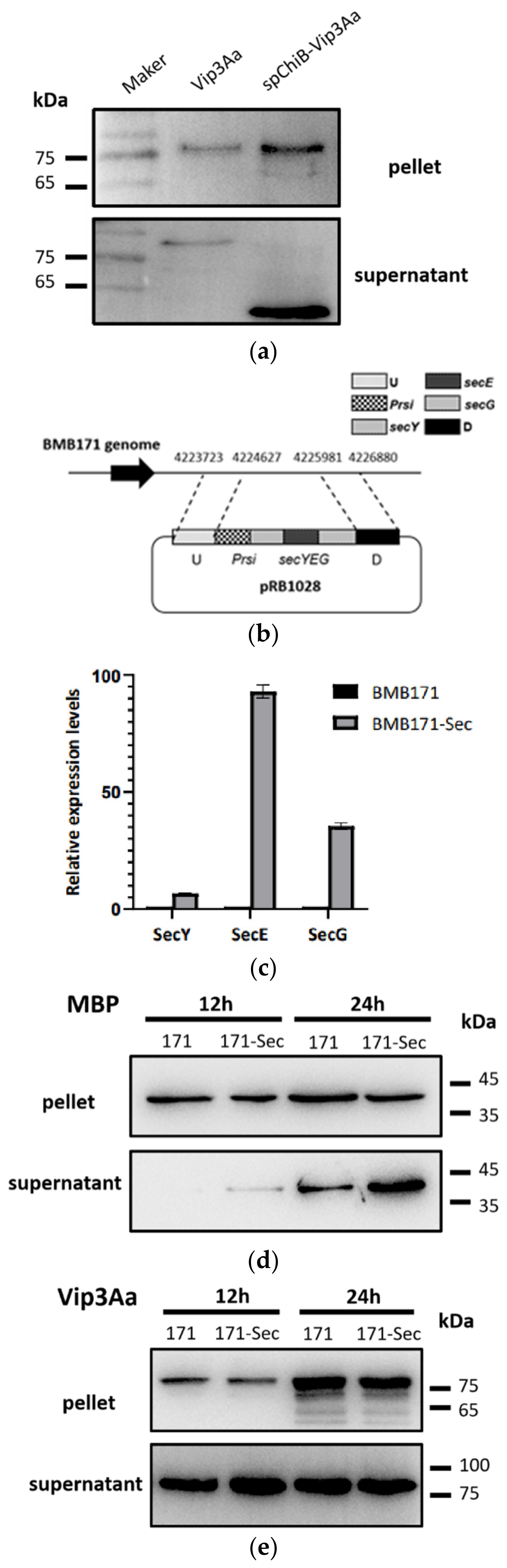
2.1. Vip3A Is Not Secreted via the Sec Secretion Pathway
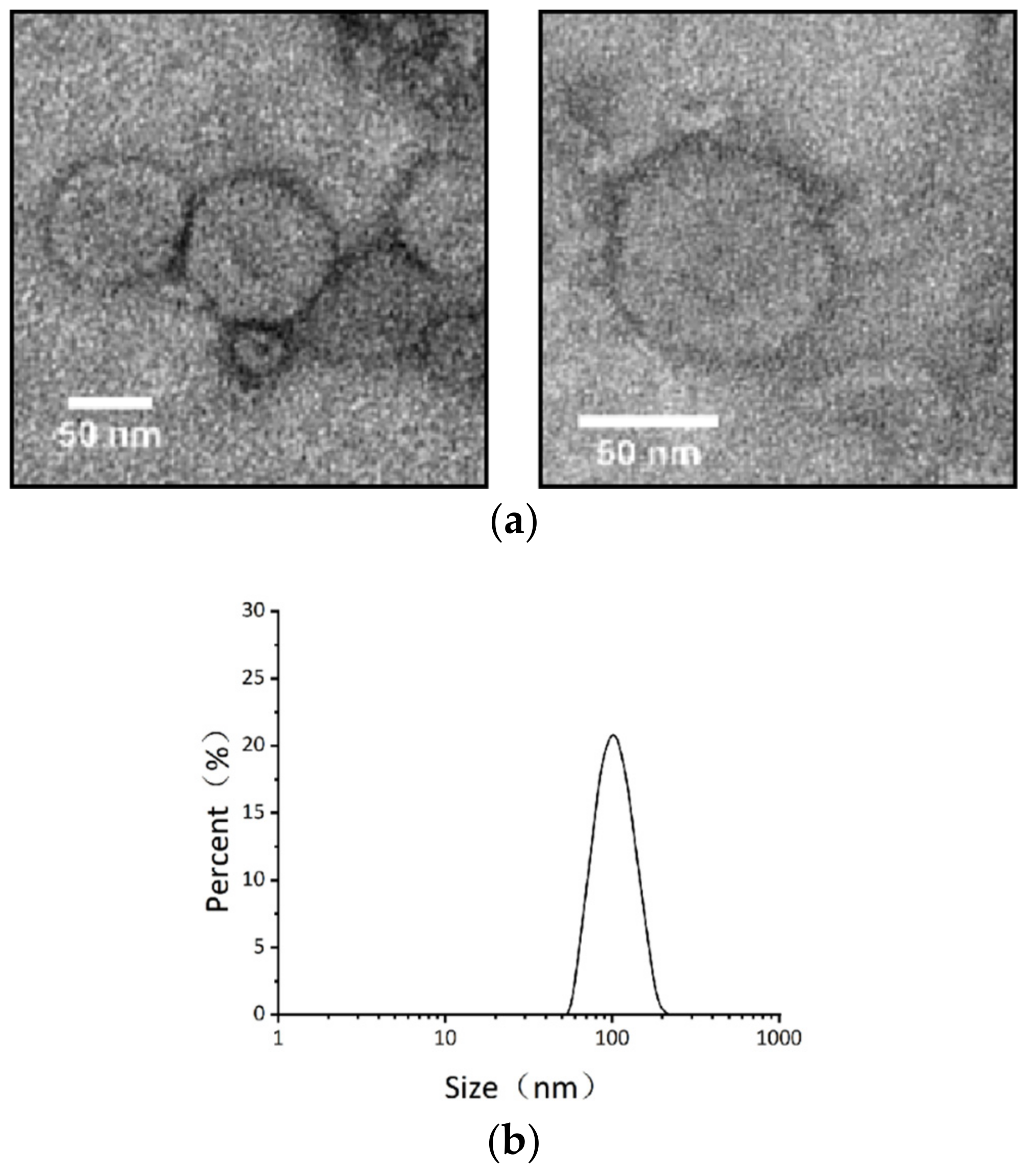
2.2. Isolation of Vesicles from B. thuringiensis BMB171 Culture Supernatants
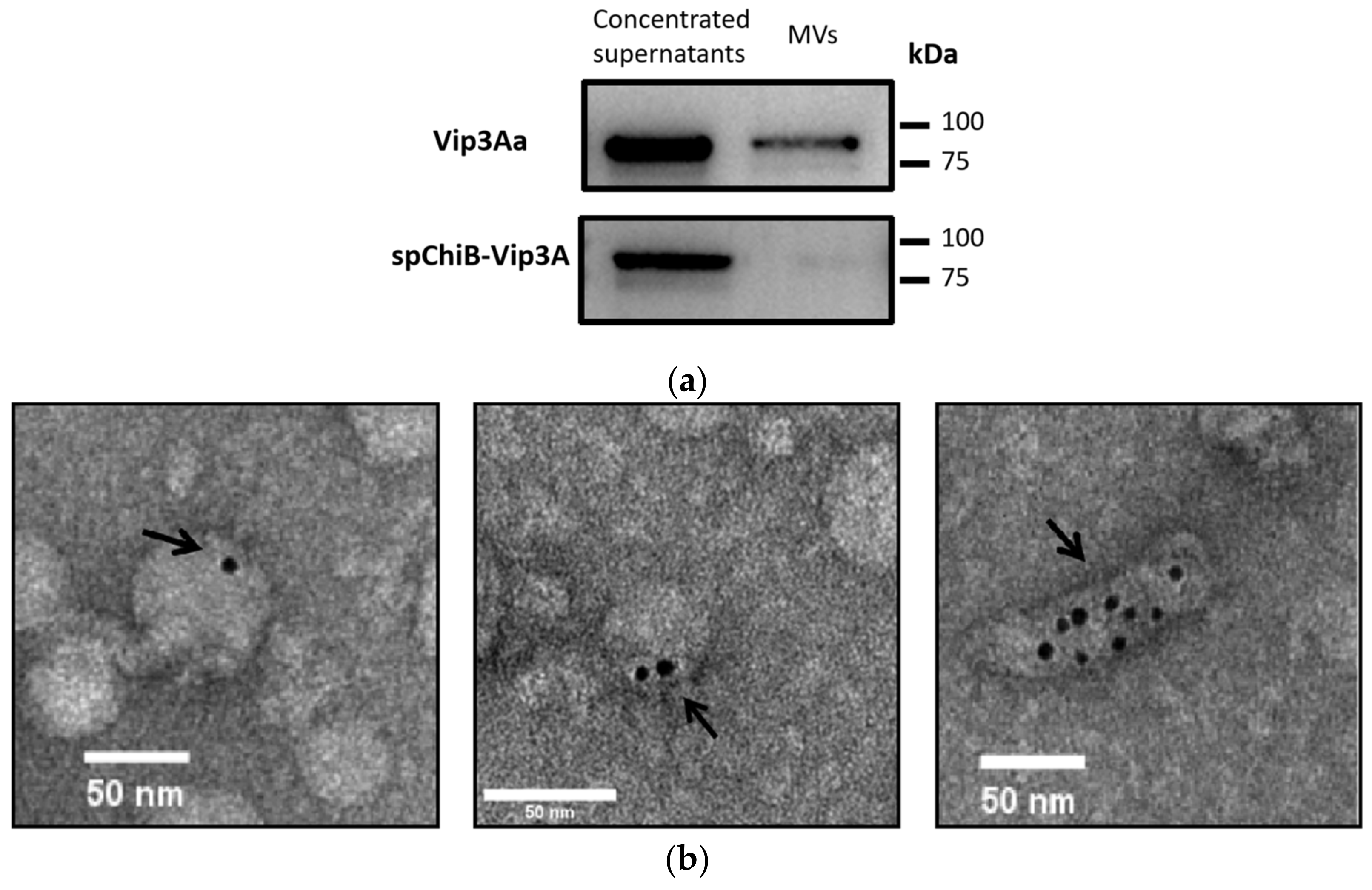
2.3. Vip3Aa Were Detected in Membrane Vesicles
2.4. Vip3Aa α-Helix Plays an Essential Role in Vip3Aa Secretion
2.5. The Signal Region of Vip3Aa Can Direct Other Proteins into Vesicles
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains and Plasmids
5.2. Media and Cultural Conditions
5.3. Gene Integration in B. thuringiensis
5.4. Construction of the Protein Expression Vector
5.5. RNA Extraction and qRT-PCR
5.6. Western Blotting
5.7. Purification of MVs
5.8. TEM Analysis
5.9. IEM Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Liu, J.G.; Yang, A.Z.; Shen, X.H.; Hua, B.G.; Shi, G.L. Specific binding of activated Vip3Aa10 to Helicoverpa armigera brush border membrane vesicles results in pore formation. J. Invertebr. Pathol. 2011, 108, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, S.; Jin, M.; Wu, C.; Chakraborty, P.; Xiao, Y. Bacillus thuringiensis vegetative insecticidal protein family Vip3A and mode of action against pest Lepidoptera. Pest. Manag. Sci. 2020, 76, 1612–1617. [Google Scholar] [CrossRef] [PubMed]
- Chakroun, M.; Bel, Y.; Caccia, S.; Abdelkefi-Mesrati, L.; Escriche, B.; Ferre, J. Susceptibility of Spodoptera frugiperda and S. exigua to Bacillus thuringiensis Vip3Aa insecticidal protein. J. Invertebr. Pathol. 2012, 110, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Kumar, H.; Kaur, S. Vegetative Insecticidal Protein (Vip): A Potential Contender From Bacillus thuringiensis for Efficient Management of Various Detrimental Agricultural Pests. Front. Microbiol. 2021, 12, 659736. [Google Scholar] [CrossRef]
- Yang, J.; Quan, Y.; Sivaprasath, P.; Shabbir, M.Z.; Wang, Z.; Ferre, J.; He, K. Insecticidal Activity and Synergistic Combinations of Ten Different Bt Toxins against Mythimna separata (Walker). Toxins 2018, 10, 454. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Fang, L.; Zhou, Z.; Pacheco, S.; Gomez, I.; Song, F.; Soberon, M.; Zhang, J.; Bravo, A. Specific binding between Bacillus thuringiensis Cry9Aa and Vip3Aa toxins synergizes their toxicity against Asiatic rice borer (Chilo suppressalis). J. Biol. Chem. 2018, 293, 11447–11458. [Google Scholar] [CrossRef] [Green Version]
- Tabashnik, B.E.; Liesner, L.R.; Ellsworth, P.C.; Unnithan, G.C.; Fabrick, J.A.; Naranjo, S.E.; Li, X.; Dennehy, T.J.; Antilla, L.; Staten, R.T.; et al. Transgenic cotton and sterile insect releases synergize eradication of pink bollworm a century after it invaded the United States. Proc. Natl. Acad. Sci. USA 2021, 118, e2019115118. [Google Scholar] [CrossRef]
- Syed, T.; Askari, M.; Meng, Z.; Li, Y.; Abid, M.A.; Wei, Y.; Guo, S.; Liang, C.; Zhang, R. Current Insights on Vegetative Insecticidal Proteins (Vip) as Next Generation Pest Killers. Toxins 2020, 12, 522. [Google Scholar] [CrossRef]
- Jiang, K.; Zhang, Y.; Chen, Z.; Wu, D.; Cai, J.; Gao, X. Structural and Functional Insights into the C-terminal Fragment of Insecticidal Vip3A Toxin of Bacillus thuringiensis. Toxins 2020, 12, 438. [Google Scholar] [CrossRef]
- Nunez-Ramirez, R.; Huesa, J.; Bel, Y.; Ferre, J.; Casino, P.; Arias-Palomo, E. Molecular architecture and activation of the insecticidal protein Vip3Aa from Bacillus thuringiensis. Nat. Commun. 2020, 11, 3974. [Google Scholar] [CrossRef]
- Byrne, M.J.; Iadanza, M.G.; Perez, M.A.; Maskell, D.P.; George, R.M.; Hesketh, E.L.; Beales, P.A.; Zack, M.D.; Berry, C.; Thompson, R.F. Cryo-EM structures of an insecticidal Bt toxin reveal its mechanism of action on the membrane. Nat. Commun. 2021, 12, 2791. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Sachdev, B.; Sharma, N.; Seth, R.; Bhatnagar, R.K. Interaction of Bacillus thuringiensis vegetative insecticidal protein with ribosomal S2 protein triggers larvicidal activity in Spodoptera frugiperda. Appl. Environ. Microbiol. 2010, 76, 7202–7209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, K.; Hou, X.Y.; Tan, T.T.; Cao, Z.L.; Mei, S.Q.; Yan, B.; Chang, J.; Han, L.; Zhao, D.; Cai, J. Scavenger receptor-C acts as a receptor for Bacillus thuringiensis vegetative insecticidal protein Vip3Aa and mediates the internalization of Vip3Aa via endocytosis. PLoS Pathog. 2018, 14, e1007347. [Google Scholar] [CrossRef]
- Jiang, K.; Hou, X.; Han, L.; Tan, T.; Cao, Z.; Cai, J. Fibroblast Growth Factor Receptor, a Novel Receptor for Vegetative Insecticidal Protein Vip3Aa. Toxins 2018, 10, 546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osman, G.H.; Soltane, R.; Saleh, I.; Abulreesh, H.H.; Gazi, K.S.; Arif, I.A.; Ramadan, A.M.; Alameldin, H.F.; Osman, Y.A.; Idriss, M. Isolation, characterization, cloning and bioinformatics analysis of a novel receptor from black cut worm (Agrotis ipsilon) of Bacillus thuringiensis vip 3Aa toxins. Saudi J. Biol. Sci. 2019, 26, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- An, B.; Zhang, Y.; Li, X.; Hou, X.; Yan, B.; Cai, J. PHB2 affects the virulence of Vip3Aa to Sf9 cells through internalization and mitochondrial stability. Virulence 2022, 13, 684–697. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Martinez, P.; Gomis-Cebolla, J.; Ferre, J.; Escriche, B. Changes in gene expression and apoptotic response in Spodoptera exigua larvae exposed to sublethal concentrations of Vip3 insecticidal proteins. Sci. Rep. 2017, 7, 16245. [Google Scholar] [CrossRef] [Green Version]
- Jiang, K.; Mei, S.Q.; Wang, T.T.; Pan, J.H.; Chen, Y.H.; Cai, J. Vip3Aa induces apoptosis in cultured Spodoptera frugiperda (Sf9) cells. Toxicon 2016, 120, 49–56. [Google Scholar] [CrossRef]
- Hou, X.; Han, L.; An, B.; Zhang, Y.; Cao, Z.; Zhan, Y.; Cai, X.; Yan, B.; Cai, J. Mitochondria and Lysosomes Participate in Vip3Aa-Induced Spodoptera frugiperda Sf9 Cell Apoptosis. Toxins 2020, 12, 116. [Google Scholar] [CrossRef] [Green Version]
- Estruch, J.J.; Warren, G.W.; Mullins, M.A.; Nye, G.J.; Craig, J.A.; Koziel, M.G. Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc. Natl. Acad. Sci. USA 1996, 93, 5389–5394. [Google Scholar] [CrossRef] [Green Version]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef] [PubMed]
- Zack, M.D.; Sopko, M.S.; Frey, M.L.; Wang, X.; Tan, S.Y.; Arruda, J.M.; Letherer, T.T.; Narva, K.E. Functional characterization of Vip3Ab1 and Vip3Bc1: Two novel insecticidal proteins with differential activity against lepidopteran pests. Sci. Rep. 2017, 7, 11112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivera, J.; Cordero, R.J.; Nakouzi, A.S.; Frases, S.; Nicola, A.; Casadevall, A. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. USA 2010, 107, 19002–19007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mekasha, S.; Linke, D. Secretion Systems in Gram-Negative Bacterial Fish Pathogens. Front. Microbiol. 2021, 12, 782673. [Google Scholar] [CrossRef] [PubMed]
- Gorasia, D.G.; Veith, P.D.; Reynolds, E.C. The Type IX Secretion System: Advances in Structure, Function and Organisation. Microorganisms 2020, 8, 1173. [Google Scholar] [CrossRef]
- Zhang, J.; Pan, Z.Z.; Xu, L.; Liu, B.; Chen, Z.; Li, J.; Niu, L.Y.; Zhu, Y.J.; Chen, Q.X. Proteolytic activation of Bacillus thuringiensis Vip3Aa protein by Spodoptera exigua midgut protease. Int. J. Biol. Macromol. 2018, 107, 1220–1226. [Google Scholar] [CrossRef]
- Lee, E.Y.; Choi, D.Y.; Kim, D.K.; Kim, J.W.; Park, J.O.; Kim, S.; Kim, S.H.; Desiderio, D.M.; Kim, Y.K.; Kim, K.P.; et al. Gram-positive bacteria produce membrane vesicles: Proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 2009, 9, 5425–5436. [Google Scholar] [CrossRef]
- Bishop, D.G.; Work, E. An extracellular glycolipid produced by Escherichia coli grown under lysine-limiting conditions. Biochem. J. 1965, 96, 567–576. [Google Scholar] [CrossRef] [Green Version]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef]
- Wang, Z.; Gan, C.; Wang, J.; Bravo, A.; Soberon, M.; Yang, Q.; Zhang, J. Nutrient conditions determine the localization of Bacillus thuringiensis Vip3Aa protein in the mother cell compartment. Microb. Biotechnol. 2021, 14, 551–560. [Google Scholar] [CrossRef]
- Choi, C.W.; Park, E.C.; Yun, S.H.; Lee, S.Y.; Lee, Y.G.; Hong, Y.; Park, K.R.; Kim, S.H.; Kim, G.H.; Kim, S.I. Proteomic characterization of the outer membrane vesicle of Pseudomonas putida KT2440. J. Proteome Res. 2014, 13, 4298–4309. [Google Scholar] [CrossRef] [PubMed]
- Orench-Rivera, N.; Kuehn, M.J. Environmentally controlled bacterial vesicle-mediated export. Cell. Microbiol. 2016, 18, 1525–1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roier, S.; Zingl, F.G.; Cakar, F.; Schild, S. Bacterial outer membrane vesicle biogenesis: A new mechanism and its implications. Microb. Cell 2016, 3, 257–259. [Google Scholar] [CrossRef]
- Roderer, D.; Glockshuber, R. Assembly mechanism of the alpha-pore-forming toxin cytolysin A from Escherichia coli. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160211. [Google Scholar] [CrossRef]
- Sartorio, M.G.; Pardue, E.J.; Feldman, M.F.; Haurat, M.F. Bacterial Outer Membrane Vesicles: From Discovery to Applications. Annu. Rev. Microbiol. 2021, 75, 609–630. [Google Scholar] [CrossRef]
- Valguarnera, E.; Scott, N.E.; Azimzadeh, P.; Feldman, M.F. Surface Exposure and Packing of Lipoproteins into Outer Membrane Vesicles Are Coupled Processes in Bacteroides. mSphere 2018, 3, e00559-18. [Google Scholar] [CrossRef] [Green Version]
- Pohl, S.; Harwood, C.R. Heterologous protein secretion by bacillus species from the cradle to the grave. Adv. Appl. Microbiol. 2010, 73, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Naskar, A.; Cho, H.; Lee, S.; Kim, K.S. Biomimetic Nanoparticles Coated with Bacterial Outer Membrane Vesicles as a New-Generation Platform for Biomedical Applications. Pharmaceutics 2021, 13, 1887. [Google Scholar] [CrossRef]
- Li, M.; Zhou, H.; Yang, C.; Wu, Y.; Zhou, X.; Liu, H.; Wang, Y. Bacterial outer membrane vesicles as a platform for biomedical applications: An update. J. Control. Release 2020, 323, 253–268. [Google Scholar] [CrossRef]
- Chen, L.; Valentine, J.L.; Huang, C.J.; Endicott, C.E.; Moeller, T.D.; Rasmussen, J.A.; Fletcher, J.R.; Boll, J.M.; Rosenthal, J.A.; Dobruchowska, J.; et al. Outer membrane vesicles displaying engineered glycotopes elicit protective antibodies. Proc. Natl. Acad. Sci. USA 2016, 113, E3609–E3618. [Google Scholar] [CrossRef] [Green Version]
- Irene, C.; Fantappie, L.; Caproni, E.; Zerbini, F.; Anesi, A.; Tomasi, M.; Zanella, I.; Stupia, S.; Prete, S.; Valensin, S.; et al. Bacterial outer membrane vesicles engineered with lipidated antigens as a platform for Staphylococcus aureus vaccine. Proc. Natl. Acad. Sci. USA 2019, 116, 21780–21788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecrivain, A.L.; Beckmann, B.M. Bacterial RNA in extracellular vesicles: A new regulator of host-pathogen interactions? Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194519. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Shao, X.; Zheng, H.; Li, M.; Wang, J.; Zhang, Q.; Li, L.; Liu, Z.; Sun, M.; Wang, S.; et al. Complete genome sequence of Bacillus thuringiensis mutant strain BMB171. J. Bacteriol. 2010, 192, 4074–4075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, C.; Ma, Y.; Wang, X.; Xie, Y.; Ali, M.K.; He, J. Functional analysis of the sporulation-specific diadenylate cyclase CdaS in Bacillus thuringiensis. Front. Microbiol. 2015, 6, 908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Rawat, S.; Arora, V.; Kottarath, S.K.; Dinda, A.K.; Vaishnav, P.K.; Nayak, B.; Mohanty, S. An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Stem Cell Res. Ther. 2018, 9, 180. [Google Scholar] [CrossRef] [Green Version]

| Strains and Plasmids | Relevant Properties | Source of Reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | F- φ80 lac ZΔM15 Δ (lacZYA-arg F) U169 endA1 recA1 hsdR17(rk−, mk+) supE44λ-thi-1 gyrA96 relA1phoA; host strain for plasmid construction | Stored in lab |
| Bacillus thuringiensis | ||
| BMB171 | An acrystalliferous mutant strain; high transformation frequency | [43] |
| BMB171-Sec | secYEG integrate into BMB171 genome, overexpression of secYEG in BMB171 | This study |
| BMB171/pHT-vip3Aa | BMB171 harboring pHT-vip3Aa | This study |
| BMB171/pPCspvip3 | BMB171 harboring pPCspvip3 | This study |
| BMB171-Sec/pHT-vip3Aa | BMB171-Sec harboringpHT-vip3Aa | This study |
| BMB171/pHT-mbp | BMB171 harboring pHT-mbp | This study |
| BMB171-Sec/pHT-mbp | BMB171-Sec harboring pHT-mbp | This study |
| BMB171/pPΔα1vip | BMB171 harboring pPΔα1vip | This study |
| BMB171/pPΔα2vip | BMB171 harboring pPΔα2vip | This study |
| BMB171/pPΔα3vip | BMB171 harboring pPΔα3vip | This study |
| BMB171/pPΔα4vip | BMB171 harboring pPΔα4vip | This study |
| BMB171/pHT-chiB | BMB171 harboringpHT-chiB | This study |
| BMB171/pHT-ΔspchiB | BMB171 harboring pHT-ΔspchiB | This study |
| BMB171/pPN1ΔspchiB | BMB171 harboring pPN1ΔspchiB | This study |
| BMB171/pPN2ΔspchiB | BMB171 harboring pPN2ΔspchiB | This study |
| BMB171/pPN3ΔspchiB | BMB171 harboring pPN3ΔspchiB | This study |
| BMB171/pPα1VΔspchiB | BMB171 harboring pPα1VΔspchiB | This study |
| Plasmids | ||
| pHT1k | E.coli and B. thuringiensis shuttle vector; AmpR, ErmR | [44] |
| pHT-vip3Aa | pHT1K + Promotor-Prsi + vip3Aa | This study |
| pPCspvip3 | pHT1K + Promotor-Prsi + signal peptide of ChiB + vip3Aa | This study |
| pHT-mbp | pHT1K + Promotor-Prsi + mbp | This study |
| pPΔα1vip | pHT1K + Promotor-Prsi + Δα1vip3Aa | This study |
| pPΔα2vip | pHT1K + Promotor-Prsi + Δα2vip3Aa | This study |
| pPΔα3vip | pHT1K + Promotor-Prsi + Δα3vip3Aa | This study |
| pPΔα4vip | pHT1K + Promotor-Prsi + Δα4vip3Aa | This study |
| pHT-chiB | pHT1K + Promotor-Prsi + chiB | This study |
| pHT-ΔspchiB | pHT1K + Promotor-Prsi + ΔspchiB | This study |
| pPN1ΔspchiB | pHT1K + Promotor-Prsi + N1 (N-terminal 77 amino acids of Vip3Aa including α1 and α2) + ΔspchiB | This study |
| pPN2ΔspchiB | pHT1K + Promotor-Prsi + N2 (N-terminal 39 amino acids of Vip3Aa including α1) + ΔspchiB | This study |
| pPN3ΔspchiB | pHT1K + Promotor-Prsi + N3 (N-terminal 22 amino acids and α2 of Vip3Aa) + ΔspchiB | This study |
| pPα1VΔspchiB | pHT1K + Promotor-Prsi + α1 of Vip3Aa + ΔspchiB | This study |
| pRB1028 | B. thuringiensis knockout vector; spcR | Stored in lab |
| pRB-secYEG | pRB1028-secYEG(U + secYEG + D); to overexpress secYEG | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, X.; Tian, H.; An, B.; Yan, B.; Cai, J. Vegetative Insecticidal Protein Vip3Aa Is Transported via Membrane Vesicles in Bacillus thuringiensis BMB171. Toxins 2022, 14, 480. https://doi.org/10.3390/toxins14070480
Zhang Y, Li X, Tian H, An B, Yan B, Cai J. Vegetative Insecticidal Protein Vip3Aa Is Transported via Membrane Vesicles in Bacillus thuringiensis BMB171. Toxins. 2022; 14(7):480. https://doi.org/10.3390/toxins14070480
Chicago/Turabian StyleZhang, Yizhuo, Xuelian Li, Hongwei Tian, Baoju An, Bing Yan, and Jun Cai. 2022. "Vegetative Insecticidal Protein Vip3Aa Is Transported via Membrane Vesicles in Bacillus thuringiensis BMB171" Toxins 14, no. 7: 480. https://doi.org/10.3390/toxins14070480
APA StyleZhang, Y., Li, X., Tian, H., An, B., Yan, B., & Cai, J. (2022). Vegetative Insecticidal Protein Vip3Aa Is Transported via Membrane Vesicles in Bacillus thuringiensis BMB171. Toxins, 14(7), 480. https://doi.org/10.3390/toxins14070480





