Extracellular Vesicles from Bothrops jararaca Venom Are Diverse in Structure and Protein Composition and Interact with Mammalian Cells
Abstract
1. Introduction
2. Results
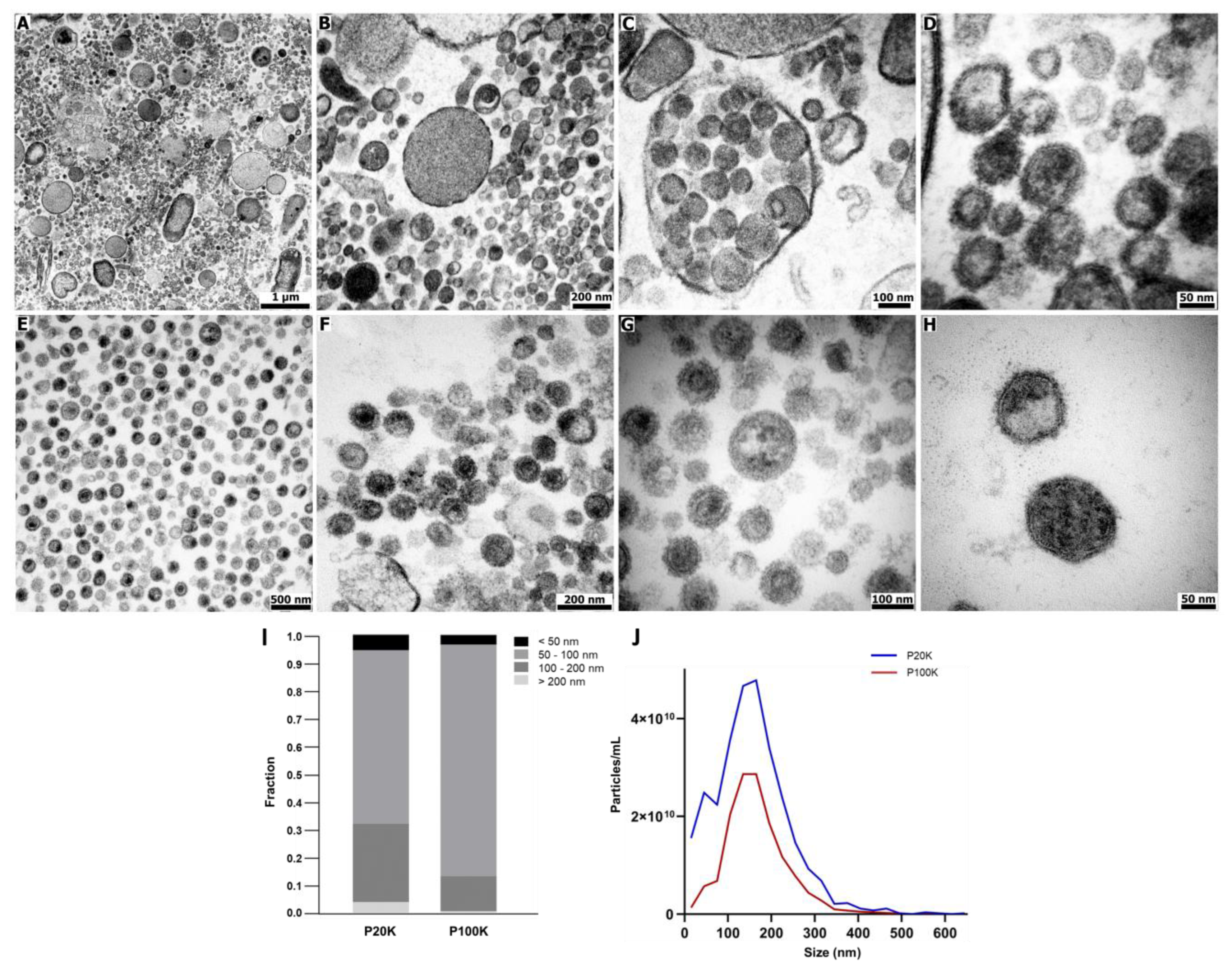
2.1. Bothrops Jararaca Venom Contains Distinct Extracellular Vesicles Populations
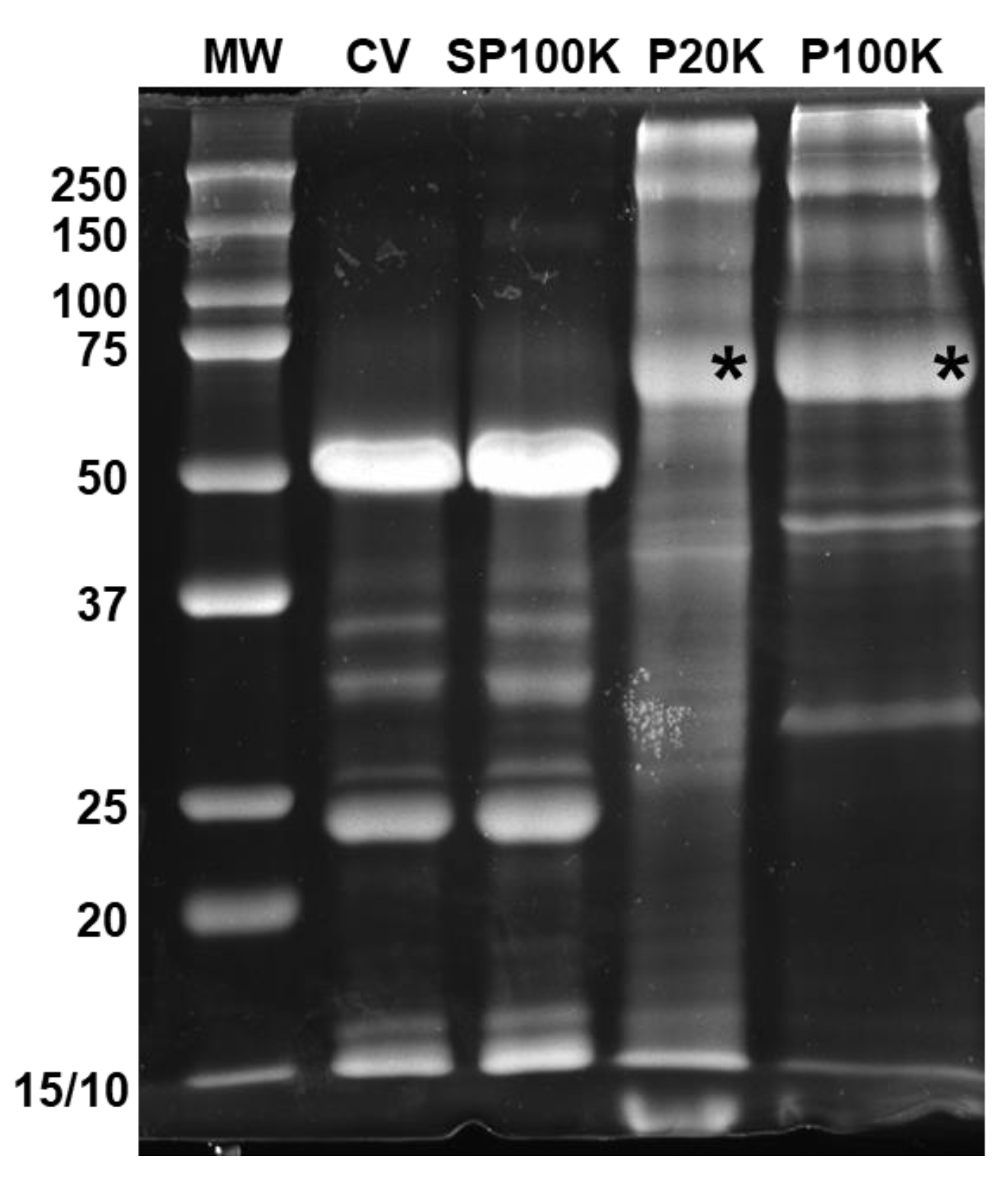
2.2. Bj-EVs Exhibit Diverse Protein Composition, Conserved EV Biomarkers, and Their Venom-Related Proteins Signature
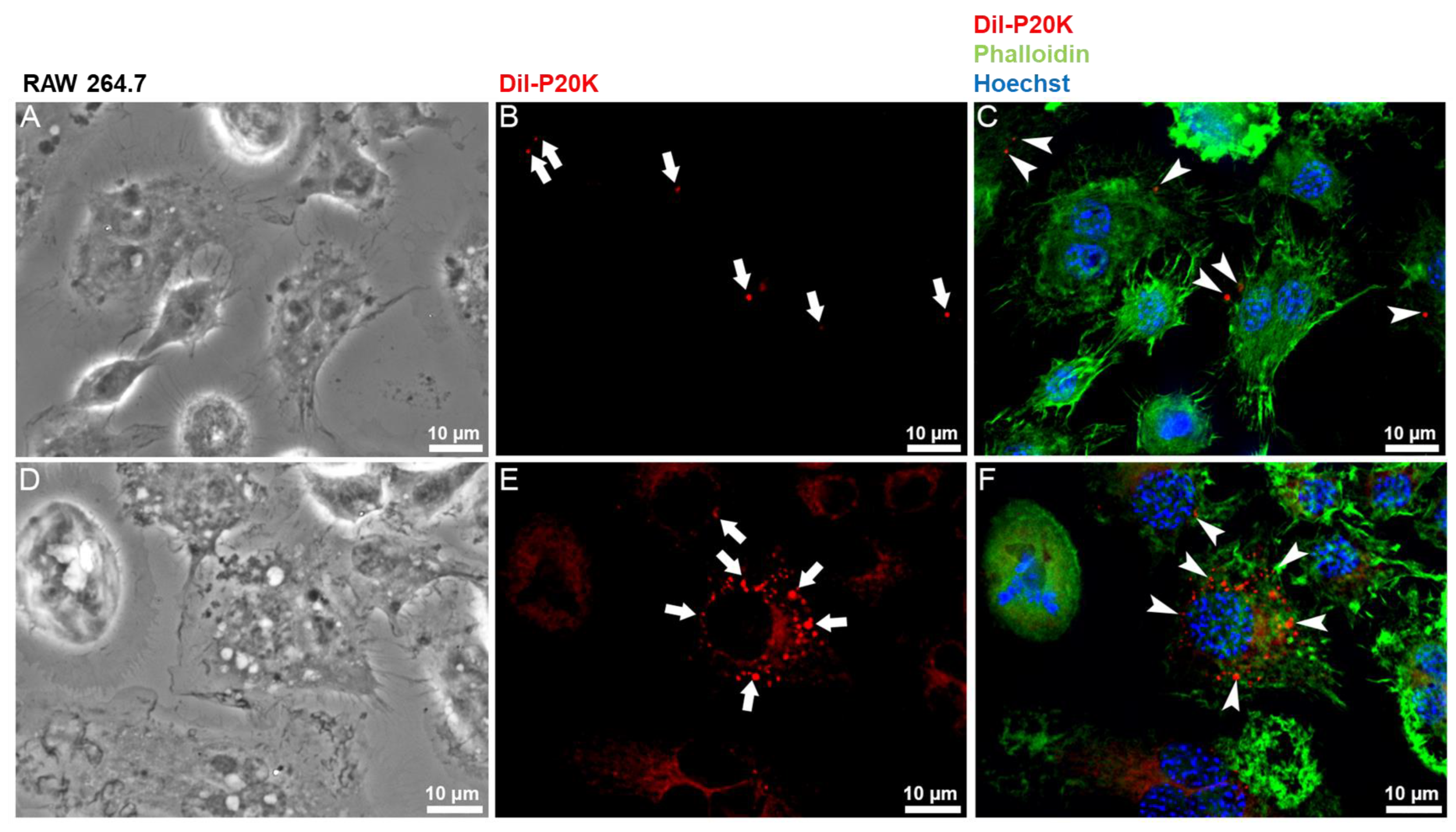
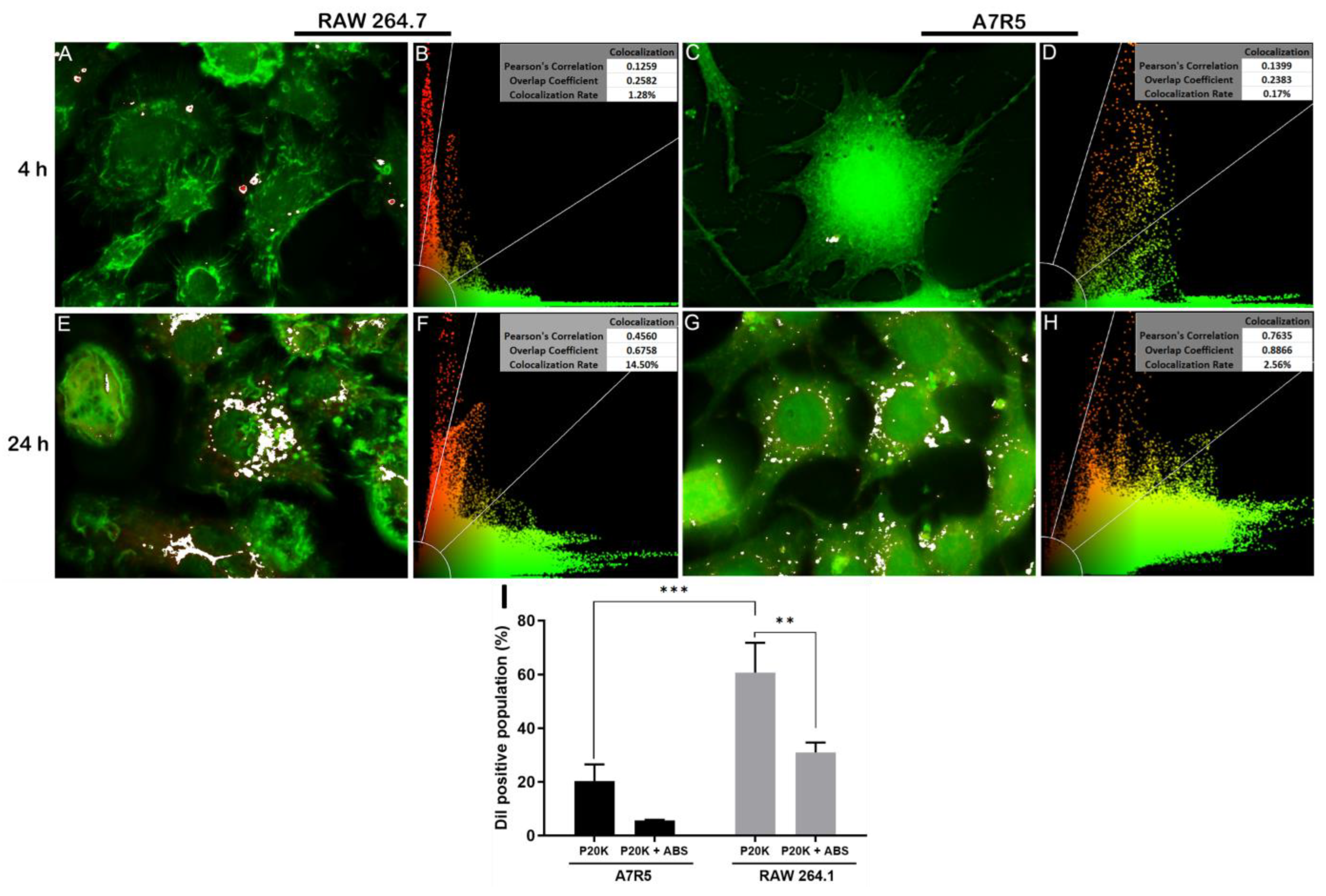
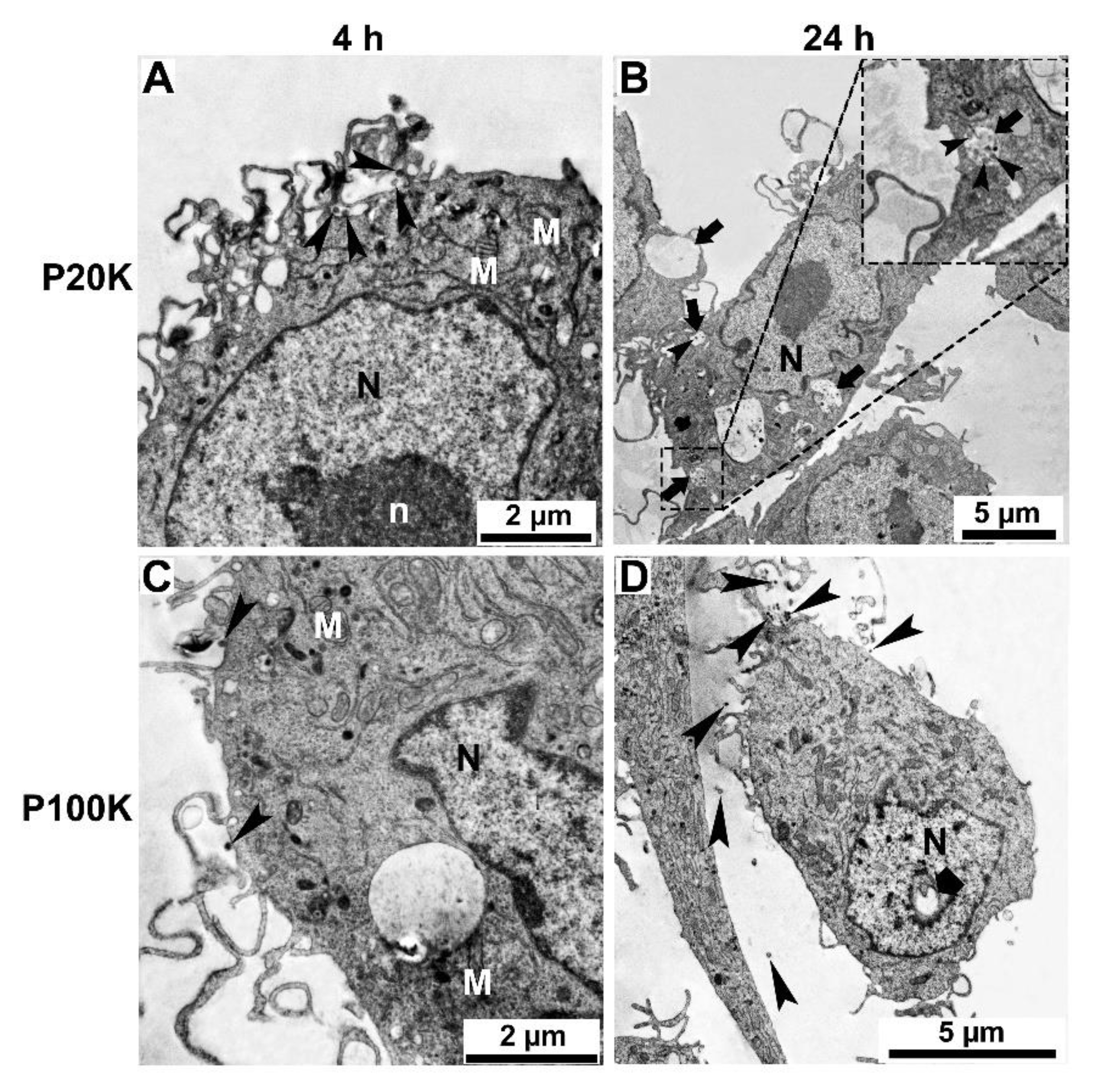
2.3. SvEVs Interact with Mammalian Cells
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bothrops jararaca Venom
5.2. Extracellular Vesicles Isolation
5.3. Transmission Electron Microscopy (TEM)
5.4. EV Size Analysis
5.5. SDS-PAGE and Western Blot
5.6. Proteomics
5.6.1. In-Gel Protein Digestion and Mass Spectrometry
5.6.2. Protein Digestion and Mass Spectrometry for Shotgun Proteomics
Data Analysis
- -
- Peptide Spectrum Matching (PSM)
- -
- Quantitative analysis
5.7. Immunofluorescence Microscopy
5.8. Bj-EV Uptake Quantification
5.9. Transmission Electron Microscopy in Scanning Electron Microscopy (STEM-IN-SEM)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Warshawsky, H.; Haddad, A.; Gonçalves, R.P.; Valeri, V.; De Lucca, F.L. Fine structure of the venom gland epithelium of the south american rattlesnake and radioautographic studies of protein formation by the secretory cells. Am. J. Anat. 1973, 138, 79–119. [Google Scholar] [CrossRef]
- Oron, U.; Bdolah, A. Intracellular transport of proteins in active and resting secretory cells of the venom gland of Vipera palaestinae. J. Cell Biol. 1978, 78, 488–502. [Google Scholar] [CrossRef]
- Villar-Briones, A.; Aird, S.D. Organic and peptidyl constituents of snake venoms: The picture is vastly more complex than we imagined. Toxins 2018, 10, 392. [Google Scholar] [CrossRef] [PubMed]
- Zancolli, G.; Casewell, N.R. Venom systems as models for studying the origin and regulation of evolutionary novelties. Mol. Biol. Evol. 2020, 37, 2777–2790. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Scheib, H.; van der Weerd, L.; Young, B.; McNaughtan, J.; Ramjan, S.F.R.; Vidal, N.; Poelmann, R.E.; Norman, J.A. Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol. Cell. Proteom. 2008, 7, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wagstaff, S.C.; Harrison, R.A.; Renjifo, C.; Wüster, W. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 2011, 28, 2637–2649. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Jackson, T.N.W.; Laustsen, A.H.; Sunagar, K. Causes and Consequences of Snake Venom Variation. Trends Pharmacol. Sci. 2020, 41, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Sazima, I. Natural History of the Jararaca Pitviper, Bothrops Jararaca, in Southeastern Brazil. In Biology of the Pitvipers; Selva Publishing: Tyler, TX, USA, 1992. [Google Scholar]
- De Sousa, L.F.; Nicolau, C.A.; Peixoto, P.S.; Bernardoni, J.L.; Oliveira, S.S.; Portes-Junior, J.A.; Mourão, R.H.V.; Lima-dos-Santos, I.; Sano-Martins, I.S.; Chalkidis, H.M.; et al. Comparison of Phylogeny, Venom Composition and Neutralization by Antivenom in Diverse Species of Bothrops Complex. PLoS Negl. Trop. Dis. 2013, 7, e2442. [Google Scholar] [CrossRef] [PubMed]
- Zelanis, A.; Tashima, A.K.; Pinto, A.F.M.; Leme, A.F.P.; Stuginski, D.R.; Furtado, M.F.; Sherman, N.E.; Ho, P.L.; Fox, J.W.; Serrano, S.M.T. Bothrops jararaca venom proteome rearrangement upon neonate to adult transition. Proteomics 2011, 11, 4218–4228. [Google Scholar] [CrossRef]
- Zelanis, A.; Menezes, M.C.; Kitano, E.S.; Liberato, T.; Tashima, A.K.; Pinto, A.F.M.; Sherman, N.E.; Ho, P.L.; Fox, J.W.; Serrano, S.M.T. Proteomic identification of gender molecular markers in Bothrops jararaca venom. J. Proteom. 2016, 139, 26–37. [Google Scholar] [CrossRef]
- Dias, G.S.; Kitano, E.S.; Pagotto, A.H.; Sant’Anna, S.S.; Rocha, M.M.T.; Zelanis, A.; Serrano, S.M.T. Individual variability in the venom proteome of juvenile bothrops jararaca specimens. J. Proteome Res. 2013, 12, 4585–4598. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves-Machado, L.; Pla, D.; Sanz, L.; Jorge, R.J.B.; Leitão-De-Araújo, M.; Alves, M.L.M.; Alvares, D.J.; De Miranda, J.; Nowatzki, J.; de Morais-Zani, K.; et al. Combined venomics, venom gland transcriptomics, bioactivities, and antivenomics of two Bothrops jararaca populations from geographic isolated regions within the Brazilian Atlantic rainforest. J. Proteom. 2016, 135, 73–89. [Google Scholar] [CrossRef]
- Moura-da-Silva, A.M.; Contreras-Bernal, J.C.; Gimenes, S.N.C.; Freitas-de-Sousa, L.A.; Portes-Junior, J.A.; da Silva Peixoto, P.; Iwai, L.K.; de Moura, V.M.; Bisnetoid, P.F.; Lacerda, M.; et al. The relationship between clinics and the venom of the causative amazon pit viper (Bothrops atrox). PLoS Negl. Trop. Dis. 2020, 14, e0008299. [Google Scholar] [CrossRef]
- Slagboom, J.; Kool, J.; Harrison, R.A.; Casewell, N.R. Haemotoxic snake venoms: Their functional activity, impact on snakebite victims and pharmaceutical promise. Br. J. Haematol. 2017, 177, 947–959. [Google Scholar] [CrossRef]
- Herrera, C.; Macêdo, J.K.A.; Feoli, A.; Escalante, T.; Rucavado, A.; Gutiérrez, J.M.; Fox, J.W. Muscle Tissue Damage Induced by the Venom of Bothrops asper: Identification of Early and Late Pathological Events through Proteomic Analysis. PLoS Negl. Trop. Dis. 2016, 10, e0004599. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Prim. 2017, 3, 17063. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.H.; Bartelt, D.C.; Greene, L.J. Isolation of Bradykinin-Potentiating Peptides from Bothrops jararaca Venom. Biochemistry 1970, 9, 2583–2593. [Google Scholar] [CrossRef] [PubMed]
- Péterfi, O.; Boda, F.; Szabó, Z.; Ferencz, E.; Bába, L. Hypotensive Snake Venom Components—A Mini-Review. Molecules 2019, 24, 2778. [Google Scholar] [CrossRef]
- Gimenes, S.N.C.; Sachett, J.A.G.; Colombini, M.; Freitas-De-sousa, L.A.; Ibiapina, H.N.S.; Costa, A.G.; Santana, M.F.; Park, J.-J.; Sherman, N.E.; Ferreira, L.C.L.; et al. Observation of bothrops atrox snake envenoming blister formation from five patients: Pathophysiological insights. Toxins 2021, 13, 800. [Google Scholar] [CrossRef]
- Mamede, C.C.N.; de Sousa Simamoto, B.B.; da Cunha Pereira, D.F.; de Oliveira Costa, J.; Ribeiro, M.S.M.; de Oliveira, F. Edema, hyperalgesia and myonecrosis induced by Brazilian bothropic venoms: Overview of the last decade. Toxicon 2020, 187, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Tchaou, B.A.; de Tové, K.-M.S.; N’Vènonfon, C.F.T.; Mfin, P.K.; Aguemon, A.-R.; Chobli, M.; Chippaux, J.-P. Acute kidney failure following severe viper envenomation: Clinical, biological and ultrasonographic aspects. J. Venom. Anim. Toxins Incl. Trop. Dis. 2020, 26, e20200059. [Google Scholar] [CrossRef]
- Boyer, L.V.; Seifert, S.A.; Clark, R.F.; McNally, J.T.; Williams, S.R.; Nordt, S.P.; Walter, F.G.; Dart, R.C. Recurrent and persistent coagulopathy following pit viper envenomation. Arch. Intern. Med. 1999, 159, 706–710. [Google Scholar] [CrossRef] [PubMed]
- Lavonas, E.J.; Khatri, V.; Daugherty, C.; Bucher-Bartelson, B.; King, T.; Dart, R.C. Medically significant late bleeding after treated Crotaline envenomation: A systematic review. Ann. Emerg. Med. 2014, 63, 71–78.e1. [Google Scholar] [CrossRef]
- Seifert, S.A.; Mascarenas, D.N.; Fullerton, L.; Warrick, B.J.; Smolinske, S.C. Unpredicted late-, new-onset thrombocytopenia and hypofibrinogenemia in Fab antivenom-treated rattlesnake envenomation. Toxicon 2020, 184, 55–56. [Google Scholar] [CrossRef] [PubMed]
- Brenes-Chacon, H.; Gutierrez, J.M.; Camacho-Badilla, K.; Soriano-Fallas, A.; Ulloa-Gutierrez, R.; Valverde, K.; Avila-Aguero, M.L. Long-term sequelae secondary to snakebite envenoming: A single centre retrospective study in a Costa Rican paediatric hospital. BMJ Paediatr. Open 2020, 4, e000735. [Google Scholar] [CrossRef]
- Lazaro, R.P. Complex regional pain syndrome following snakebite: A putatively rare complication of envenomation and review of the literature. Int. Med. Case Rep. J. 2020, 13, 603–607. [Google Scholar] [CrossRef]
- Carneiro, S.M.; Fernandes, W.; Sant’Anna, S.S.; Yamanouye, N. Microvesicles in the venom of Crotalus durissus terrificus (Serpentes, Viperidae). Toxicon 2007, 49, 106–110. [Google Scholar] [CrossRef]
- Ogawa, Y.; Kanai-Azuma, M.; Akimoto, Y.; Kawakami, H.; Yanoshita, R. Exosome-like vesicles in Gloydius blomhoffii blomhoffii venom. Toxicon 2008, 51, 984–993. [Google Scholar] [CrossRef]
- Souza-Imberg, A.; Carneiro, S.M.; Giannotti, K.C.; Sant’Anna, S.S.; Yamanouye, N. Origin and characterization of small membranous vesicles present in the venom of Crotalus durissus terrificus. Toxicon 2017, 136, 27–33. [Google Scholar] [CrossRef]
- Carregari, V.C.; Rosa-Fernandes, L.; Baldasso, P.; Bydlowski, S.P.; Marangoni, S.; Larsen, M.R.; Palmisano, G. Snake Venom Extracellular vesicles (SVEVs) reveal wide molecular and functional proteome diversity. Sci. Rep. 2018, 8, 12067. [Google Scholar] [CrossRef]
- Willard, N.K.; Salazar, E.; Oyervides, F.A.; Wiebe, C.S.; Ocheltree, J.S.; Cortez, M.; Perez, R.P.; Markowitz, H.; Iliuk, A.; Sanchez, E.E.; et al. Proteomic identification and quantification of snake venom biomarkers in venom and plasma extracellular vesicles. Toxins 2021, 13, 654. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- van Niel, G.; Carter, D.R.F.; Clayton, A.; Lambert, D.W.; Raposo, G.; Vader, P. Challenges and directions in studying cell–cell communication by extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2022, 23, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Chávez, A.S.O.; O’Neal, A.; Santambrogio, L.; Kotsyfakis, M.; Pedra, J.H.F. Message in a vesicle-trans-kingdom intercommunication at the vector-host interface. J. Cell Sci. 2019, 132, jcs224212. [Google Scholar] [CrossRef] [PubMed]
- Woith, E.; Fuhrmann, G.; Melzig, M.F. Extracellular vesicles—Connecting kingdoms. Int. J. Mol. Sci. 2019, 20, 5695. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.-M.; Palmquist, J.; Huang, S.-D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef]
- Zonneveld, M.I.; van Herwijnen, M.J.C.; Fernandez-Gutierrez, M.M.; Giovanazzi, A.; de Groot, A.M.; Kleinjan, M.; van Capel, T.M.M.; Sijts, A.J.A.M.; Taams, L.S.; Garssen, J.; et al. Human milk extracellular vesicles target nodes in interconnected signalling pathways that enhance oral epithelial barrier function and dampen immune responses. J. Extracell. Vesicles 2021, 10, e12071. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef]
- Vanaja, S.K.; Russo, A.J.; Behl, B.; Banerjee, I.; Yankova, M.; Deshmukh, S.D.; Rathinam, V.A.K. Bacterial Outer Membrane Vesicles Mediate Cytosolic Localization of LPS and Caspase-11 Activation. Cell 2016, 165, 1106–1119. [Google Scholar] [CrossRef] [PubMed]
- Babatunde, K.A.; Yesodha Subramanian, B.; Ahouidi, A.D.; Martinez Murillo, P.; Walch, M.; Mantel, P.Y. Role of Extracellular Vesicles in Cellular Cross Talk in Malaria. Front. Immunol. 2020, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Toda, H.; Diaz-Varela, M.; Segui-Barber, J.; Roobsoong, W.; Baro, B.; Garcia-Silva, S.; Galiano, A.; Gualdrón-López, M.; Almeida, A.C.G.; Brito, M.A.M.; et al. Plasma-derived extracellular vesicles from Plasmodium vivax patients signal spleen fibroblasts via NF-kB facilitating parasite cytoadherence. Nat. Commun. 2020, 11, 2761. [Google Scholar] [CrossRef] [PubMed]
- Wan, B.; Goguet, E.; Ravallec, M.; Pierre, O.; Lemauf, S.; Volkoff, A.-N.; Gatti, J.-L.; Poirié, M. Venom atypical extracellular vesicles as interspecies vehicles of virulence factors involved in host specificity: The case of a drosophilaparasitoid wasp. Front. Immunol. 2019, 10, 1688. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, L.M.; Cardenas, G.A.C.; Jiacomini, I.G.; Ramírez, M.I. A new insight into the cellular mechanisms of envenomation: Elucidating the role of extracellular vesicles in Loxoscelism. Toxicol. Lett. 2021, 350, 202–212. [Google Scholar] [CrossRef]
- Xun, C.; Wang, L.; Yang, H.; Xiao, Z.; Deng, M.; Xu, R.; Zhou, X.; Chen, P.; Liu, Z. Origin and characterization of extracellular vesicles present in the spider venom of ornithoctonus hainana. Toxins 2021, 13, 579. [Google Scholar] [CrossRef] [PubMed]
- Ofir-Birin, Y.; Regev-Rudzki, N. Extracellular vesicles in parasite survival. Science 2019, 363, 817–818. [Google Scholar] [CrossRef] [PubMed]
- Coakley, G.; Maizels, R.M.; Buck, A.H. Exosomes and Other Extracellular Vesicles: The New Communicators in Parasite Infections. Trends Parasitol. 2015, 31, 477–489. [Google Scholar] [CrossRef]
- Wey, B.; Heavner, M.E.; Wittmeyer, K.T.; Briese, T.; Hopper, K.R.; Govind, S. Immune suppressive extracellular vesicle proteins of Leptopilina heterotoma are encoded in the wasp genome. G3 Genes Genomes Genet. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Ramroop, J.R.; Heavner, M.E.; Razzak, Z.H.; Govind, S. A parasitoid wasp of Drosophila employs preemptive and reactive strategies to deplete its host’s blood cells. PLoS Pathog. 2021, 17, e1009615. [Google Scholar] [CrossRef] [PubMed]
- Wan, B.; Yang, L.; Zhang, J.; Qiu, L.; Fang, Q.; Yao, H.; Poirié, M.; Gatti, J.-L.; Ye, G. The Venom of the Ectoparasitoid Wasp Pachycrepoideus vindemiae (Hymenoptera: Pteromalidae) Induces Apoptosis of Drosophila melanogaster Hemocytes. Insects 2020, 11, 363. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P. Morphology and ultrastructure of the venom glands of the northern pacific rattlesnake Crotalus viridis oreganus. J. Morphol. 1991, 208, 109–128. [Google Scholar] [CrossRef]
- Ogawa, Y.; Mamura, Y.; Murayama, N.; Yanoshita, R. Characterization and cDNA cloning of dipeptidyl peptidase IV from the venom of Gloydius blomhoffi brevicaudus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2006, 145, 35–42. [Google Scholar] [CrossRef]
- Ogawa, Y.; Murayama, N.; Fujita, Y.; Yanoshita, R. Characterization and cDNA cloning of aminopeptidase A from the venom of Gloydius blomhoffi brevicaudus. Toxicon 2007, 49, 1172–1181. [Google Scholar] [CrossRef]
- Gandham, S.; Su, X.; Wood, J.; Nocera, A.L.; Alli, S.C.; Milane, L.; Zimmerman, A.; Amiji, M.; Ivanov, A.R. Technologies and Standardization in Research on Extracellular Vesicles. Trends Biotechnol. 2020, 38, 1066–1098. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, C.; Di Vizio, D.; Sahoo, S.; Théry, C.; Witwer, K.W.; Wauben, M.; Hill, A.F. Techniques used for the isolation and characterization of extracellular vesicles: Results of a worldwide survey. J. Extracell. Vesicles 2016, 5, 32945. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-K.; Lee, J.; Kim, S.R.; Choi, D.-S.; Yoon, Y.J.; Kim, J.H.; Go, G.; Nhung, D.; Hong, K.; Jang, S.C.; et al. EVpedia: A community web portal for extracellular vesicles research. Bioinformatics 2015, 31, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef] [PubMed]
- Melani, R.D.; Araujo, G.D.T.; Carvalho, P.C.; Goto, L.; Nogueira, F.C.S.; Junqueira, M.; Domont, G.B. Seeing beyond the tip of the iceberg: A deep analysis of the venome of the Brazilian Rattlesnake, Crotalus durissus terrificus. EuPA Open Proteom. 2015, 8, 144–156. [Google Scholar] [CrossRef]
- Viala, V.L.; Hildebrand, D.; Trusch, M.; Fucase, T.M.; Sciani, J.M.; Pimenta, D.C.; Arni, R.K.; Schlüter, H.; Betzel, C.; Mirtschin, P.; et al. Venomics of the Australian eastern brown snake (Pseudonaja textilis): Detection of new venom proteins and splicing variants. Toxicon 2015, 107, 252–265. [Google Scholar] [CrossRef]
- Shelke, G.V.; Yin, Y.; Jang, S.C.; Lässer, C.; Wennmalm, S.; Hoffmann, H.J.; Li, L.; Gho, Y.S.; Nilsson, J.A.; Lötvall, J. Endosomal signalling via exosome surface TGFβ-1. J. Extracell. Vesicles 2019, 8, 1650458. [Google Scholar] [CrossRef]
- Bonsergent, E.; Grisard, E.; Buchrieser, J.; Schwartz, O.; Théry, C.; Lavieu, G. Quantitative characterization of extracellular vesicle uptake and content delivery within mammalian cells. Nat. Commun. 2021, 12, 5721. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, H.M.; Masoumi, N.; Witwer, K.W.; Pegtel, D.M. Extracellular Vesicles Exploit Viral Entry Routes for Cargo Delivery. Microbiol. Mol. Biol. Rev. 2016, 80, 369–386. [Google Scholar] [CrossRef] [PubMed]
- Nabhan, J.F.; Hu, R.; Oh, R.S.; Cohen, S.N.; Lu, Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc. Natl. Acad. Sci. USA 2012, 109, 4146–4151. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, Y.; Peng, J.; Wu, D.; Zhao, X.; Cui, Y.; Chen, L.; Yan, X.; Du, Y.; Yu, L. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 2015, 25, 24–38. [Google Scholar] [CrossRef] [PubMed]
- D’Acunzo, P.; Pérez-González, R.; Kim, Y.; Hargash, T.; Miller, C.; Alldred, M.J.; Erdjument-Bromage, H.; Penikalapati, S.C.; Pawlik, M.; Saito, M.; et al. Mitovesicles are a novel population of extracellular vesicles of mitochondrial origin altered in down syndrome. Sci. Adv. 2021, 7, eabe5085. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Zhang, Q.; Jeppesen, D.K.; Higginbotham, J.N.; Graves-Deal, R.; Trinh, V.Q.; Ramirez, M.A.; Sohn, Y.; Neininger, A.C.; Taneja, N.; McKinley, E.T.; et al. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat. Cell Biol. 2021, 23, 1240–1254. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed]
- Kugeratski, F.G.; Hodge, K.; Lilla, S.; McAndrews, K.M.; Zhou, X.; Hwang, R.F.; Zanivan, S.; Kalluri, R. Quantitative Proteomics Identifies the Core Proteome of Exosomes with Syntenin-1 as the Highest Abundant Protein and a Putative Universal Biomarker. Nat. Cell Biol. 2021, 23, 631–641. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Ghossoub, R.; Lembo, F.; Rubio, A.; Gaillard, C.B.; Bouchet, J.; Vitale, N.; Slavík, J.; Machala, M.; Zimmermann, P. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 2014, 5, 3477. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [PubMed]
- Sales, P.B.V.; Santoro, M.L. Nucleotidase and DNase activities in Brazilian snake venoms. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2008, 147, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W. A brief review of the scientific history of several lesser-known snake venom proteins: L-amino acid oxidases, hyaluronidases and phosphodiesterases. Toxicon 2013, 62, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Isbister, G.K. A review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef]
- Melani, R.D.; Skinner, O.S.; Fornelli, L.; Domont, G.B.; Compton, P.D.; Kelleher, N.L. Mapping proteoforms and protein complexes from king cobra venom using both denaturing and native top-down proteomics. Mol. Cell. Proteom. 2016, 15, 2423–2434. [Google Scholar] [CrossRef]
- Gren, E.C.K.; Kitano, E.S.; Andrade-Silva, D.; Iwai, L.K.; Reis, M.S.; Menezes, M.C.; Serrano, S.M.T. Comparative analysis of the high molecular mass subproteomes of eight Bothrops snake venoms. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 30, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, C.; Huang, T.F. Inhibition of platelet aggregation by 5′ -nucleotidase purified from Trimeresurus gramineus snake venom. Toxicon 1983, 21, 491–501. [Google Scholar] [CrossRef]
- Dhananjaya, B.L.; Nataraju, A.; Rajesh, R.; Gowda, C.D.R.; Sharath, B.K.; Vishwanath, B.S.; D’Souza, C.J.M. Anticoagulant effect of Naja naja venom 5′nucleotidase: Demonstration through the use of novel specific inhibitor, vanillic acid. Toxicon 2006, 48, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Hart, M.L.; Köhler, D.; Eckle, T.; Kloor, D.; Stahl, G.L.; Eltzschig, H.K. Direct treatment of mouse or human blood with soluble 5′-nucleotidase inhibits platelet aggregation. Arter. Thromb. Vasc. Biol. 2008, 28, 1477–1483. [Google Scholar] [CrossRef] [PubMed]
- Sajevic, T.; Leonardi, A.; Križaj, I. Haemostatically active proteins in snake venoms. Toxicon 2011, 57, 627–645. [Google Scholar] [CrossRef]
- Trummal, K.; Samel, M.; Aaspõllu, A.; Tõnismägi, K.; Titma, T.; Subbi, J.; Siigur, J.; Siigur, E. 5′-Nucleotidase from Vipera lebetina venom. Toxicon 2015, 93, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Uzair, B.; Khan, B.A.; Sharif, N.; Shabbir, F.; Menaa, F. Phosphodiesterases (PDEs) from Snake Venoms: Therapeutic Applications. Protein Pept. Lett. 2018, 25, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Aird, S.D. Ophidian envenomation strategies and the role of purines. Toxicon 2002, 40, 335–393. [Google Scholar] [CrossRef]
- Aird, S.D. Taxonomic distribution and quantitative analysis of free purine and pyrimidine nucleosides in snake venoms. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2005, 140, 109–126. [Google Scholar] [CrossRef]
- Dhananjaya, B.L.; D’Souza, C.J.M. The pharmacological role of nucleotidases in snake venoms. Cell Biochem. Funct. 2010, 28, 171–177. [Google Scholar] [CrossRef]
- Caccin, P.; Pellegatti, P.; Fernandez, J.; Vono, M.; Cintra-Francischinelli, M.; Lomonte, B.; Gutiérrez, J.M.; Di Virgilio, F.; Montecucco, C. Why myotoxin-containing snake venoms possess powerful nucleotidases? Biochem. Biophys. Res. Commun. 2013, 430, 1289–1293. [Google Scholar] [CrossRef] [PubMed]
- Simmons, W.H.; Orawski, A.T. Membrane-bound Aminopeptidase P from Bovine Lung. Its purification, properties, and degradation of bradykinin. J. Biol. Chem. 1992, 267, 4897–4903. [Google Scholar] [CrossRef]
- Harris, J.; Schwinn, N.; Mahoney, J.A.; Lin, H.-H.; Shaw, M.; Howard, C.J.; Da Silva, R.P.; Gordon, S. A vitellogenic-like carboxypeptidase expressed by human macrophages is localized in endoplasmic reticulum and membrane ruffles. Int. J. Exp. Pathol. 2006, 87, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Mucha, A.; Drag, M.; Dalton, J.P.; Kafarski, P. Metallo-aminopeptidase inhibitors. Biochimie 2010, 92, 1509–1529. [Google Scholar] [CrossRef]
- Malovan, G.; Hierzberger, B.; Suraci, S.; Schaefer, M.; Santos, K.; Jha, S.; Macheroux, P. The emerging role of dipeptidyl peptidase 3 in pathophysiology. FEBS J. 2022, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P.; Baxter, L.M. Bioweapons synthesis and storage: The venom gland of front-fanged snakes. Zool. Anz. J. Comp. Zool. 2006, 245, 147–159. [Google Scholar] [CrossRef]
- Mackessy, S.P. Venom production and secretion in reptiles. J. Exp. Biol. 2022, 225, jeb227348. [Google Scholar] [CrossRef]
- Serrano, S.M.T.; Maroun, R.C. Snake venom serine proteinases: Sequence homology vs. substrate specificity, a paradox to be solved. Toxicon 2005, 45, 1115–1132. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M.T. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008, 275, 3016–3030. [Google Scholar] [CrossRef] [PubMed]
- Moura-da-Silva, A.M.; Almeida, M.T.; Portes-Junior, J.A.; Nicolau, C.A.; Gomes-Neto, F.; Valente, R.H. Processing of snake venom metalloproteinases: Generation of toxin diversity and enzyme inactivation. Toxins 2016, 8, 183. [Google Scholar] [CrossRef] [PubMed]
- Murayama, N.; Hayashi, M.A.F.; Ohi, H.; Ferreira, L.A.F.; Hermann, V.V.; Saito, H.; Fujita, Y.; Higuchi, S.; Fernandes, B.L.; Yamane, T.; et al. Cloning and sequence analysis of a Bothrops jararaca cDNA encoding a precursor of seven bradykinin-potentiating peptides and a C-type natriuretic peptide. Proc. Natl. Acad. Sci. USA 1997, 94, 1189–1193. [Google Scholar] [CrossRef] [PubMed]
- Lameu, C.; Neiva, M.; Hayashi, M.A.F. Venom Bradykinin-Related Peptides (BRPs) and Its Multiple Biological Roles. In An Integrated View of the Molecular Recognition and Toxinology; Baptista, G.R., Ed.; IntechOpen: London, UK, 2013. [Google Scholar]
- Almeida, D.D.; Viala, V.L.; Nachtigall, P.G.; Broe, M.; Lisle Gibbs, H.; De Toledo Serrano, S.M.; Moura-Da-Silva, A.M.; Ho, P.L.; Nishiyama, M.Y.; Junqueira-De-Azevedo, I.L.M. Tracking the recruitment and evolution of snake toxins using the evolutionary context provided by the Bothrops jararaca genome. Proc. Natl. Acad. Sci. USA 2021, 118, e2015159118. [Google Scholar] [CrossRef] [PubMed]
- Tashima, A.K.; Zelanis, A.; Kitano, E.S.; Ianzer, D.; Melo, R.L.; Rioli, V.; Sant’Anna, S.S.; Schenberg, A.C.G.; Camargo, A.C.M.; Serrano, S.M.T. Peptidomics of three bothrops snake venoms: Insights into the molecular diversification of proteomes and peptidomes. Mol. Cell. Proteom. 2012, 11, 1245–1262. [Google Scholar] [CrossRef] [PubMed]
- Cidade, D.A.P.; Wermelinger, L.S.; Lôbo-Hajdu, G.; Dávila, A.M.R.; Bon, C.; Zingali, R.B.; Albano, R.M. Molecular diversity of disintegrin-like domains within metalloproteinase precursors of Bothrops jararaca. Toxicon 2006, 48, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Portes-Junior, J.A.; Yamanouye, N.; Carneiro, S.M.; Knittel, P.S.; Sant’Anna, S.S.; Nogueira, F.C.S.; Junqueira, M.; Magalhaes, G.S.; Domont, G.B.; Moura-Da-Silva, A.M. Unraveling the processing and activation of snake venom metalloproteinases. J. Proteome Res. 2014, 13, 3338–3348. [Google Scholar] [CrossRef] [PubMed]
- Viana, L.G.; Valente, R.H.; Heluany, C.S.; Souza-Imberg, A.; Luna, M.S.; Perales, J.; Yamanouye, N. Bothrops jararaca venom gland secretory cells in culture: Effects of noradrenaline on toxin production and secretion. Toxicon 2017, 133, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Murayama, N.; Yanoshita, R. Molecular cloning and characterization of ecto-5′-nucleotidase from the venoms of Gloydius blomhoffi. Toxicon 2009, 54, 408–412. [Google Scholar] [CrossRef]
- Rokyta, D.R.; Lemmon, A.R.; Margres, M.J.; Aronow, K. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genom. 2012, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- Alcedo, K.P.; Bowser, J.L.; Snider, N.T. The elegant complexity of mammalian ecto-5′-nucleotidase (CD73). Trends Cell Biol. 2021, 31, 829–842. [Google Scholar] [CrossRef]
- Trams, E.G.; Lauter, C.J.; Salem, N., Jr.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Marar, C.; Starich, B.; Wirtz, D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021, 22, 560–570. [Google Scholar] [CrossRef]
- Chiarella, A.M.; Ryu, Y.K.; Manji, G.A.; Rustgi, A.K. Extracellular ATP and Adenosine in Cancer Pathogenesis and Treatment. Trends Cancer 2021, 7, 731–750. [Google Scholar] [CrossRef] [PubMed]
- Boison, D.; Yegutkin, G.G. Adenosine Metabolism: Emerging Concepts for Cancer Therapy. Cancer Cell 2019, 36, 582–596. [Google Scholar] [CrossRef]
- Schneider, E.; Winzer, R.; Rissiek, A.; Ricklefs, I.; Meyer-Schwesinger, C.; Ricklefs, F.L.; Bauche, A.; Behrends, J.; Reimer, R.; Brenna, S.; et al. CD73-mediated adenosine production by CD8 T cell-derived extracellular vesicles constitutes an intrinsic mechanism of immune suppression. Nat. Commun. 2021, 12, 5911. [Google Scholar] [CrossRef] [PubMed]
- Di Virgilio, F.; Adinolfi, E. Extracellular purines, purinergic receptors and tumor growth. Oncogene 2017, 36, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Cronstein, B.N.; Sitkovsky, M. Adenosine and adenosine receptors in the pathogenesis and treatment of rheumatic diseases. Nat. Rev. Rheumatol. 2017, 13, 41–51. [Google Scholar] [CrossRef]
- Perrot, I.; Michaud, H.A.; Giraudon-Paoli, M.; Augier, S.; Docquier, A.; Gros, L.; Courtois, R.; Déjou, C.; Jecko, D.; Becquart, O.; et al. Blocking Antibodies Targeting the CD39/CD73 Immunosuppressive Pathway Unleash Immune Responses in Combination Cancer Therapies. Cell Rep. 2019, 27, 2411–2425.e9. [Google Scholar] [CrossRef]
- Syn, N.; Wang, L.; Sethi, G.; Thiery, J.-P.; Goh, B.-C. Exosome-Mediated Metastasis: From Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharmacol. Sci. 2016, 37, 606–617. [Google Scholar] [CrossRef]
- Salimu, J.; Webber, J.; Gurney, M.; Al-Taei, S.; Clayton, A.; Tabi, Z. Dominant immunosuppression of dendritic cell function by prostate-cancer-derived exosomes. J. Extracell. Vesicles 2017, 6, 1368823. [Google Scholar] [CrossRef] [PubMed]
- Morandi, F.; Marimpietri, D.; Horenstein, A.L.; Bolzoni, M.; Toscani, D.; Costa, F.; Castella, B.; Faini, A.C.; Massaia, M.; Pistoia, V.; et al. Microvesicles released from multiple myeloma cells are equipped with ectoenzymes belonging to canonical and non-canonical adenosinergic pathways and produce adenosine from ATP and NAD+. Oncoimmunology 2018, 7, e1458809. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Li, R.; Yang, Y.; Shi, C.; Shen, Y.; Lu, C.; Chen, Y.; Zhou, W.; Lin, A.; Yu, L.; et al. Specific Decrease in B-Cell-Derived Extracellular Vesicles Enhances Post-Chemotherapeutic CD8+ T Cell Responses. Immunity 2019, 50, 738–750.e7. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Walle, T.; Cornish, A.E.; Basu, S.; Anandhan, S.; Fernandez, I.; Vence, L.; Blando, J.; Zhao, H.; Yadav, S.S.; et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat. Med. 2020, 26, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Dhananjaya, B.L.; Vishwanath, B.S.; D’souza, C.J. Snake Venom Nucleases, Nucleotidases, and Phosphomonoesterases. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press Taylor & Francis: Boca Raton, FL, USA, 2010; ISBN 9780849391651. [Google Scholar]
- Saini, A.; Patel, R.; Gaba, S.; Singh, G.; Gupta, G.D.; Monga, V. Adenosine receptor antagonists: Recent advances and therapeutic perspective. Eur. J. Med. Chem. 2022, 227, 113907. [Google Scholar] [CrossRef]
- Thompson, L.F.; Eltzschig, H.K.; Ibla, J.C.; Van De Wiele, C.J.; Resta, R.; Morote-Garcia, J.C.; Colgan, S.P. Crucial role for ecto-5′-nucleotidase (CD73) in vascular leakage during hypoxia. J. Exp. Med. 2004, 200, 1395–1405. [Google Scholar] [CrossRef]
- Chaves, M.M.; Savio, L.E.B.; Coutinho-Silva, R. Purinergic signaling: A new front-line determinant of resistance and susceptibility in leishmaniasis. Biomed. J. 2021, 45, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Tan, N.H.; Fung, S.Y.; Tan, K.Y.; Yap, M.K.K.; Gnanathasan, C.A.; Tan, C.H. Functional venomics of the Sri Lankan Russell’s viper (Daboia russelii) and its toxinological correlations. J. Proteom. 2015, 128, 403–423. [Google Scholar] [CrossRef]
- Laustsen, A.H.; Lomonte, B.; Lohse, B.; Fernández, J.; Gutiérrez, J.M. Unveiling the nature of black mamba (Dendroaspis polylepis) venom through venomics and antivenom immunoprofiling: Identification of key toxin targets for antivenom development. J. Proteom. 2015, 119, 126–142. [Google Scholar] [CrossRef] [PubMed]
- Dhananjaya, B.L.; D’Souza, C.J.M. An overview on nucleases (DNase, RNase, and phosphodiesterase) in snake venoms. Biochemistry (Moscow) 2010, 75, 1–6. [Google Scholar] [CrossRef]
- Jones, E.J.; Booth, C.; Fonseca, S.; Parker, A.; Cross, K.; Miquel-Clopés, A.; Hautefort, I.; Mayer, U.; Wileman, T.; Stentz, R.; et al. The Uptake, Trafficking, and Biodistribution of Bacteroides thetaiotaomicron Generated Outer Membrane Vesicles. Front. Microbiol. 2020, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Verweij, F.J.; Hyenne, V.; Van Niel, G.; Goetz, J.G. Extracellular Vesicles: Catching the Light in Zebrafish. Trends Cell Biol. 2019, 29, 770–776. [Google Scholar] [CrossRef]
- Verweij, F.J.; Balaj, L.; Boulanger, C.M.; Carter, D.R.F.; Compeer, E.B.; D’Angelo, G.; El Andaloussi, S.; Goetz, J.G.; Gross, J.C.; Hyenne, V.; et al. The power of imaging to understand extracellular vesicle biology in vivo. Nat. Methods 2021, 18, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- da Costa Galizio, N.; Serino-Silva, C.; Stuginski, D.R.; Abreu, P.A.E.; Sant’Anna, S.S.; Grego, K.F.; Tashima, A.K.; Tanaka-Azevedo, A.M.; de Morais-Zani, K. Compositional and functional investigation of individual and pooled venoms from long-term captive and recently wild-caught Bothrops jararaca snakes. J. Proteom. 2018, 186, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Furtado, M.F.D.; Travaglia-Cardoso, S.R.; Rocha, M.M.T. Sexual dimorphism in venom of Bothrops jararaca (Serpentes: Viperidae). Toxicon 2006, 48, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Antunes, T.C.; Yamashita, K.M.; Barbaro, K.C.; Saiki, M.; Santoro, M.L. Comparative analysis of newborn and adult Bothrops jararaca snake venoms. Toxicon 2010, 56, 1443–1458. [Google Scholar] [CrossRef] [PubMed]
- Verweij, F.J.; Revenu, C.; Arras, G.; Dingli, F.; Loew, D.; Pegtel, D.M.; Follain, G.; Allio, G.; Goetz, J.G.; Zimmermann, P.; et al. Live Tracking of Inter-organ Communication by Endogenous Exosomes In Vivo. Dev. Cell 2019, 48, 573–589.e4. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Takahashi, Y.; Nishikawa, M.; Kato, K.; Morishita, M.; Yamashita, T.; Matsumoto, A.; Charoenviriyakul, C.; Takakura, Y. Macrophage-dependent clearance of systemically administered B16BL6-derived exosomes from the blood circulation in mice. J. Extracell. Vesicles 2015, 4, 26238. [Google Scholar] [CrossRef]
- Yang, L.; Wan, B.; Wang, B.-B.; Liu, M.-M.; Fang, Q.; Song, Q.-S.; Ye, G.-Y. The Pupal Ectoparasitoid Pachycrepoideus vindemmiae Regulates Cellular and Humoral Immunity of Host Drosophila melanogaster. Front. Physiol. 2019, 10, 1282. [Google Scholar] [CrossRef]
- Wan, B.; Poirié, M.; Gatti, J.-L. Parasitoid wasp venom vesicles (venosomes) enter Drosophila melanogaster lamellocytes through a flotillin/lipid raft-dependent endocytic pathway. Virulence 2020, 11, 1512–1521. [Google Scholar] [CrossRef]
- Escalante, T.; Rucavado, A.; Fox, J.W.; Gutiérrez, J.M. Key events in microvascular damage induced by snake venom hemorrhagic metalloproteinases. J. Proteom. 2011, 74, 1781–1794. [Google Scholar] [CrossRef]
- Rucavado, A.; Escalante, T.; Kalogeropoulos, K.; Camacho, E.; Gutiérrez, J.M.; Fox, J.W. Analysis of wound exudates reveals differences in the patterns of tissue damage and inflammation induced by the venoms of Daboia russelii and Bothrops asper in mice. Toxicon 2020, 186, 94–104. [Google Scholar] [CrossRef]
- Fabbiano, F.; Corsi, J.; Gurrieri, E.; Trevisan, C.; Notarangelo, M.; D’Agostino, V.G. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J. Extracell. Vesicles 2020, 10, e12043. [Google Scholar] [CrossRef]
- He, B.; Cai, Q.; Qiao, L.; Huang, C.-Y.; Wang, S.; Miao, W.; Ha, T.; Wang, Y.; Jin, H. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 2021, 7, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Currier, R.B.; Calvete, J.J.; Sanz, L.; Harrison, R.A.; Rowley, P.D.; Wagstaff, S.C. Unusual stability of messenger RNA in snake venom reveals gene expression dynamics of venom replenishment. PLoS ONE 2012, 7, e41888. [Google Scholar] [CrossRef] [PubMed]
- Modahl, C.M.; Mackessy, S.P. Full-Length Venom Protein cDNA Sequences from Venom-Derived mRNA: Exploring Compositional Variation and Adaptive Multigene Evolution. PLoS Negl. Trop. Dis. 2016, 10, e0004587. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, G.; Logan, R.A.E.; Leung, K.-Y.D.; Newberry, F.J.; Rowley, P.D.; Dunbar, J.P.; Wagstaff, S.C.; Casewell, N.R.; Harrison, R.A. Stabilising the Integrity of Snake Venom mRNA Stored under Tropical Field Conditions Expands Research Horizons. PLoS Negl. Trop. Dis. 2016, 10, e0004615. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martin, R.; Wang, G.; Brandão, B.B.; Zanotto, T.M.; Shah, S.; Patel, S.K.; Schilling, B.; Kahn, C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2022, 601, 446–451. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Ferraz, C.R.; Arrahman, A.; Xie, C.; Casewell, N.R.; Lewis, R.J.; Kool, J.; Cardoso, F.C. Multifunctional toxins in snake venoms and therapeutic implications: From pain to hemorrhage and necrosis. Front. Ecol. Evol. 2019, 7, 218. [Google Scholar] [CrossRef]
- Carvalho, P.C.; Lima, D.B.; Leprevost, F.V.; Santos, M.D.M.; Fischer, J.S.G.; Aquino, P.F.; Moresco, J.J.; Yates, J.R., III; Barbosa, V.C. Integrated analysis of shotgun proteomic data with PatternLab for proteomics 4.0. Nat. Protoc. 2016, 11, 102–117. [Google Scholar] [CrossRef]
- Valente, R.H.; Luna, M.S.; de Oliveira, U.C.; Nishiyama-Junior, M.Y.; Junqueira-de-Azevedo, I.D.L.; Portes-Junior, J.A.; Clissa, P.B.; Viana, L.G.; Sanches, L.; Moura-da-Silva, A.M.; et al. Bothrops jararaca accessory venom gland is an ancillary source of toxins to the snake. J. Proteom. 2018, 177, 137–147. [Google Scholar] [CrossRef]
- Junqueira-De-Azevedo, I.L.M.; Bastos, C.M.V.; Ho, P.L.; Luna, M.S.; Yamanouye, N.; Casewell, N.R. Venom-related transcripts from bothrops jararaca tissues provide novel molecular insights into the production and evolution of snake venom. Mol. Biol. Evol. 2015, 32, 754–766. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, P.C.; Fischer, J.S.G.; Xu, T.; Yates, J.R.; Barbosa, V.C. PatternLab: From mass spectra to label-free differential shotgun proteomics. Curr. Protoc. Bioinform. 2012, 40, 13.19.1–13.19.18. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, P.C.; Yates, J.R.; Barbosa, V.C. Improving the TFold test for differential shotgun proteomics. Bioinformatics 2012, 28, 1652–1654. [Google Scholar] [CrossRef] [PubMed]
- Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, Ö.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; Bertier, L.; et al. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 2017, 14, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Roucourt, B.; Meeussen, S.; Bao, J.; Zimmermann, P.; David, G. Heparanase activates the syndecan-syntenin-ALIX exosomepathway. Cell Res. 2015, 25, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Zhuang, X.; Shen, J.; Jiang, L. Plant ESCRT Complexes: Moving Beyond Endosomal Sorting. Trends Plant Sci. 2017, 22, 986–998. [Google Scholar] [CrossRef]
- Choi, D.; Go, G.; Kim, D.K.; Lee, J.; Park, S.M.; Di Vizio, D.; Gho, Y.S. Quantitative proteomic analysis of trypsin-treatedextracellular vesicles to identify the real-vesicular proteins. J. Extracell. Vesicles 2020, 9, 1757209. [Google Scholar] [CrossRef]
- Gradilla, A.C.; González, E.; Seijo, I.; Andrés, G.; Bischoff, M.; González-Mendez, L.; Sánchez, V.; Callejo, A.; Ibáñez, C.; Guerra, M.; et al. Exosomes as Hedgehog carriers in cytoneme-mediated transport and secretion. Nat. Commun. 2014, 5, 5649. [Google Scholar] [CrossRef]
- Koles, K.; Nunnari, J.; Korkut, C.; Barria, R.; Brewer, C.; Li, Y.; Leszyk, J.; Zhang, B.; Budnik, V. Mechanism of evennessinterrupted (Evi)-exosome release at synaptic boutons. J. Biol. Chem. 2012, 287, 16820–16834. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Karfilis, K.V.; Ri, S.; Schekman, R. Y-box protein 1 is required to sort microRNAs intoexosomes in cells and in a cell-free reaction. eLife 2016, 5, e19276. [Google Scholar] [CrossRef] [PubMed]
- Sork, H.; Corso, G.; Krjutskov, K.; Johansson, H.J.; Nordin, J.Z.; Wiklander, O.P.B.; Lee, Y.X.F.; Westholm, J.O.; Lehtiö, J.; Wood, M.J.A.; et al. Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci. Rep. 2018, 8, 10813. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Koutras, C.; Donnelier, J.; Alshehri, M.; Fotouhi, M.; Girard, M.; Casha, S.; McPherson, P.S.; Robbins, S.M.; Braun, J.E.A. Neurons Export Extracellular Vesicles Enriched in Cysteine String Protein and Misfolded Protein Cargo. Sci. Rep. 2017, 7, 956. [Google Scholar] [CrossRef]
- Rodríguez-Vega, A.; Losada-Barragán, M.; Berbert, L.R.; Mesquita-Rodrigues, C.; Bombaça, A.C.S.; Menna-Barreto, R.; Aquino, P.; Carvalho, P.C.; Padrón, G.; de Jesus, J.B.; et al. Quantitative analysis of proteins secreted by Leishmania (Viannia)braziliensis strains associated to distinct clinical manifestations of American Tegumentary Leishmaniasis. J. Proteom. 2021, 232, 104077. [Google Scholar] [CrossRef] [PubMed]



| Bj-EVs | Crude Venom | |
|---|---|---|
| Major venom toxins | 5NTD ▲ | 5NTD |
| CTL | CTL ▲ | |
| LAO | LAO ▲ | |
| PDE ▲ | PDE | |
| PLA2 | PLA2 ▲ | |
| SVMP | SVMP ▲ | |
| SVSP | SVSP ▲ | |
| X | svVEGF | |
| Minor venom proteins | Aminopeptidase A | X |
| Aminopeptidase P ▲ | Aminopeptidase P | |
| Dipeptidase-2 ▲ | Dipeptidase-2 | |
| X | Glutaminyl-peptide cyclotransferase | |
| X | Hyaluronidase |
| Possible Function | Venom-Related EV Markers | Enriched Samples |
|---|---|---|
| Toxic or venom production | Ecto-5′-nucleotidase—5NTD | P20K, P100K |
| Toxic or venom production | Phosphodiesterase—PDE | P20K, P100K |
| Toxic or venom processing | Aminopeptidases (A, P) | P20K, P100K |
| Toxic or venom processing | Dipeptidases (DPEP-2, CNDP) | P20K, P100K |
| Toxic or venom processing | Dipeptidyl peptidase 4—DPP-IV | P20K, P100K |
| Toxic or venom processing | Dipeptidyl peptidase 3—DPP-3 | P20K |
| Toxic or venom processing | Peptidase D | P20K |
| Toxic or venom processing | Serine carboxypeptidase CPVL | P20K |
| Venom maturation and stability | Peptidylprolyl isomerase | P20K, P100K |
| Venom maturation and stability | Disulfide-isomerase | P20K, P100K |
| Venom stability | PLA2 inhibitor | P20K |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves-Machado, L.; Verçoza, B.R.F.; Nogueira, F.C.S.; Melani, R.D.; Domont, G.B.; Rodrigues, S.P.; Rodrigues, J.C.F.; Zingali, R.B. Extracellular Vesicles from Bothrops jararaca Venom Are Diverse in Structure and Protein Composition and Interact with Mammalian Cells. Toxins 2022, 14, 806. https://doi.org/10.3390/toxins14110806
Gonçalves-Machado L, Verçoza BRF, Nogueira FCS, Melani RD, Domont GB, Rodrigues SP, Rodrigues JCF, Zingali RB. Extracellular Vesicles from Bothrops jararaca Venom Are Diverse in Structure and Protein Composition and Interact with Mammalian Cells. Toxins. 2022; 14(11):806. https://doi.org/10.3390/toxins14110806
Chicago/Turabian StyleGonçalves-Machado, Larissa, Brunno Renato Farias Verçoza, Fábio César Sousa Nogueira, Rafael Donadélli Melani, Gilberto Barbosa Domont, Silas Pessini Rodrigues, Juliany Cola Fernandes Rodrigues, and Russolina Benedeta Zingali. 2022. "Extracellular Vesicles from Bothrops jararaca Venom Are Diverse in Structure and Protein Composition and Interact with Mammalian Cells" Toxins 14, no. 11: 806. https://doi.org/10.3390/toxins14110806
APA StyleGonçalves-Machado, L., Verçoza, B. R. F., Nogueira, F. C. S., Melani, R. D., Domont, G. B., Rodrigues, S. P., Rodrigues, J. C. F., & Zingali, R. B. (2022). Extracellular Vesicles from Bothrops jararaca Venom Are Diverse in Structure and Protein Composition and Interact with Mammalian Cells. Toxins, 14(11), 806. https://doi.org/10.3390/toxins14110806






