A Spider Toxin Exemplifies the Promises and Pitfalls of Cell-Free Protein Production for Venom Biodiscovery
Abstract
:1. Introduction
2. Results and Discussion
2.1. Architecture of the Gene Construct
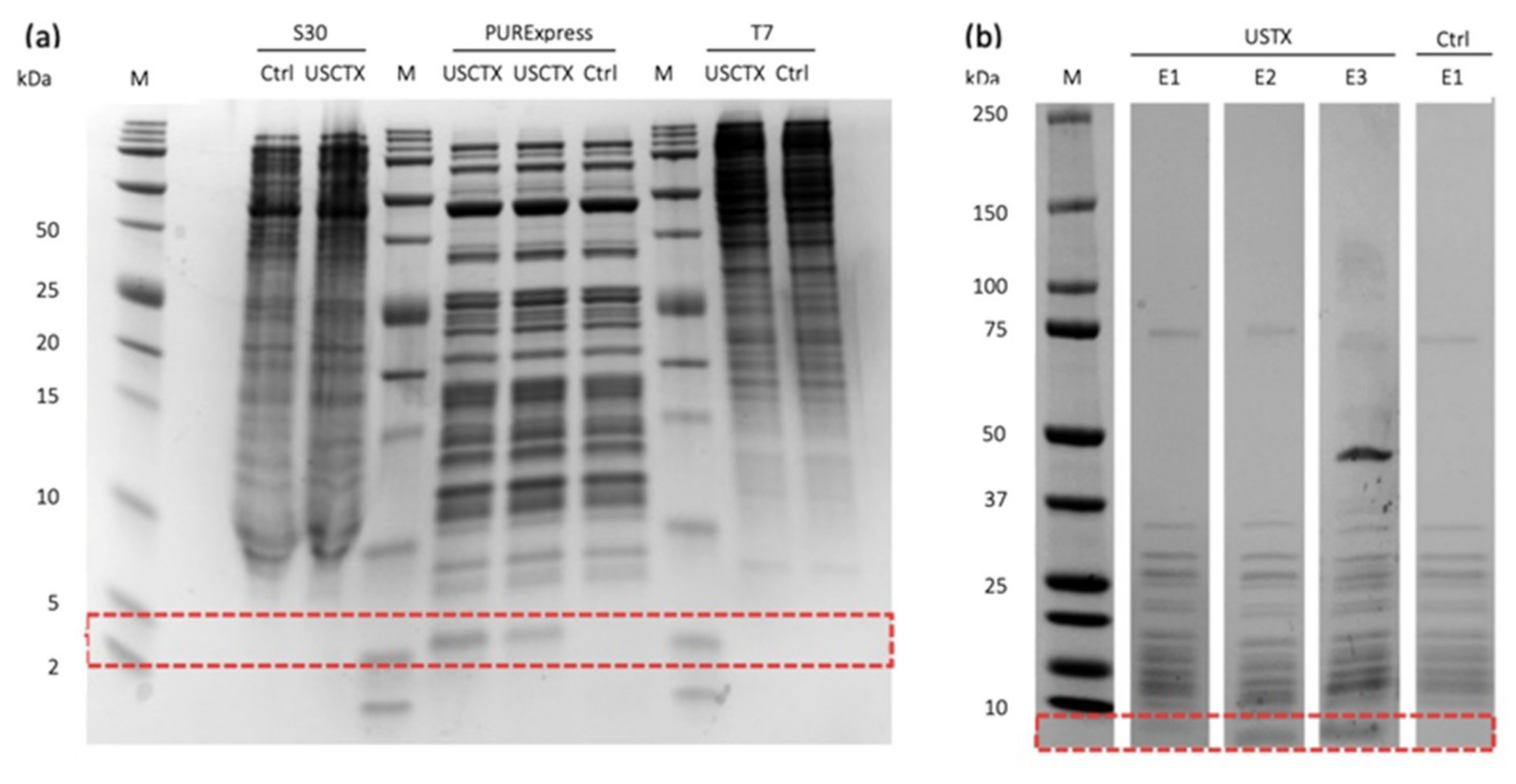
2.2. Only One Cell-Free System Produces USCTX
2.3. Purification of USCTX
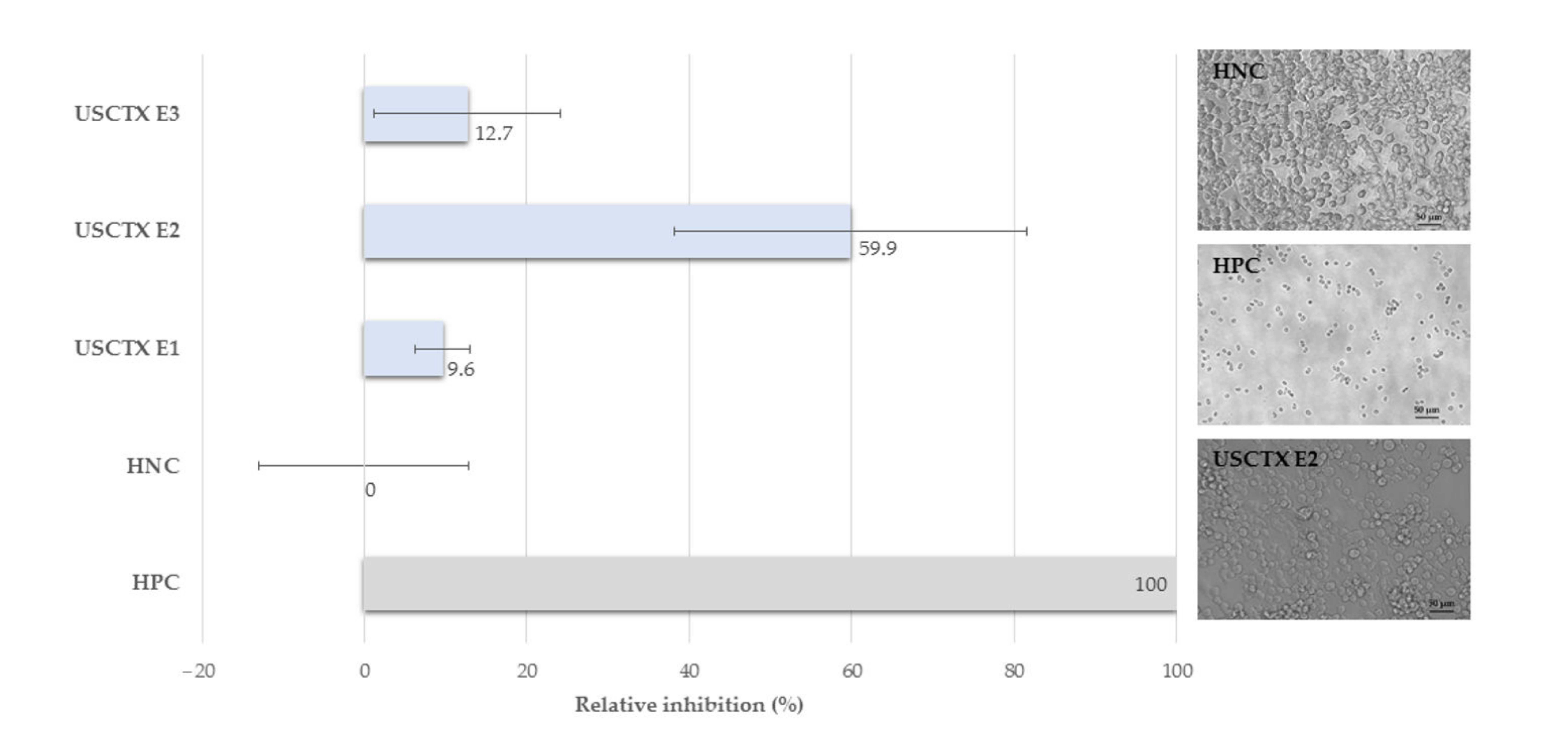
2.4. USCTX Is Active against Neuroblasts
2.5. Problematic Disulfide Crosslinking in Cell-Free-Expressed USCTX
2.6. Cell-Free Protein Production for Venom Bioprospecting?
3. Conclusions
4. Materials and Methods
4.1. Sequence Selection and Construct Preparation
4.2. Cell-Free Production and SDS-PAGE
4.3. IMAC Purification
4.4. MALDI-TOF Mass Spectrometry
4.5. LC-ESI Mass Spectrometry
4.6. Bioassays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.A.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The Toxicogenomic Multiverse: Convergent Recruitment of Proteins Into Animal Venoms. Annu. Rev. Genomics Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [Green Version]
- Herzig, V.; Cristofori-Armstrong, B.; Israel, M.R.; Nixon, S.A.; Vetter, I.; King, G.F. Animal toxins—Nature’s evolutionary-refined toolkit for basic research and drug discovery. Biochem. Pharmacol. 2020, 181, 114096. [Google Scholar] [CrossRef]
- Windley, M.J.; Herzig, V.; Dziemborowicz, S.A.; Hardy, M.C.; King, G.F.; Nicholson, G.M. Spider-venom peptides as bioinsecticides. Toxins 2012, 4, 191–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, G.F.; Hardy, M.C. Spider-venom peptides: Structure, pharmacology, and potential for control of insect pests. Annu. Rev. Entomol. 2013, 58, 475–496. [Google Scholar] [CrossRef]
- Herzig, V. Arthropod assassins: Crawling biochemists with diverse toxin pharmacopeias. Toxicon 2019, 158, 33–37. [Google Scholar] [CrossRef] [Green Version]
- Von Reumont, B.M.; Campbell, L.I.; Jenner, R.A. Quo Vadis venomics? A roadmap to neglected venomous invertebrates. Toxins 2014, 6, 3488–3551. [Google Scholar] [CrossRef] [PubMed]
- Lüddecke, T.; Vilcinskas, A.; Lemke, S. Phylogeny-guided selection of priority groups for venom bioprospecting: Harvesting toxin sequences in tarantulas as a case study. Toxins 2019, 11, 488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzig, V.; King, G.F.; Undheim, E.A.B. Can we resolve the taxonomic bias in spider venom research? Toxicon X 2019, 1, 100005. [Google Scholar] [CrossRef]
- Zobel-Thropp, P.A.; Mullins, J.; Kristensen, C.; Kronmiller, B.A.; David, C.L.; Breci, L.A.; Binford, G.J. Not so Dangerous After All? Venom Composition and Potency of the Pholcid (Daddy Long-Leg) Spider Physocyclus mexicanus. Front. Ecol. Evol. 2019, 7, 256. [Google Scholar] [CrossRef] [Green Version]
- Drukewitz, S.H.; von Reumont, B.M. The Significance of Comparative Genomics in Modern Evolutionary Venomics. Front. Ecol. Evol. 2019, 7, 163. [Google Scholar] [CrossRef] [Green Version]
- Luna-Ramirez, K.; Tonk, M.; Rahnamaeian, M.; Vilcinskas, A. Bioactivity of natural and engineered antimicrobial peptides from venom of the scorpions Urodacus yaschenkoi and U. Manicatus. Toxins 2017, 9, 22. [Google Scholar] [CrossRef] [Green Version]
- Heep, J.; Skaljac, M.; Grotmann, J.; Kessel, T.; Seip, M.; Schmidtberg, H.; Vilcinskas, A. Identification and functional characterization of a novel insecticidal decapeptide from the myrmicine ant Manica rubida. Toxins 2019, 11, 562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chassagnon, I.R.; McCarthy, C.A.; Chin, Y.K.Y.; Pineda, S.S.; Keramidas, A.; Mobli, M.; Pham, V.; De Silva, T.M.; Lynch, J.W.; Widdop, R.E.; et al. Potent neuroprotection after stroke afforded by a double-knot spider-venom peptide that inhibits acid-sensing ion channel 1a. Proc. Natl. Acad. Sci. USA 2017, 114, 3750–3755. [Google Scholar] [CrossRef] [Green Version]
- Escoubas, P.; Bernard, C.; Lambeau, G.; Lazdunski, M.; Darbon, H. Recombinant production and solution structure of PcTx1, the specific peptide inhibitor of ASIC1a proton-gated cation channels. Protein Sci. 2003, 12, 1332–1343. [Google Scholar] [CrossRef] [Green Version]
- Herzig, V.; Sunagar, K.; Wilson, D.T.R.; Pineda, S.S.; Israel, M.R.; Dutertre, S.; McFarland, B.S.; Undheim, E.A.B.; Hodgson, W.C.; Alewood, P.F.; et al. Australian funnel-web spiders evolved human-lethal δ-hexatoxins for defense against vertebrate predators. Proc. Natl. Acad. Sci. USA 2020, 117, 24920–24928. [Google Scholar] [CrossRef] [PubMed]
- Carlson, E.D.; Gan, R.; Hodgman, C.E.; Jewett, M.C. Cell-free protein synthesis: Applications come of age. Biotechnol. Adv. 2012, 30, 1185–1194. [Google Scholar] [CrossRef] [Green Version]
- Tinafar, A.; Jaenes, K.; Pardee, K. Synthetic Biology Goes Cell-Free. BMC Biol. 2019, 17, 64. [Google Scholar] [CrossRef] [Green Version]
- Krinsky, N.; Kaduri, M.; Shainsky-Roitman, J.; Goldfeder, M.; Ivanir, E.; Benhar, I.; Shoham, Y.; Schroeder, A. A simple and rapid method for preparing a cell-free bacterial lysate for protein synthesis. PLoS ONE 2016, 11, e0165137. [Google Scholar] [CrossRef]
- Silverman, A.D.; Karim, A.S.; Jewett, M.C. Cell-free gene expression: An expanded repertoire of applications. Nat. Rev. Genet. 2020, 21, 151–170. [Google Scholar] [CrossRef] [PubMed]
- Loening, N.M.; Wilson, Z.N.; Zobel-Thropp, P.A.; Binford, G.J. Solution Structures of Two Homologous Venom Peptides from Sicarius dolichocephalus. PLoS ONE 2013, 8, e54401. [Google Scholar] [CrossRef]
- Vlasak, R.; Kreil, G. Nucleotide sequence of cloned cDNAs coding for preprosecapin, a major product of queen-bee venom glands. Eur. J. Biochem. 1984, 145, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, W.; Kou, X.; Luo, Y.; Zhang, Y.; Ma, B.; Wang, M.; Huang, K. Establishment and optimization of a wheat germ cell-free protein synthesis system and its application in venom kallikrein. Protein Expr. Purif. 2012, 84, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Codon Usage Database. Available online: https://www.kazusa.or.jp/codon/ (accessed on 25 June 2021).
- Langenegger, N.; Nentwig, W.; Kuhn-Nentwig, L. Spider venom: Components, modes of action, and novel strategies in transcriptomic and proteomic analyses. Toxins 2019, 11, 611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matos, C.F.R.O.; Robinson, C.; Alanen, H.I.; Prus, P.; Uchida, Y.; Ruddock, L.W.; Freedman, R.B.; Keshavarz-Moore, E. Efficient export of prefolded, disulfide-bonded recombinant proteins to the periplasm by the Tat pathway in Escherichia coli CyDisCo strains. Biotechnol. Prog. 2014, 30, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Duracova, M.; Klimentova, J.; Fucikova, A.; Dresler, J. Proteomic methods of detection and quantification of protein toxins. Toxins 2018, 10, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, A.A.; Robinson, S.D.; Hamilton, B.F.; Undheim, E.A.B.; King, G.F. Deadly Proteomes: A Practical Guide to Proteotranscriptomics of Animal Venoms. Proteomics 2020, 20, 1900324. [Google Scholar] [CrossRef]
- Mouchbahani-Constance, S.; Sharif-Naeini, R. Proteomic and Transcriptomic Techniques to Decipher the Molecular Evolution of Venoms. Toxins 2021, 13, 154. [Google Scholar] [CrossRef]
- Hatahet, F.; Boyd, D.; Beckwith, J. Disulfide bond formation in prokaryotes: History, diversity and design. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 1402–1414. [Google Scholar] [CrossRef] [Green Version]
- Murakami, S.; Matsumoto, R.; Kanamori, T. Constructive approach for synthesis of a functional IgG using a reconstituted cell-free protein synthesis system. Sci. Rep. 2019, 9, 671. [Google Scholar] [CrossRef] [Green Version]
- Chiba, C.H.; Knirsch, M.C.; Azzoni, A.R.; Moreira, A.R.; Stephano, M.A. Cell-free protein synthesis: Advances on production process for biopharmaceuticals and immunobiological products. Biotechniques 2021, 70, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Zietek, B.M.; Still, K.B.M.; Jaschusch, K.; Bruyneel, B.; Ariese, F.; Brouwer, T.J.F.; Luger, M.; Limburg, R.J.; Rosier, J.C.; Iperen, D.J.V.; et al. Bioactivity Profiling of Small-Volume Samples by Nano Liquid Chromatography Coupled to Microarray Bioassaying Using High-Resolution Fractionation. Anal. Chem. 2019, 91, 10458–10466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zietek, B.M.; Mayar, M.; Slagboom, J.; Bruyneel, B.; Vonk, F.J.; Somsen, G.W.; Casewell, N.R.; Kool, J. Liquid chromatographic nanofractionation with parallel mass spectrometric detection for the screening of plasmin inhibitors and (metallo)proteinases in snake venoms. Anal. Bioanal. Chem. 2018, 410, 5751–5763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, C.; Slagboom, J.; Albulescu, L.O.; Somsen, G.W.; Vonk, F.J.; Casewell, N.R.; Kool, J. Neutralising effects of small molecule toxin inhibitors on nanofractionated coagulopathic Crotalinae snake venoms. Acta Pharm. Sin. B 2020, 10, 1835–1845. [Google Scholar] [CrossRef] [PubMed]
- Still, K.B.M.; Slagboom, J.; Kidwai, S.; Xie, C.; Zhao, Y.; Eisses, B.; Jiang, Z.; Vonk, F.J.; Somsen, G.W.; Casewell, N.R.; et al. Development of high-throughput screening assays for profiling snake venom phospholipase A2 activity after chromatographic fractionation. Toxicon 2020, 184, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Slagboom, J.; Mladić, M.; Xie, C.; Kazandjian, T.D.; Vonk, F.; Somsen, G.W.; Casewell, N.R.; Kool, J. High throughput screening and identification of coagulopathic snake venom proteins and peptides using nanofractionation and proteomics approaches. PLoS Negl. Trop. Dis. 2020, 14, e0007802. [Google Scholar] [CrossRef] [Green Version]
- Pineda, S.S.; Chaumeil, P.A.; Kunert, A.; Kaas, Q.; Thang, M.W.C.; Le, L.; Kuhn, M.; Herzig, V.; Saez, N.J.; Cristofori-Armstrong, B.; et al. ArachnoServer 3.0: An online resource for automated discovery, analysis and annotation of spider toxins. Bioinformatics 2018, 34, 1074–1076. [Google Scholar] [CrossRef] [Green Version]
- Von Reumont, B.M.; Lüddecke, T.; Timm, T.; Lochnit, G.; Vilcinskas, A.; von Döhren, J.; Nilsson, M.A. Proteo-transcriptomic analysis identifies potential novel toxins secreted by the predatory, orey-piercing ribbon worm Amphiporus lactifloreus. Mar. Drugs 2020, 18, 407. [Google Scholar] [CrossRef]
- Lüddecke, T.; von Reumont, B.M.; Förster, F.; Billion, A.; Timm, T.; Lochnit, G.; Vilcinskas, A.; Lemke, S. An Economic Dilemma Between Molecular Weapon Systems May Explain an Arachnological-atypical Venom in Wasp Spiders (Argiope bruennichi). Biomolecules 2020, 10, 978. [Google Scholar] [CrossRef] [PubMed]

| Component | Organism | System | Expression | Activity |
|---|---|---|---|---|
| Kallikrein [23] | Snake (not determined) | Wheat germ | Yes | Yes |
| Preprosecapin [22] | A. mellifera queens | Wheat germ | Yes | Not tested |
| USCTX | H. dolichocephala | NEB PURExpress (E. coli) | Yes | Yes |
| USCTX | H. dolichocephala | S30 Extract (E. coli) | No | – |
| USCTX | H. dolichocephala | TnT T7 (Spodoptera frugiperda) | No | – |
| Fraction | Toxin Yield | Purification Step | Solvent |
|---|---|---|---|
| E1 | Low | Flow-through | Cell-free extract |
| E2 | High | First washing | Water |
| E3 | High | Second washing | Washing buffer |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lüddecke, T.; Paas, A.; Talmann, L.; Kirchhoff, K.N.; von Reumont, B.M.; Billion, A.; Timm, T.; Lochnit, G.; Vilcinskas, A. A Spider Toxin Exemplifies the Promises and Pitfalls of Cell-Free Protein Production for Venom Biodiscovery. Toxins 2021, 13, 575. https://doi.org/10.3390/toxins13080575
Lüddecke T, Paas A, Talmann L, Kirchhoff KN, von Reumont BM, Billion A, Timm T, Lochnit G, Vilcinskas A. A Spider Toxin Exemplifies the Promises and Pitfalls of Cell-Free Protein Production for Venom Biodiscovery. Toxins. 2021; 13(8):575. https://doi.org/10.3390/toxins13080575
Chicago/Turabian StyleLüddecke, Tim, Anne Paas, Lea Talmann, Kim N. Kirchhoff, Björn M. von Reumont, André Billion, Thomas Timm, Günter Lochnit, and Andreas Vilcinskas. 2021. "A Spider Toxin Exemplifies the Promises and Pitfalls of Cell-Free Protein Production for Venom Biodiscovery" Toxins 13, no. 8: 575. https://doi.org/10.3390/toxins13080575
APA StyleLüddecke, T., Paas, A., Talmann, L., Kirchhoff, K. N., von Reumont, B. M., Billion, A., Timm, T., Lochnit, G., & Vilcinskas, A. (2021). A Spider Toxin Exemplifies the Promises and Pitfalls of Cell-Free Protein Production for Venom Biodiscovery. Toxins, 13(8), 575. https://doi.org/10.3390/toxins13080575







