Abstract
Snakebite envenomation is a serious neglected tropical disease, and its management is often complicated by the diversity of snake venoms. In Asia, pit vipers of the Ovophis species complex are medically important venomous snakes whose venom properties have not been investigated in depth. This study characterized the venom proteomes of Ovophis convictus (West Malaysia), Ovophis tonkinensis (northern Vietnam, southern China), and Ovophis okinavensis (Okinawa, Japan) by applying liquid chromatography-tandem mass spectrometry, which detected a high abundance of snake venom serine proteases (SVSP, constituting 40–60% of total venom proteins), followed by phospholipases A2, snake venom metalloproteinases of mainly P-III class, L-amino acid oxidases, and toxins from other protein families which were less abundant. The venoms exhibited different procoagulant activities in human plasma, with potency decreasing from O. tonkinensis > O. okinavensis > O. convictus. The procoagulant nature of venom confirms that consumptive coagulopathy underlies the pathophysiology of Ovophis pit viper envenomation. The hetero-specific antivenoms Gloydius brevicaudus monovalent antivenom (GbMAV) and Trimeresurus albolabris monovalent antivenom (TaMAV) were immunoreactive toward the venoms, and cross-neutralized their procoagulant activities, albeit at variably limited efficacy. In the absence of species-specific antivenom, these hetero-specific antivenoms may be useful in treating coagulotoxic envenomation caused by the different snakes in their respective regions.
Key Contribution:
This study investigated the proteomes and coagulotoxicity of the Ovophis pit viper venom of three species from different geographical locales, elucidating the venom variability and pathophysiology of envenomation. The findings regarding the immunoreactivity and cross-neutralization activities of hetero-specific antivenoms provided insights into the treatment of snakebite envenomation caused by these Asiatic pit viper species.
1. Introduction
Snakebite envenomation affects poor and marginalized populations in the tropics and subtropics most heavily, killing more than 100,000 people annually, and with three times as many suffer chronic complications [1,2,3,4]. In 2017, the World Health Organization formally recognized snakebite envenomation as one of the most neglected tropical diseases and called for a global strategy to overcome the problem [5]. This global health crisis remains to be adequately addressed with a comprehensive solution.
The management of snakebite envenomation tends to be complicated by the great diversity and variable distribution of venomous snakes in different regions. Knowledge gaps in the identity, geographical distribution, and venom properties of specific species limit our understanding of the pathophysiology of envenomation, and this consequently hinders appropriate treatment [3]. Studies have extensively shown that venom composition between closely related species, or even within a single species from different populations, can vary considerably due to their adaptation to different ecological niches [6,7,8]. Snake venom variation has deep implications for the utility of antivenom against snakes (either of the same species or different populations) whose venoms are not included in the immunization process during antivenom production. Moreover, the supply and species coverage of antivenom products in most regions are inadequate, as antivenoms are produced by only a handful of domestic manufacturers in a few countries, with products indicated for selected important species in their respective countries [9].
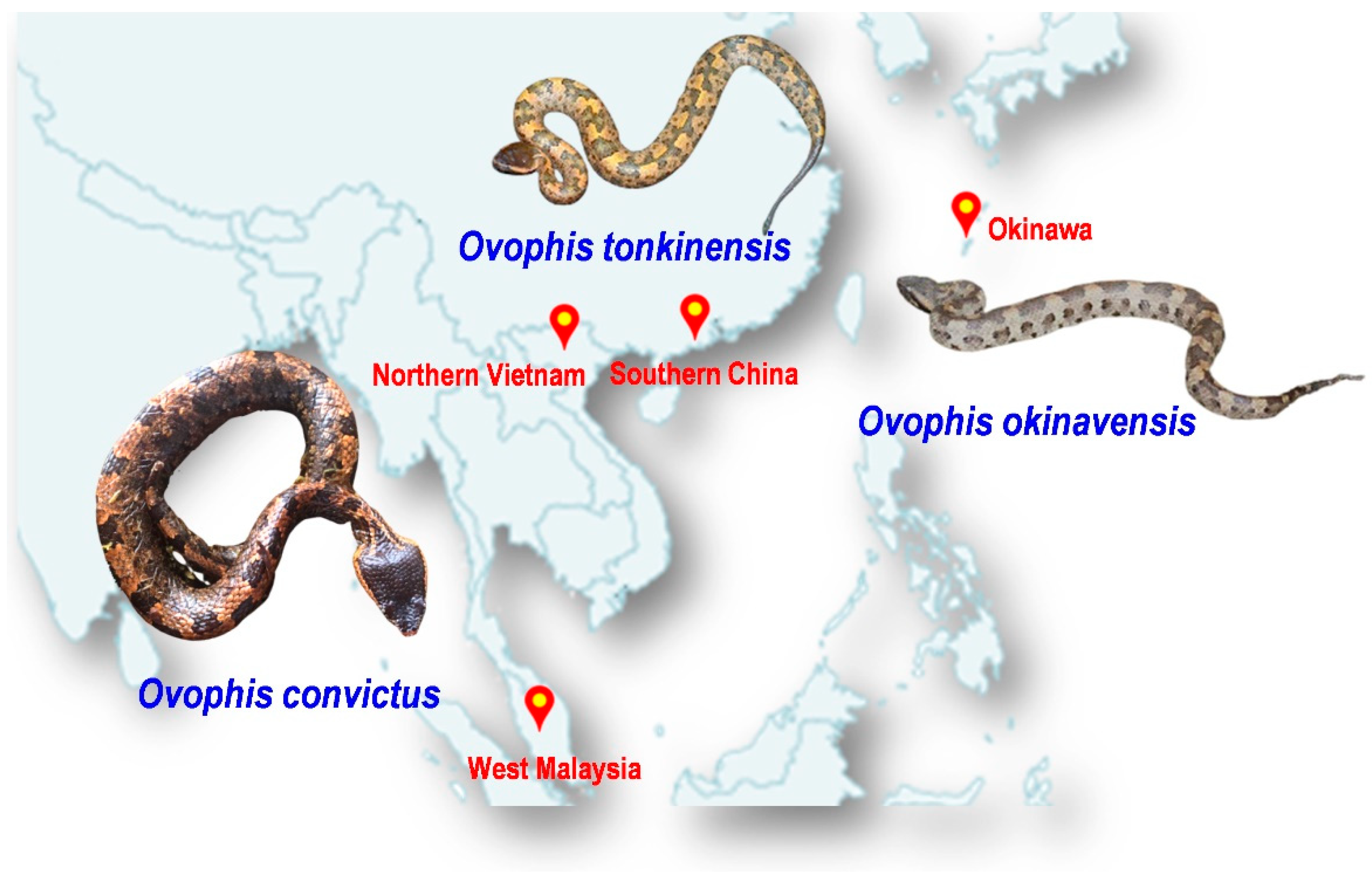
The abovementioned issues undermine the effort to improve the management of snakebite envenomation in prevalent areas, including Southeast Asia and East Asia, where agricultural activities are intensive, venomous snakes are abundant and diverse, yet suitable antivenom products are limited [10]. One of the prime examples of medically important yet lesser-known venomous snakes in the region is the Ovophis species complex, which comprises at least five morphologically similar but distinct species, i.e., Ovophis makazayazaya, Ovophis tonkinensis, Ovophis monticola, Ovophis zayuensis, and Ovophis convictus, and, in addition, a long-recognized distinct islandic species, i.e., Ovophis okinavensis [11]. The former five species were considered representative of various geographical montane subspecies distributed across West Malaysia (Peninsular Malaya), Indochina (including Vietnam), the Indian subcontinent, southern China, and Taiwan Island, with some overlapping ranges. Locally, they are known by various names including the Indo-Malayan Mountain Pit Viper, Mountain Pit Viper, Blotched Pit Viper, Brown Pit Viper, and Mountain Iron-head Snake, all of which are distributed across Asia. The more easterly dispersed islandic population of Ovophis okinavensis (Hime Habu) is endemic to the Ryukyu Islands (including Okinawa) in the southern part of Japan [12]. Taking O. convictus from the Malayan Peninsula as an example, adult male snakes can grow up to 0.5 m while full-grown females may be over 1 m. The body is stout and the triangular head, which is blackish-brown in color, is relatively bigger than the neck. The small head and body scales are smooth, and the body is usually brown or greyish-brown with one or two dorsal series of large, squarish, dark-brown blotches (Figure 1). Ovophis pit vipers are ground-dwelling and terrestrial, and most of the montane species are found at elevations of over 700 m [12].
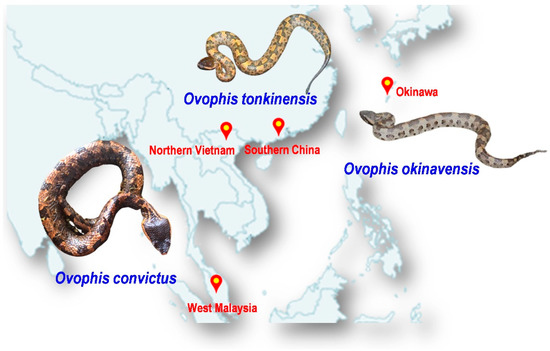
Figure 1.
Illustration of three Ovophis pit viper species studied in the current work: Ovophis convictus from West Malaysia, Ovophis tonkinensis from northern Vietnam and southern China, and Ovophis okinavensis from Okinawa, Japan. Photo credits: Choo Hock Tan (O. convictus), Abdel Bizid (O. tonkinensis), Wikimedia (O. okinavensis).
In the Malaysian experience, envenomation by Ovophis pit viper typically affects agricultural and rural communities in highland areas, in particular those in plantations and vegetable farms along the central mountain range of Peninsular Malaysia [13]. This phenomenon supports the notion that envenomation by these species is an occupational hazard to agricultural workers and their families residing near the habitat of the snake. Clinically, the envenomation caused by pit vipers can result in hemostatic derangement whereby victims develop hemorrhagic syndrome, characterized by prolonged bleeding time and coagulopathy [14,15]. Epidemiological data on envenomation caused by the Ovophis spp., however, is scarce in most parts of the world, presumably due to under-reporting. Bites were usually reported anecdotally or in isolated cases based on limited hospital records [14,16]. The venom of at least one of the species, O. okinavensis, demonstrated plasma procoagulant activity [17], while its lethal effect was approximately 3-fold weaker compared with the venom of Protobothrops flavoviridis (Okinawa Habu) [18]. Details of the venom properties, however, have not been investigated comprehensively and compared across different species. This hampers our understanding of the composition, antigenicity, and toxic activities of their venoms. Moreover, there is no specific antivenom available for this genus of medically important pit vipers, creating potentially life-threatening challenges for patients envenomed by these species, while the use of para-specific antivenom(s) as potential treatments has not yet been evaluated. Therefore, by applying a proteomic approach, this study aimed to unravel the venom composition of montane pit vipers from selected geographical origins. Hetero-specific antivenoms available in the region were also investigated for their immunological cross-reactivities toward the different Ovophis pit viper venoms. Given that coagulopathy is the predominant toxicity of Ovophis pit viper envenomation, the differential coagulotoxic activities of the venoms were studied, and the efficacy of the most immunoreactive hetero-specific antivenom in cross-neutralizing the toxicity was evaluated for insights into potential therapies.
2. Results and Discussion
2.1. Gel Electrophoretic Profiling of Ovophis Pit Viper Venoms
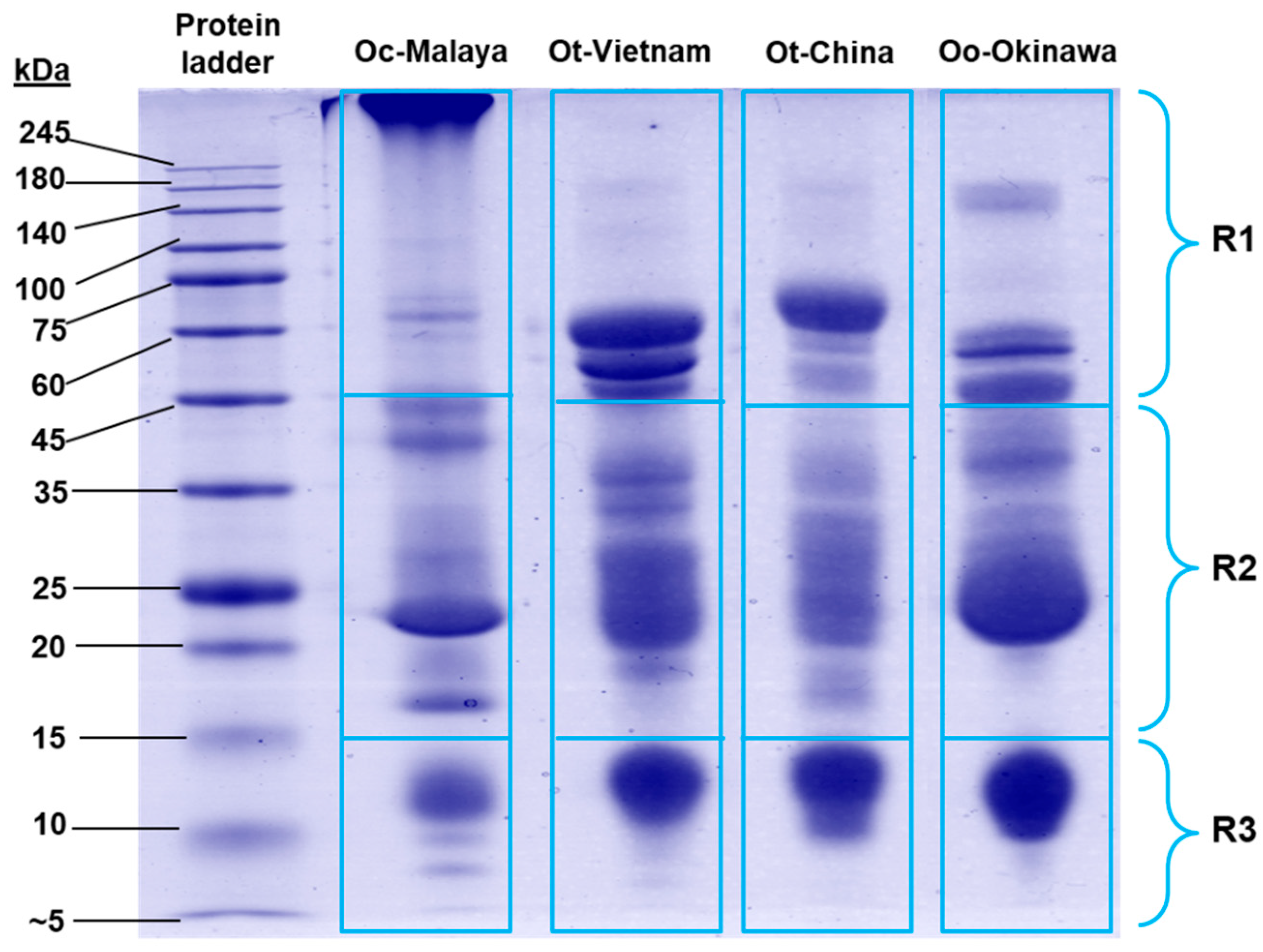
On SDS-PAGE, Ovophis venom proteins were separated according to molecular weights under reducing conditions (Figure 2). The protein composition of each venom was heterogenous, with multiple protein bands of varying intensity distributed across a wide range of molecular weights. The majority of proteins in the venoms were between 15 and 45 kDa, constituting 47.27–58.38% of total venom proteins (estimated by relative gel band intensity) (Supplementary Table S1). Along with the high molecular weight (>45 kDa) proteins, variable banding patterns were observed in SDS-PAGE of the different venoms. Meanwhile, the low molecular weight proteins (between 10 and 14 kDa) were present more uniformly across the samples.
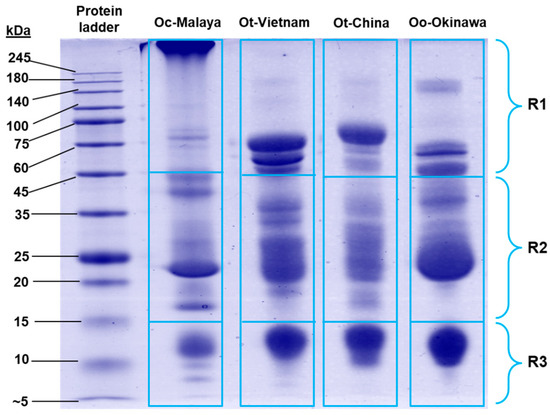
Figure 2.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of Ovophis pit viper venoms under reducing conditions. Proteins migrated on the gel were categorized into three molecular weight ranges, R1–R3 (<15 kDa, 15–45 kDa, and >45 kDa, respectively), and the gel band intensities were estimated with Pierce myImage Analysis Software. Abbreviations: Oc-Malaya, Ovophis convictus (West Malaysia); Ot-Vietnam, Ovophis tonkinensis (northern Vietnam); Ot-China, Ovophis tonkinensis (southern China); Oo-Okinawa, Ovophis okinavensis (Okinawa, Japan).
2.2. Venom Proteomics of Ovophis Pit Viper Venoms
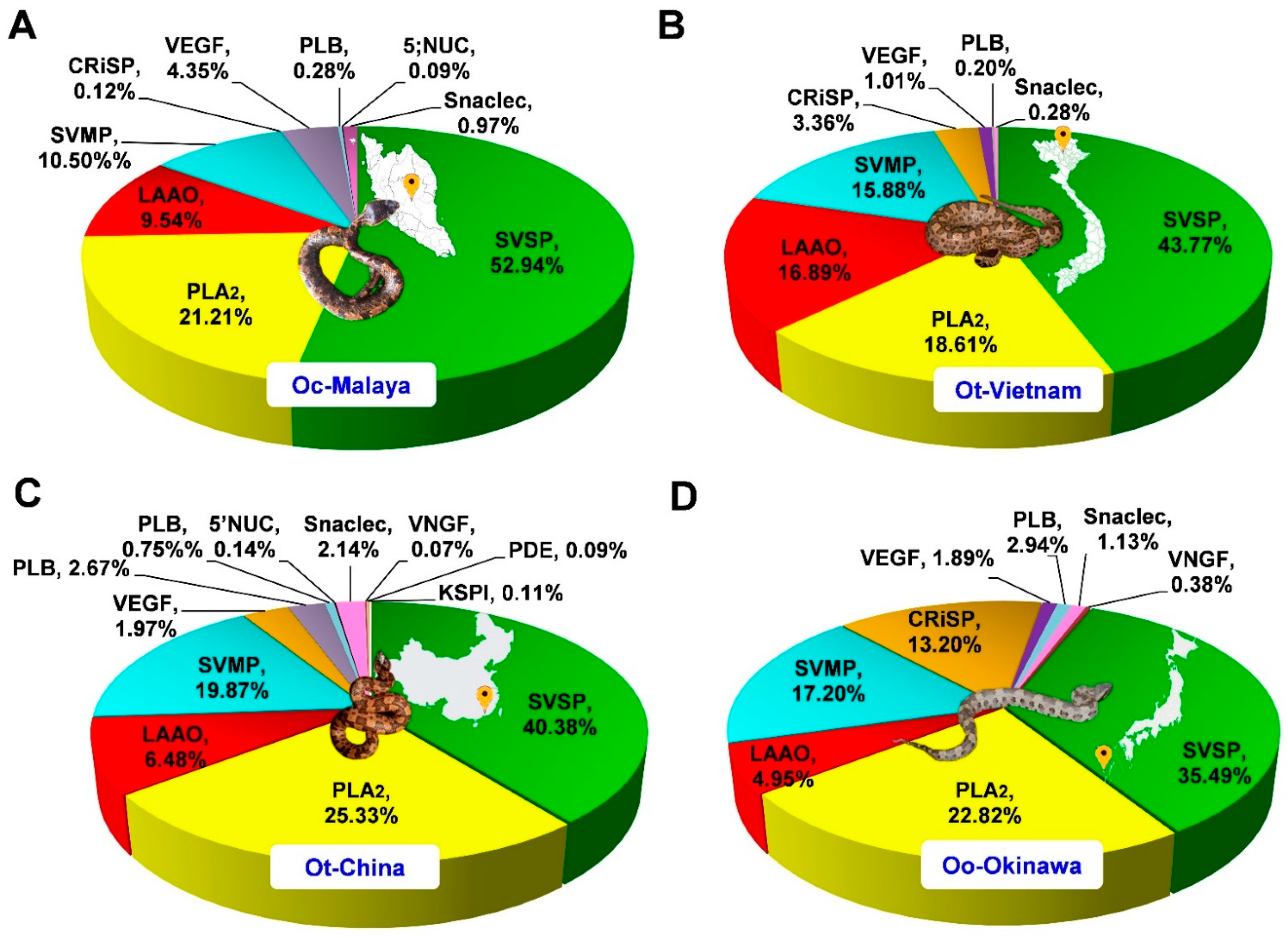
The venom proteins were identified and analyzed by nano-LC-MS/MS, followed by data mining. The venom proteomes were assembled and sorted by the toxin families, as shown in Figure 3. Four major toxin protein families were identified in the Ovophis pit viper venom proteomes, i.e., snake venom serine protease (SVSP), phospholipase A2 (PLA2), snake venom metalloproteinase (SVMP), and L-amino acid oxidase (LAAO). SVSP were the most diversely and abundantly expressed proteins in all venoms, comprising multiple distinct forms and constituting approximately 35–53% of total venom proteins (Table 1). PLA2 were the second most abundant proteins in all venom proteomes, representing 21.21%, 18.61%, 25.33%, and 22.82% of total proteins in the venoms of Oc-Malaya, Ot-Vietnam, Ot-China, and Oo-Okinawa, respectively. This was followed by SVMP and LAAO, with protein abundances varying between 4.95% and 19.87% in the four snake venom proteomes. Other proteins detected were mostly of lower abundances, including families of cysteine-rich secretory proteins, venom endothelial growth factor, phospholipase B-like protein, 5′ nucleotidase, snaclecs, venom nerve growth factor, phosphodiesterase, and Kunitz-type serine protease inhibitor (Figure 3). Mass spectrometry results of the tryptic peptides for protein identification (including protein scores, mass-to-charge ratios, spectral counts and intensities, and sequences) are provided in Supplementary File S2.
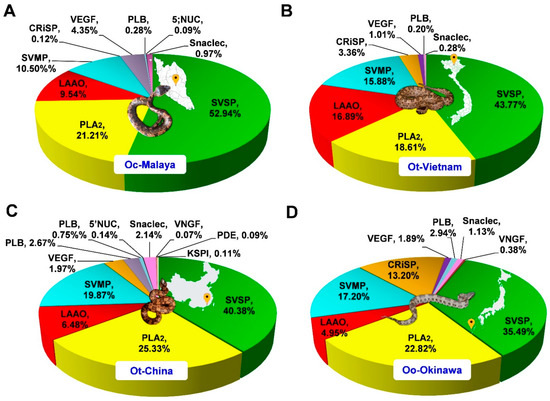
Figure 3.
Venom proteomes of Ovophis pit viper venoms. (A) Ovophis convictus from West Malaysia (Oc-Malaya), (B) Ovophis tonkinensis from North Vietnam (Ot-Vietnam), (C) Ovophis tonkinensis from Southern China (Ot-China), (D) Ovophis okinavensis from Okinawa, Japan (Oo-Okinawa). Relative abundance is expressed as percentage of total venom proteins for individual protein families. Abbreviations: SVSP, snake venom serine proteases; PLA2, phospholipase A2; SVMP, snake venom metalloproteinase; LAAO, L-amino acid oxidase; Snaclec, snake venom C-type lectin; CRiSP, cysteine-rich secretory venom protein; VEGF, venom endothelial growth factor; PDE, phosphodiesterase; PLB, phospholipase B; 5′NUC, 5′ nucleotidase; VNGF, venom nerve growth factor; KSPI, Kunitz-type protease inhibitor. Photo credits: Choo Hock Tan (Ot-Malaya), Henry Lee (Ot-Vietnam), Abdel Bizid (Ot-China), Wikimedia (Oo-Okinawa).

Table 1.
Snake venom protein families and subtypes in Ovophis pit viper venoms.
In all four Ovophis pit viper venom proteomes, SVSP made up virtually half the bulk of proteins, indicating their crucial role in the biological activity of the venom. The domination of SVSP in these venoms was consistent with a previously reported venom-gland transcriptomic study of O. okinavensis which demonstrated that SVSP transcripts were highly expressed, accounting for as much as 93.11% of toxin transcription in the transcriptome [19]. SVSPs are classified within the clan PA as subclan S, family S1 (chymotrypsin), or subfamily A of the proteolytic enzymes (MEROPS classification, http://merops.sanger.ac.uk, accessed on 31 May 2021). Such proteases are characterized by a typical chymotrypsin fold and two six-stranded β-barrels, with active sites that lie in the cleft between the latter and include the canonical catalytic triad His-Asp-Ser [20]. SVSPs demonstrate substrate specificity and thus vary in their pharmacological activities, with virtually all affecting hemostasis. A number of SVSPs have only fibrinogenolytic activity and are called ‘thrombin-like’ enzymes for their ‘fibrinogen clotting’ activity [21,22]. Snake venom thrombin-like serine proteases preferentially release fibrinopeptide A and/or fibrinopeptide B from fibrinogen to produce friable fibrin clots composed of short polymers that are rapidly dispersed and no longer cross-linked by activated factor XIII [23]. The continuous event exhausts the normal fibrinogens in the plasma, resulting in consumptive coagulopathy and, consequently, systemic bleeding, hypovolemic shock, and death [15]. A few SVSPs, known as kallikrein-like proteases have the ability to release bradykinin from kininogen, and may cause a reduction in blood pressure as well as the inhibition of blood coagulation in victims [21]. In the Ovophis pit viper venoms, the diversity of SVSPs expressed implies possible deployment of different mechanisms in the pathogenesis of coagulopathy. Further studies should aim to delineate the substrate specificity and structure–activity relationship of the individual proteins. In this regard, de novo venom-gland transcriptomics and genomics of the different Ovophis pit vipers should be the subject of future research to unveil the authentic sequences of the diverse SVSPs belonging to each species. This will also provide crucial insights into the evolutionary significance of their venom phenotypes, in which SVSPs dominate the expression of toxins.
PLA2 present in the Ovophis pit viper venom proteomes were, in majority, matched by homology to mountain or highland pit vipers in Asia, e.g., Ovophis species (including Ovophis makazayaya), Trimeresurus sabahi, and Trimeresurus cardamonensis. PLA2 with sequences specific to O. tonkinensis in the database were identified in both Ot-Vietnam and Ot-China venoms, while PLA2 specific to O. convictus was matched to Oc-Malaya, besides other PLA2 proteoforms. Snake venom PLA2 are known to exhibit a wide array of pharmacological activities, but the major putative functions in the context of Ovophis pit viper envenomation are likely related to anticoagulant and proinflammatory effects [24,25]. All subtypes identified in the proteomes belonged to Group II secretory PLA2 typically present in viperid snake venoms. The PLA2 forms to which they were matched are of acidic type (theoretical pI 4–5) and contained aspartic acid as the 49th amino acid residue, which is critical for enzymatic activity (phospholipid hydrolysis) [26]. The catalytically active viperid D49 PLA2 are usually more prevailing in anticoagulant activity compared with the non-catalytic variants of myotoxic and/or neurotoxic K49 PLA2 [27].
LAAO is another protein family which may contribute to significant inflammatory and cytotoxic activities of the venom, with effects seemingly related, at least in part, to hydrogen peroxide, a secondary product formed during the chemical reaction catalyzed by LAAOs [28,29]. The number of LAAO forms detected varied from one to seven among the different venoms, implying potentially divergent LAAO functions in the different lineages. Snake venom LAAO proteins are known to be heavily glycosylated, resulting in glycoforms with varying physicochemical properties [30]. The proteomic approach per se, however, is limited in regard to profiling the glycosylation of LAAO. Glycomic and glycoproteomic studies would be needed in this context to shed light on the diversity of glycosylated LAAO.
While SVMPs are generally abundantly present in the venom proteomes of Asiatic viperids (for examples see [31,32,33]), the abundances of this enzymatic protein family in the Ovophis pit viper venoms studied were relatively low (<20% of total venom proteins). The majority of SVMP detected belong to Class P-III of SVMP, which are high molecular weight proteins containing disintegrin-like and cysteine-rich domains following the ADAM proteinase domain [34,35]. The structural complexity of P-III enzymes allows more diverse functions including hemorrhagic, proinflammatory, cytotoxic, procoagulant, and platelet aggregation-inhibiting activities, which are relevant in the context of Ovophis pit viper envenomation, although the relatively lower SVMP abundance likely indicates a lesser role in comparison with the dominant SVSP in the venoms. In the previously reported O. okinavensis venom-gland transcriptomics, SVMP transcripts likewise showed a low abundance of 4.2% toxin transcript abundance, with P-III transcripts being more abundantly expressed than P-II transcripts, while P-I was not detected [19]. This finding is largely in agreement with the present proteomic finding on Ovophis pit viper venoms.
Other venom proteins detected were variably low in abundances (<5% of total venom proteins), with the exception of CRiSP (13.2%) in O. okinavensis venom. Some of the CRiSPs derived from snake venoms are L-type Ca2+ or cyclic nucleotide-gated channel-blocking toxins, but their roles in snakebite envenomation remain largely unknown [36]. VEGFs may exhibit proinflammatory activity and cause vascular permeability increment [37], while snaclecs including C-type lectins can modulate and compromise platelet function [38], thus potentiating the hemotoxic effect of the envenomation. PLB-like proteins are probably the least investigated snake venom toxins; earlier studies suggested that this enzyme exhibited in vitro hemolytic activity [39,40]. Other proteins present in trace amounts were more variably detected in different venom samples. 5′NT (detected in Oc-Malaya and Ot-China), VNGF (detected in Ot-China and Oo-Okinawa), and PDE (detected in Oc-China) may induce local vasodilation and facilitate venom spread from the bite site [41,42]. KSPI was solely found in the venom of Ot-China with a negligible abundance, although this protein may induce in vivo anticoagulation [43]. These minor proteins, present in small amounts in the venoms, are usually not the lethal or principal venom components. Nevertheless, given their almost ubiquitous occurrence and evolutionary conservation in snake venoms, these proteins may play ancillary roles which are integral to the species’ survival and are thus worth investigating.
2.3. Immunoreactivity of Antivenoms toward Ovophis Pit Viper Venoms
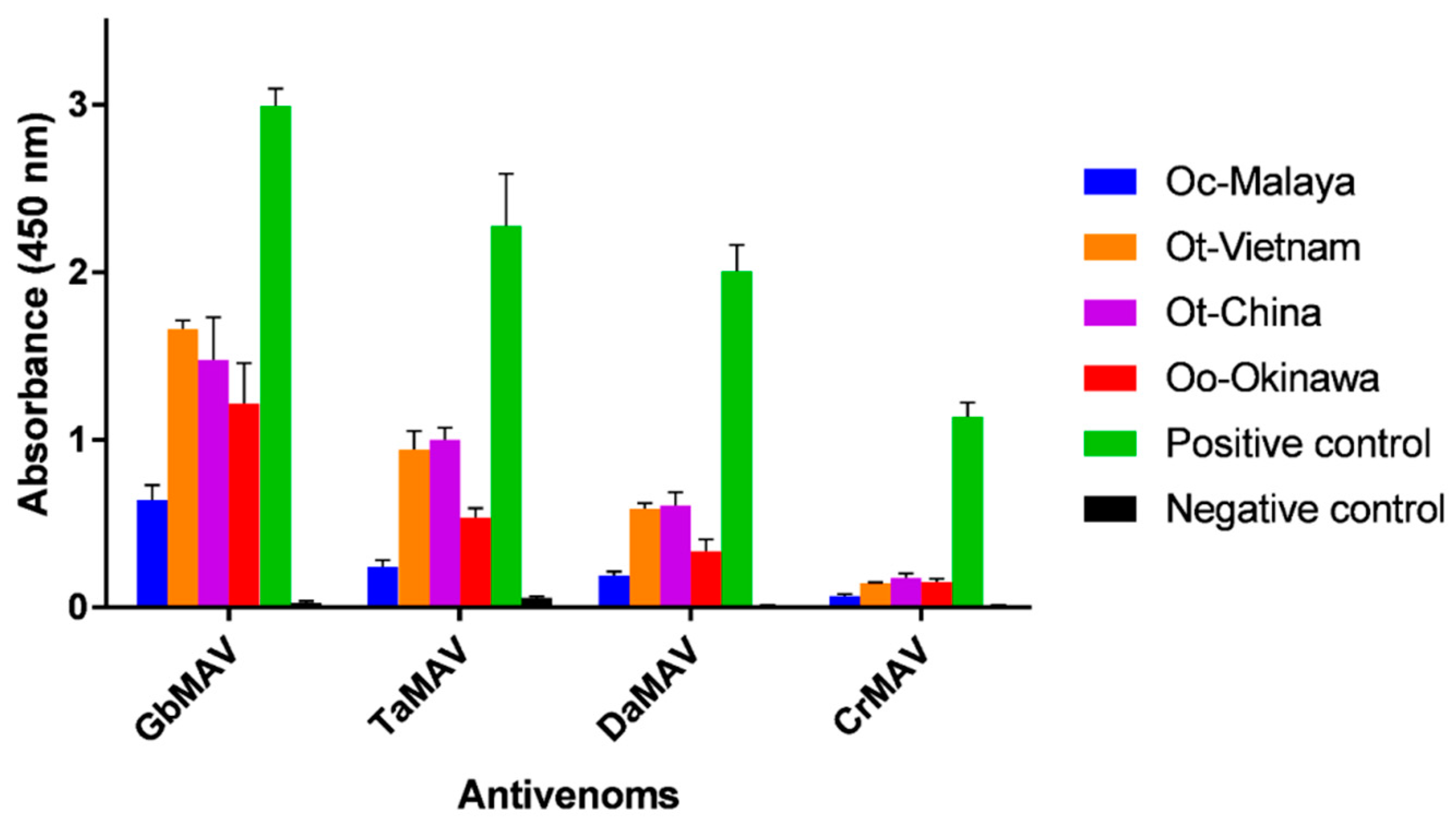
Considering that there is neither species-specific nor genus-specific antivenom available against Ovophis pit vipers, four heterologous monovalent antivenom products (related to pit vipers) available in the regions of Southeast and East Asia were evaluated for their cross-reactivities. On the whole, the antivenoms showed limited immunoreactivity toward the Ovophis pit viper venoms compared with the respective homologous venoms (Figure 4). Among the different antivenoms, GbMAV and TaMAV were more immunoreactive than DaMAV and CrMAV by a approximately two to three-folds. This indicated that the venom antigenicity of Ovophis pit vipers was more similar to those of G. brevicaudus and T. albolabris than D. acutus and C. rhodostoma, which are phylogenetically less related basal crotalids [44].
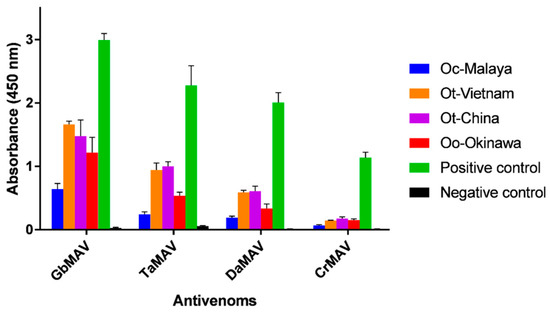
Figure 4.
Immunological binding activity of four antivenoms toward Ovophis venoms. The antivenom dose used was 1:3000 dilution from a stock of 10 mg/mL. The absorbance values are means ± S.E.M. of triplicates. The positive and negative controls of the respective antivenoms used were as stated in Materials and Methods. Abbreviations: Oc-Malaya, Ovophis convictus from West Malaysia; Ot-Vietnam, Ovophis tonkinensis from northern Vietnam; Ot-China, Ovophis tonkinensis from southern China; Oo-Okinawa, Ovophis okinavensis from Okinawa, Japan.
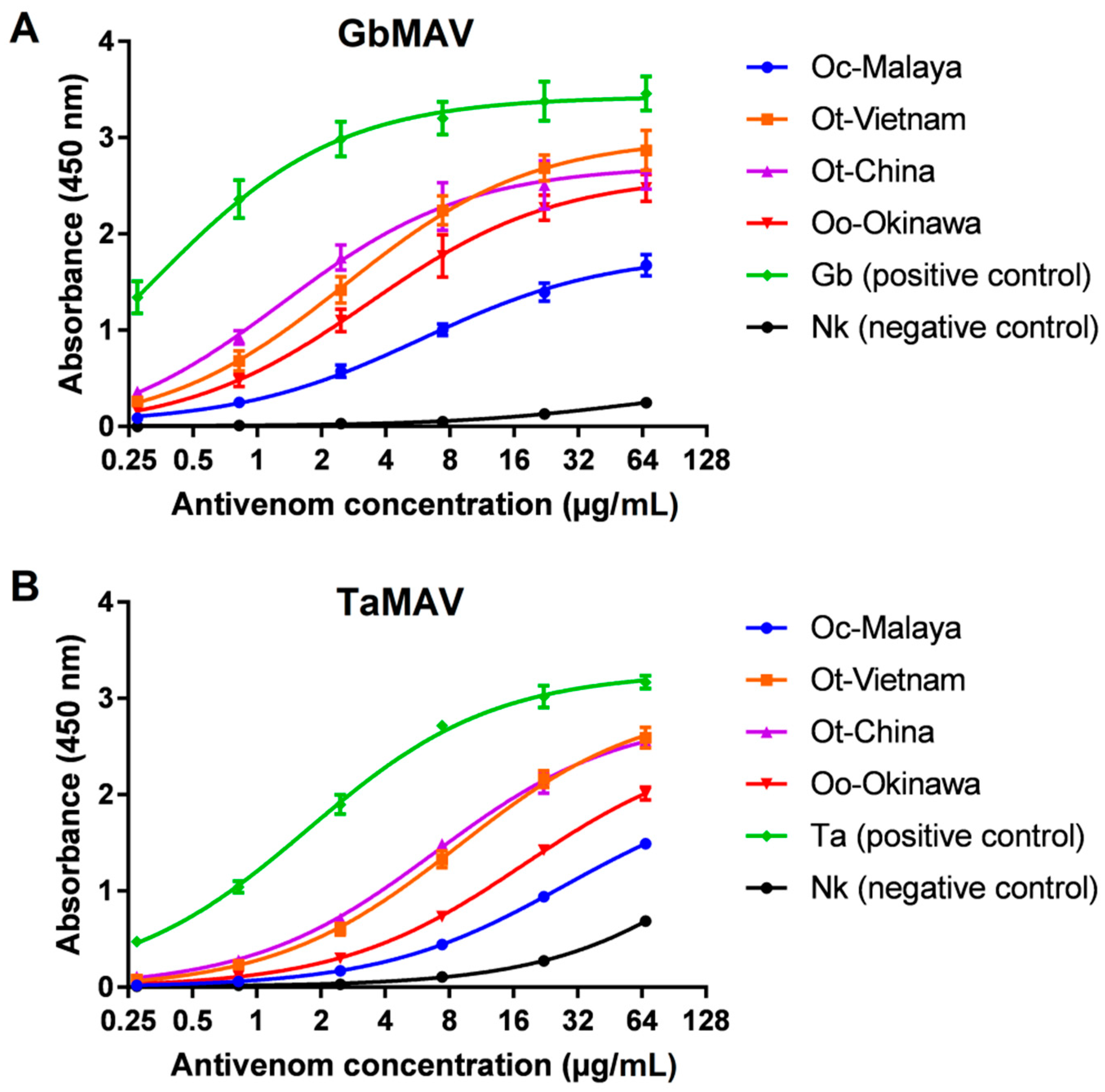
The immunological binding activities of GbMAV and TaMAV were then examined at serial concentrations. The two antivenoms showed increasing immunoreactivity in a dose-dependent manner toward all Ovophis pit viper venoms tested, with GbMAV appearing to be more efficacious than TaMAV in binding (Figure 5). The serial immunoreactivities of GbMAV toward Ot-Vietnam, Ot-China, and Oo-Okinawa were comparable, while its immunoreactivity was significantly lower toward Oc-Malaya, with a higher half-maximal concentration (EC50) (Table 2). The immunoreactivities of TaMAV toward Ot-Vietnam and Ot-China were comparable, low toward Oo-Okinawa, and much lower toward Oc-Malaya. The finding suggests that Ot-Vietnam and Ot-China venoms shared rather conserved protein epitopes despite their separated geographical origins, and this is, in a way, reflective of the single species status for the two according to the revised taxonomy proposed by Malhotra et al. [11]. Of note, the southern and southwestern Chinese species was previously known as Ovophis orientalis, which was regarded to be distinct from O. tonkinensis in northern Vietnam. Comparing Ot-Vietnam, Ot-China, and Oo-Okinawa, all three venoms were antigenically similar to the Gloydius species, while Oo-Okinawa was less antigenically related than the former two to Trimeresurus species. Phylogenetically, O. okinavensis sits basal to Gloydius rather than being closely related to other Ovophis species and Trimeresurus pit vipers [44]. On the other hand, both antivenoms showed markedly lower immunoreactivities toward Oc-Malaya venom, suggesting that the venom of the O. convictus from West Malaysia is antigenically varied from Ot-Vietnam, Ot-China, and Oo-Okinawa from the northern subtropics closer to the Tropic of Cancer. The interspecies discrepancy suggests a possible venom phenotypic variation associated with the geographical distribution of Oc-Malaya, being further to the south near the Equator where a tropical climate predominates.
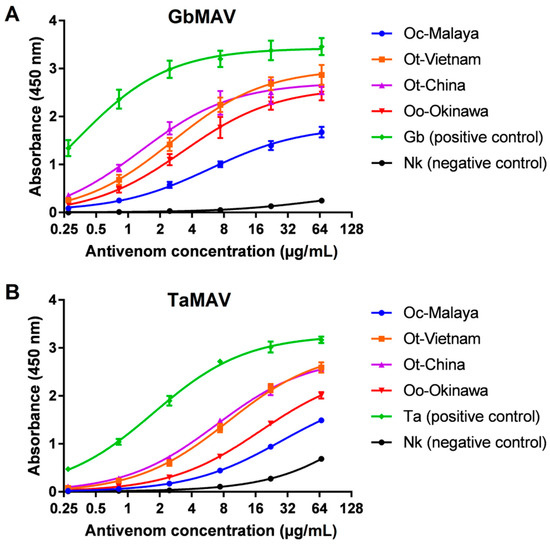
Figure 5.
Concentration-dependent immunoreactivity of antivenoms toward Ovophis pit viper venoms. (A) Chinese Gloydius brevicaudus monovalent antivenom (GbMAV), (B) Thai Trimeresurus albolabris monovalent antivenom (TaMAV). Absorbance values are means ± S.E.M. of triplicates. Abbreviations: Oc-Malaya, Ovophis convictus from West Malaysia; Ot-Vietnam, Ovophis tonkinensis from northern Vietnam; Ot-China, Ovophis tonkinensis from southern China; Oo-Okinawa, Ovophis okinavensis from Okinawa, Japan; Ta, Trimeresurus albolabris; Nk, Naja kaouthia.

Table 2.
Half-maximal concentrations (EC50) of Gloydius brevicaudus monovalent antivenom (GbMAV) and Thai Trimeresurus albolabris monovalent antivenom (TaMAV) toward Ovophis venoms.
On the other hand, all Ovophis pit viper venoms showed weak-to-moderate binding with the antivenoms tested, in comparison with the homologous venoms used in the production of the respective products (Figure 5). The immunological cross-reactivity of antivenom toward venoms of different genera is anticipated to be low, since venom compositions are likely to vary significantly across different genera of snakes. Apparently, while Ovophis pit viper venoms were characterized by highly abundant SVSP and relatively less SVMP, the converse was observed in the venom proteomes of G. brevicaudus, T. albolabris, D. acutus, and C. rhodostoma, in which a higher proportion of SVMP instead of SVSP was reported [31,45,46,47]. The remarkable differences in venom proteomes are likely accompanied by variations in the overall antigenicity of the venom proteins. Moreover, SVSP (the predominant toxins in Ovophis venoms) may have glycan chains that can alter the surface epitope and thus compromise the immunoreactivity of the antivenoms [48,49].
2.4. Procoagulant Effect of Ovophis Venoms and Its Neutralization by Antivenoms
Existing immunoassays including ELISA, immunoblotting, and affinity chromatography-based antivenomics elucidate the immunological binding activity of antivenom to venom proteins, but these assays do not determine if the antivenom is functionally effective in neutralizing the pathological activities of the venom. Hence, the neutralizing efficacy of GbMAV and TaMAV were further evaluated against the procoagulant activity of Ovophis pit viper venoms in human plasma. The procoagulant activity is putatively induced by the principal toxin SVSP, which dominated the venom proteomes (Figure 3) and is responsible for venom-induced consumptive coagulopathy in most envenomation caused by pit vipers, including clinically observed envenomation by Ovophis spp. [14,15]. Tested in human plasma, the venoms showed moderate-to-strong procoagulant activity (minimum coagulant dose, MCD = 4–10 µg/mL), with differential potency in the decreasing order of Ot-Vietnam > Oo-Okinawa > Oc-Malaya (Table 3). The venom of Ot-China, on the other hand, demonstrated a remarkably higher procoagulant activity in which its MCD was estimated to be lower than 1 µg/mL. In the Ovophis pit viper venoms, the plasma-clotting effect is consistent with the dominance of SVSP in their proteomes, as also observed in several Asiatic pit vipers which have convergently evolved the coagulopathic mechanism, causing similar hemotoxic manifestations in envenomation [31,32,50]. Different toxin components in the respective venoms, however, may have varying potency and synergistic interaction. Further works with purification and characterization of the individual procoagulant toxins will be needed to elucidate the differential potency of venom procoagulant activity.

Table 3.
Procoagulation and neutralization of Ovophis venoms by Gloydius brevicaudus monovalent antivenom (GbMAV) and Trimeresurus albolabris monovalent antivenom (TaMAV).
In the neutralization assay, GbMAV and TaMAV were able to mitigate the procoagulant effects of the venoms to varying degrees. The cross-neutralization effects of GbMAV and TaMAV were generally comparable, with both being most efficacious against the Ot-Vietnam venom (effective dose, ED, of approximately 3 µL), while efficacy against Oo-Okinawa and Oc-Malaya were moderate (ED of approximately 5–6 µL) (Table 3). The cross-neutralization finding is corroboratively supported by the immunological cross-reactivity, which indicated that GbMAV and TaMAV bound to most of the procoagulant toxins in the venoms, thus inhibiting the coagulotoxic effect. The efficacy of the cross-neutralization, however, might not be congruent with the degree of immunoreactivity, as the immunoreactivity was a reflection of antivenom binding to all antigenic proteins including nontoxic components in the venom. The functional neutralization is therefore a more robust and valid assessment for antivenom efficacy and is especially essential when exploring the therapeutic potential of hetero-specific antivenoms. The current finding suggests that both GbMAV which is available in China, and TaMAV in Southeast Asia, are potentially alternative choices of antivenom that may be repurposed to treat the coagulopathic envenomation caused by Ovophis pit vipers, pending dosage optimization and clinical trials.
On a side note, the neutralization of venom-induced lethality is normally a standard test of antivenom efficacy [51,52]; however, the Ovophis pit viper venoms were not found to be lethal to mice at a high dose of 4 µg/g in the current work. The lack of venom lethality is clinically possible, as fatality due to envenomation by these species has not been reported, although patients can develop hemotoxic complication with consumptive coagulopathy which is potentially life-threatening without proper treatment.
3. Conclusions
The present study characterized the venom proteomes of three Ovophis pit viper species from four geographical locales, demonstrating protein compositions that were dominated by SVSP (40–60% of total venom proteins) with putative procoagulant activity. The venoms induced moderate-to-strong clotting effect in citrated human plasma, indicating the role of consumptive coagulopathy in the pathophysiology of envenomation. The hetero-specific antivenoms GbMAV and TaMAV were both immunoreactive toward the venoms, and were variably effective in cross-neutralizing their procoagulant activities. Taken together, the study suggests the potential para-specific utility of the antivenoms in the treatment of coagulotoxic envenomation caused by Ovophis pit vipers in their respective regions.
4. Methods and Materials
4.1. Venoms and Antivenoms
Venom samples of Ovophis species complex were sourced from specimens originated from West Malaysia (O. convictus), northern Vietnam, the southern Chinese mainland (O. tonkinensis), and Okinawa Island of Japan (Ovophis okinavensis). For simplicity, the venom samples were labeled with abbreviations in this report as follows: Oc-Malaya, Ot-Vietnam, Ot-China, Oo-Okinawa, respectively, as per above. Venoms were stored in lyophilized form at −20 °C until use. Other samples used as controls in the immunoreactivity study were venoms of Naja kaouthia, Trimeresurus albolabris, and Calloselasma rhodostoma, supplied by the Queen Saovabha Memorial Institute (QSMI, Bangkok, Thailand), and Deinagkistrodon acutus, Gloydius brevicaudus, and Protobothrops mucrosquamatus, obtained from Latoxan (Valence, France).
The hetero-specific antivenoms used in the present study were: (1) Gloydius brevicaudus monovalent antivenom (GbMAV, liquid antivenom, raised against G. brevicaudus venom of Chinese origin, batch no. 20141001); (2) Deinagkistrodon acutus monovalent antivenom (DaMAV, liquid antivenom, raised against D. acutus venom of Chinese origin, batch no. 20140501); (3) green pit viper (Trimeresurus albolabris) monovalent antivenom (TaMAV, lyophilized antivenom, raised against Thai T. albolabris venom, batch no. TA00116); (4) Calloselasma rhodostoma monovalent antivenom (CrMAV, lyophilized antivenom, raised against Thai C. rhodostoma venom, batch no. TA00116). Lyophilized antivenoms were reconstituted in 10 mL of ultrapure water prior to use as per the manufacturer’s instructions. Protein concentration of all antivenoms was determined using Pierce Bicinchoninic Acid Protein Assay Kit (ThermoFisher Scientific, Waltham, MA, USA).
4.2. Chemicals and Materials
All chemicals and reagents used were of analytical grade. Ammonium bicarbonate, dithiothreitol (DTT), and iodoacetamide were purchased from Sigma-Aldrich (St. Louis, MO, USA). MS-grade trypsin protease was purchased from ThermoFisher Scientific (Waltham, MA, USA). Millipore ZipTip C18 pipette tips were purchased from Merck (Kenilworth, NJ, USA). PM2700 ExcelBand 3-color Broad Range Protein Marker (~5–245 kDa) was obtained from SMOBIO (Hsinchu, Taiwan).
4.3. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
Forty micrograms of the respective Ovophis venoms were electrophoresed with 15% SDS-PAGE under reducing conditions in accordance with the Laemmli protocol [53], calibrated with the prestained protein ladder (5–245 kDa). The electrophoretic separation was conducted at 90 V for 120 min. Proteins were stained with Coomassie Brilliant Blue R-250 for visualization. The gel was then scanned using Image Scanner III Labscan 6.0 (GE Healthcare, Chicago, IL, USA). The relative intensity of gel regions (I, II, and III) were determined using myImage analysis software (Thermo Scientific, Waltham, MA, USA).
4.4. In-Solution Tryptic Digestion and Tandem Mass Spectrometry (Nano-ESI-LCMS/MS)
Ten micrograms of each venom sample of Ovophis species were subjected to reduction with DTT and alkylation with iodoacetamide, and were digested in-solution with mass spectrometry-grade trypsin proteases as described previously [54]. The digested peptides were desalted and extracted with Millipore ZipTip C18 pipette tips in preparation for mass-spectrometry analysis.
For protein identification, the extracted peptides were reconstituted in 7 μL of 0.1% formic acid in water. Peptides were subjected to nano-electrospray ionization-liquid chromatography-tandem mass spectrometry (nano-ESI-LCMS/MS) with an Agilent 1200 HPLC-Chip/MS Interface coupled with the Agilent 6520 Accurate-Mass Q-TOF LC/MS system. The peptides were loaded (1 μL) in a 300 Å, C18 enrichment column, followed by a 75 μm × 150 mm analytical column (Agilent No.: G4240-62010). Solvent A (0.1% formic acid in water) and solution B (90% acetonitrile in water with 0.1% formic acid) were used for elution of the peptides at the stepwise linear gradients: 3–50% solution B for 30 min, 50–95% solution B for 2 min, and 95% solution B for 5 min. The ion polarity was set to positive ionization polarity mode. Positive ionization mode was selected for ion polarity. Flow rate and temperature of drying gas were set to 11 L/min and 290 °C, respectively. Fragmentor voltage was configured to 175 V and the capillary voltage was set to 1800 V. Spectra were obtained in MS/MS mode, with an MS scan range of 200–3000 m/z and MS/MS scan range of 50–3200 m/z. Precursor charge selection was set as doubly charged state and above, with the exclusion of precursors 1221.9906 m/z (z = 1) and 299.2944 (z = 1) set as reference ions. Data were generated with MH (protonated peptide ion) mass span between 50 and 3200 Da and sorted with Agilent Spectrum Mill MS Proteomics Workbench software packages version B.06.00.201. Carbamidomethylation of cysteine residues was set as a fixed modification and oxidation of methionine residues as a variable modification. The spectra were searched against a nonredundant protein sequence database from the National Center for Biotechnology Information (NCBI) (taxonomy: Serpentes, taxid: 8570). The identified proteins or peptides were validated with the following filters: protein score >10, peptide score >10, and scored peak intensity (SPI) >70%. The relative abundance of each protein was estimated based on the ratio of mean spectral intensity (MSI) of the protein relative to the total MSI of all proteins identified within a molecular weight range indicated by SDS-PAGE. The MSI ratio of the protein was then multiplied by the gel band intensity accordingly, with the calculation formula as shown below:
4.5. Data Availability
The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org, dataset uploaded on 1 June 2021) via the iProX partner repository [55] with the dataset identifier PXD: PXD026416.
4.6. Enzyme-Linked Immunosorbent Assay (ELISA)
The immunoreactivity of the antivenoms toward the four Ovophis venoms was examined through an indirect enzyme-linked immunosorbent assay (ELISA), optimized in the laboratory [56]. N. kaouthia venom was used as a negative control for all assays. Positive controls were the venoms of G. brevicaudus (for GbMAV), D. acutus (for DaMAV), T. albolabris (for TaMAV), and C. rhodostoma (for CrMAV), respectively. Briefly, 96-well immunoplates were coated overnight with 10 ng of Ovophis venom in 100 µL coating buffer at 4 °C. The plates were washed four times with phosphate-buffered saline (Oxiod, Hampshire, UK) containing 0.5% Tween 20 (Sigma-Aldrich, St. Louis, MO, USA) (PBST). Then, 100 µL of each antivenom dilution (1:3000) from a stock of 10 mg/mL were added to each well and incubated for one hour at room temperature. The wash step was repeated and 100 µL of 1:10,000 diluted horseradish peroxidase-conjugated anti-horse IgG (Jackson ImmunoResearch Inc., West Grove, PA, USA) in PBST was added to each well and incubated for 1 h at room temperature. After another cycle of washing, 50 µL of TMB substrate (3,3′,5,5′-Tetramethylbenzidine) solution was added to each well and incubated in the dark for 25 min at room temperature. Finally, 50 µL of 12.5% sulfuric acid was added to terminate the reaction before the absorbance reading at 450 nm. The experiments were performed in triplicate and absorbance values were expressed as means ± S.E.M.
Following this, the two monovalent antivenoms (GbMAV and TaMAV) which showed relatively high immunoreactivity toward Ovophis venoms were further examined in the concentration-dependent study. Serially diluted antivenoms (1:150, 1:450, 1:1350, 1:4050, 1:12,150, and 1:36,450), prepared in 100 µL from a stock of 10 mg/mL, were used. All procedures of indirect ELISA proceeded as described above. The half-maximal effective concentration (EC50, in µg/mL), defined as the antivenom dose at which half-maximal binding occurred, was determined using GraphPad Prism (version 7.04).
4.7. Procoagulant Activity of Venom and Neutralization by Antivenom
The procoagulant activity of Ovophis venoms was assessed with a modified turbidimetric method using citrated human plasma as previously described [57]. In brief, 100 μL of citrated human plasma, containing 0.4 M CaCl2, was added to 100 μL of venom of various concentrations in a 96-well microplate at 37 °C. The procoagulant activity was assessed by monitoring absorbance at 405 nm for 15 min, with readings taken in 15 s intervals, using the Infinite M1000 Pro Multimode plate reader (Männedorf, Switzerland). An increase in the absorbance by 0.02 units from the mean of the two preceding absorbance measurements indicates plasma clotting. The minimum clotting dose (MCD) was defined as the minimal venom dose (µg) required to induce clotting of plasma in 5 min.
The neutralization of procoagulant activity was conducted by incubating a venom dose of 2MCD of respective venoms with different doses of antivenom (GbMAV and TaMAV), to make up a total volume of 100 µL, at 37 °C for 30 min. Following that, the mixture was added to 100 μL of citrated human plasma, containing 0.4 M CaCl2. The absorbance was monitored and the clotting time was determined as described above. The effective dose (ED) of antivenom was defined as the dose of antivenom (µL) required to prolong the clotting time of the plasma by three times that of the control (2MCD with no antivenom).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/toxins13080514/s1, Table S1: Relative gel band intensity of Ovophis pit viper venoms, File S2: Mass spectrometry data of Ovophis pit viper venom proteomics.
Author Contributions
Conceptualization, C.H.T. and K.Y.T.; methodology, C.H.T. and K.Y.T.; validation, C.H.T., K.Y.T. and P.P.; formal analysis, P.P. and K.Y.T.; investigation, C.H.T., K.Y.T. and P.P.; resources, C.H.T.; data curation, C.H.T. and K.Y.T.; writing—original draft preparation, C.H.T.; writing—review and editing, C.H.T., K.Y.T. and P.P.; visualization, C.H.T., K.Y.T. and P.P.; project administration, C.H.T.; funding acquisition, C.H.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fundamental Research Grant Scheme from the Ministry of Higher Education, Government of Malaysia (FRGS/1/2019/SKK08/UM/02/19).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org, dataset uploaded on 1 June 2021) via the iProX partner repository with the dataset identifier PXD: PXD026416.
Acknowledgments
The authors acknowledge Pei Wan Ooi for technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. Prevalence of Snakebite Envenoming. Available online: https://www.who.int/snakebites/epidemiology/en/ (accessed on 2 June 2021).
- World Health Organization. Guidelines for the Management of Snakebites, 2nd ed.; WHO Regional Office for Southeast Asia: Geneva, Switzerland, 2016. [Google Scholar]
- Gutierrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Primers 2017, 3, 17079. [Google Scholar] [CrossRef]
- Kasturiratne, A.; Wickremasinghe, A.R.; de Silva, N.; Gunawardena, N.K.; Pathmeswaran, A.; Premaratna, R.; Savioli, L.; Lalloo, D.G.; de Silva, H.J. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008, 5, e218. [Google Scholar] [CrossRef]
- Chippaux, J.P. Snakebite envenomation turns again into a neglected tropical disease! J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 38. [Google Scholar] [CrossRef]
- Casewell, N.R.; Jackson, T.N.W.; Laustsen, A.H.; Sunagar, K. Causes and Consequences of Snake Venom Variation. Trends Pharmacol. Sci. 2020, 41, 570–581. [Google Scholar] [CrossRef]
- Tan, K.Y.; Ng, T.S.; Bourges, A.; Ismail, A.K.; Maharani, T.; Khomvilai, S.; Sitprija, V.; Tan, N.H.; Tan, C.H. Geographical variations in king cobra (Ophiophagus hannah) venom from Thailand, Malaysia, Indonesia and China: On venom lethality, antivenom immunoreactivity and in vivo neutralization. Acta Trop. 2020, 203, 105311. [Google Scholar] [CrossRef]
- Pla, D.; Sanz, L.; Quesada-Bernat, S.; Villalta, M.; Baal, J.; Chowdhury, M.A.W.; Leon, G.; Gutierrez, J.M.; Kuch, U.; Calvete, J.J. Phylovenomics of Daboia russelii across the Indian subcontinent. Bioactivities and comparative in vivo neutralization and in vitro third-generation antivenomics of antivenoms against venoms from India, Bangladesh and Sri Lanka. J. Proteom. 2019, 207, 103443. [Google Scholar] [CrossRef]
- Williams, D.; Gutiérrez, J.; Calvete, J.; Wüster, W.; Ratanabanangkoon, K.; Paiva, O.; Brown, N.; Casewell, N.; Harrison, R.; Rowley, P.; et al. Ending the drought: New strategies for improving the flow of affordable, effective antivenoms in Asia and Africa. J. Proteom. 2011, 74, 1735–1767. [Google Scholar] [CrossRef] [PubMed]
- Tan, N.H.; Tan, K.Y.; Tan, C.H. Snakebite in Southeast Asia: Envenomation and Clinical Management. In Handbook of Venoms and Toxins of Reptiles, 2nd ed.; Mackessy, S.P., Ed.; CRC Press: Boca Raton, FL, USA, 2021; p. 22. [Google Scholar]
- Malhotra, A.; Dawson, K.; Guo, P.; Thorpe, R.S. Phylogenetic structure and species boundaries in the mountain pitviper Ovophis monticola (Serpentes: Viperidae: Crotalinae) in Asia. Mol. Phylogenet. Evol. 2011, 59, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Uetz, P.; Freed, P.; Aguilar, R.; Hošek, J. The Reptile Database. Available online: http://www.reptile-database.org (accessed on 2 June 2021).
- Tan, C.H.; Tan, N.H. Toxinology of Snake Venoms: The Malaysian Context. In Snake Venoms; Gopalakrishnakone, P., Inagaki, H., Mukherjee, A.K., Rahmy, T.R., Vogel, C.-W., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 1–37. [Google Scholar] [CrossRef]
- Sivaganabalan, R.; Ismail, A.K.; Salleh, M.S.; Mohan, K.; Choo, T.C.; Adnan, A.; Ariff, A.M.; Mohamed, Z.; Thevarajah, N.; Daud, R.; et al. Guideline: Management of Snakebite Ministry of Health Malaysia, 1st ed.; Ministry of Health Malaysia: Putrajaya, Malaysia, 2017.
- Slagboom, J.; Kool, J.; Harrison, R.A.; Casewell, N.R. Haemotoxic snake venoms: Their functional activity, impact on snakebite victims and pharmaceutical promise. Br. J. Haematol. 2017, 177, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Pandey, D.P.; Chaudhary, B.; Ram Shrestha, B. Documentation of a proven Mountain Pitviper (Ovophis monticola) envenomation in Kathmandu, Nepal, with its distribution ranges: Implications for prevention and control of pitviper bites in Asia. J. Venom Res. 2021, 11, 1–6. [Google Scholar] [PubMed]
- Nielsen, V.G.; Frank, N.; Matika, R.W. Effects of Heme Modulation on Ovophis and Trimeresurus Venom Activity in Human Plasma. Toxins 2018, 10, 322. [Google Scholar] [CrossRef]
- Hirosh, U.; Masatosh, N. Skin injuries due to poisonous snake bites. Ryukyu Med. J. 2004, 23, 11–20. [Google Scholar]
- Aird, S.D.; Watanabe, Y.; Villar-Briones, A.; Roy, M.C.; Terada, K.; Mikheyev, A.S. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis). BMC Genom. 2013, 14, 790. [Google Scholar] [CrossRef] [PubMed]
- Parry, M.A.; Jacob, U.; Huber, R.; Wisner, A.; Bon, C.; Bode, W. The crystal structure of the novel snake venom plasminogen activator TSV-PA: A prototype structure for snake venom serine proteinases. Structure 1998, 6, 1195–1206. [Google Scholar] [CrossRef][Green Version]
- Matsui, T.; Fujimura, Y.; Titani, K. Snake venom proteases affecting hemostasis and thrombosis. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2000, 1477, 146–156. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Mackessy, S.P. Biochemical and pharmacological properties of a new thrombin-like serine protease (Russelobin) from the venom of Russell’s Viper (Daboia russelii russelii) and assessment of its therapeutic potential. Biochim. Biophys. Acta 2013, 1830, 3476–3488. [Google Scholar] [CrossRef] [PubMed]
- Swenson, S.; Markland, F.S., Jr. Snake venom fibrin(ogen)olytic enzymes. Toxicon 2005, 45, 1021–1039. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Lomonte, B. Phospholipases A2: Unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon 2013, 62, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Saikia, D.; Mukherjee, A.K. Anticoagulant and Membrane Damaging Properties of Snake Venom Phospholipase A2 Enzymes. In Snake Venoms; Inagaki, H., Vogel, C.-W., Mukherjee, A.K., Rahmy, T.R., Gopalakrishnakone, P., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 87–104. [Google Scholar] [CrossRef]
- Damico, D.C.; Vassequi-Silva, T.; Torres-Huaco, F.D.; Nery-Diez, A.C.; de Souza, R.C.; Da Silva, S.L.; Vicente, C.P.; Mendes, C.B.; Antunes, E.; Werneck, C.C.; et al. LmrTX, a basic PLA(2) (D49) purified from Lachesis muta rhombeata snake venom with enzymatic-related antithrombotic and anticoagulant activity. Toxicon 2012, 60, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Tonello, F.; Rigoni, M. Cellular Mechanisms of Action of Snake Phospholipase A2 Toxins. In Snake Venoms; Inagaki, H., Vogel, C.-W., Mukherjee, A.K., Rahmy, T.R., Gopalakrishnakone, P., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 49–65. [Google Scholar] [CrossRef]
- Tan, K.K.; Bay, B.H.; Gopalakrishnakone, P. L-amino acid oxidase from snake venom and its anticancer potential. Toxicon 2018, 144, 7–13. [Google Scholar] [CrossRef]
- Paloschi, M.V.; Pontes, A.S.; Soares, A.M.; Zuliani, J.P. An Update on Potential Molecular Mechanisms Underlying the Actions of Snake Venom L-amino Acid Oxidases (LAAOs). Curr. Med. Chem. 2018, 25, 2520–2530. [Google Scholar] [CrossRef] [PubMed]
- Izidoro, L.F.; Sobrinho, J.C.; Mendes, M.M.; Costa, T.R.; Grabner, A.N.; Rodrigues, V.M.; da Silva, S.L.; Zanchi, F.B.; Zuliani, J.P.; Fernandes, C.F.; et al. Snake venom L-amino acid oxidases: Trends in pharmacology and biochemistry. BioMed. Res. Int. 2014, 2014, 196754. [Google Scholar] [CrossRef]
- Liew, J.L.; Tan, N.H.; Tan, C.H. Proteomics and preclinical antivenom neutralization of the mangrove pit viper (Trimeresurus purpureomaculatus, Malaysia) and white-lipped pit viper (Trimeresurus albolabris, Thailand) venoms. Acta Trop. 2020, 209, 105528. [Google Scholar] [CrossRef]
- Tan, C.H.; Tan, K.Y.; Ng, T.S.; Quah, E.S.H.; Ismail, A.K.; Khomvilai, S.; Sitprija, V.; Tan, N.H. Venomics of Trimeresurus (Popeia) nebularis, the Cameron Highlands Pit Viper from Malaysia: Insights into Venom Proteome, Toxicity and Neutralization of Antivenom. Toxins 2019, 11, 95. [Google Scholar] [CrossRef]
- Lingam, T.M.C.; Tan, K.Y.; Tan, C.H. Proteomics and antivenom immunoprofiling of Russell’s viper (Daboia siamensis) venoms from Thailand and Indonesia. J. Venom. Anim. Toxins Incl. Trop. Dis. 2020, 26, e20190048. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M. Exploring snake venom proteomes: Multifaceted analyses for complex toxin mixtures. Proteomics 2008, 8, 909–920. [Google Scholar] [CrossRef]
- Fox, J.W.; Serrano, S.M. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon 2005, 45, 969–985. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Morita, T. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon 2004, 44, 227–231. [Google Scholar] [CrossRef]
- Rucavado, A.; Escalante, T.; Camacho, E.; Gutiérrez, J.M.; Fox, J.W. Systemic vascular leakage induced in mice by Russell’s viper venom from Pakistan. Sci. Rep. 2018, 8, 16088. [Google Scholar] [CrossRef] [PubMed]
- Morita, T. Structures and functions of snake venom CLPs (C-type lectin-like proteins) with anticoagulant-, procoagulant-, and platelet-modulating activities. Toxicon 2005, 45, 1099–1114. [Google Scholar] [CrossRef]
- Bernheimer, A.W.; Linder, R.; Weinstein, S.A.; Kim, K.S. Isolation and characterization of a phospholipase B from venom of Collett’s snake, Pseudechis colletti. Toxicon 1987, 25, 547–554. [Google Scholar] [CrossRef]
- Bernheimer, A.W.; Weinstein, S.A.; Linder, R. Isoelectric analysis of some Australian elapid snake venoms with special reference to phospholipase B and hemolysis. Toxicon 1986, 24, 841–849. [Google Scholar] [CrossRef]
- Dhananjaya, B.L.; D’Souza, C.J. An overview on nucleases (DNase, RNase, and phosphodiesterase) in snake venoms. Biochem. Biokhimiia 2010, 75, 1–6. [Google Scholar] [CrossRef]
- Lavin, M.F.; Earl, S.; Birrell, G.; St. Pierre, L.; Guddat, L.; de Jersey, J.; Masci, P. Snake venom nerve growth factors. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press: Boca Raton, FL, USA, 2009; pp. 377–391. [Google Scholar]
- Mukherjee, A.K.; Mackessy, S.P.; Dutta, S. Characterization of a Kunitz-type protease inhibitor peptide (Rusvikunin) purified from Daboia russelii russelii venom. Int. J. Biol. Macromol. 2014, 67, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Alencar, L.R.V.; Quental, T.B.; Grazziotin, F.G.; Alfaro, M.L.; Martins, M.; Venzon, M.; Zaher, H. Diversification in vipers: Phylogenetic relationships, time of divergence and shifts in speciation rates. Mol. Phylogenet. Evol. 2016, 105, 50–62. [Google Scholar] [CrossRef]
- Tang, E.L.H.; Tan, N.H.; Fung, S.Y.; Tan, C.H. Comparative proteomes, immunoreactivities and neutralization of procoagulant activities of Calloselasma rhodostoma (Malayan pit viper) venoms from four regions in Southeast Asia. Toxicon 2019, 169, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.F.; Wang, J.; He, Y.; Qu, Y.F.; Lin, L.H.; Ma, X.M.; Ji, X. Proteomic and biochemical analyses of short-tailed pit viper (Gloydius brevicaudus) venom: Age-related variation and composition-activity correlation. J. Proteom. 2014, 105, 307–322. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.C.; Huang, M.N.; Chang, J.F.; Liu, C.C.; Chen, C.K.; Hsieh, C.H. Snake venom proteome and immuno-profiling of the hundred-pace viper, Deinagkistrodon acutus, in Taiwan. Acta Trop. 2019, 189, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Kuniyoshi, A.K.; Rocha, M.; Carvalho, D.C.; Juliano, M.A.; Neto, L.J.; Tambourgi, D.V.; Portaro, F.C.V. Angiotensin-degrading serine peptidase: A new chymotrypsin-like activity in the venom of Bothrops jararaca partially blocked by the commercial antivenom. Toxicon 2012, 59, 124–131. [Google Scholar] [CrossRef]
- Lee, L.P.; Tan, K.Y.; Tan, C.H. Snake venom proteomics and antivenomics of two Sundaic lance-headed pit vipers: Trimeresurus wiroti (Malaysia) and Trimeresurus puniceus (Indonesia). Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 100875. [Google Scholar] [CrossRef]
- Debono, J.; Bos, M.H.A.; Do, M.S.; Fry, B.G. Clinical implications of coagulotoxic variations in Mamushi (Viperidae: Gloydius) snake venoms. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2019, 225, 108567. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Solano, G.; Pla, D.; Herrera, M.; Segura, Á.; Vargas, M.; Villalta, M.; Sánchez, A.; Sanz, L.; Lomonte, B.; et al. Preclinical Evaluation of the Efficacy of Antivenoms for Snakebite Envenoming: State-of-the-Art and Challenges Ahead. Toxins 2017, 9, 163. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Liew, J.L.; Tan, K.Y.; Tan, N.H. Assessing SABU (Serum Anti Bisa Ular), the sole Indonesian antivenom: A proteomic analysis and neutralization efficacy study. Sci. Rep. 2016, 6, 37299. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, K.Y.; Tan, N.H. Revisiting Notechis scutatus venom: On shotgun proteomics and neutralization by the “bivalent” Sea Snake Antivenom. J. Proteom. 2016, 144, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Chen, T.; Wu, S.; Yang, C.; Bai, M.; Shu, K.; Li, K.; Zhang, G.; Jin, Z.; He, F.; et al. iProX: An integrated proteome resource. Nucleic Acids Res. 2019, 47, D1211–D1217. [Google Scholar] [CrossRef] [PubMed]
- Lingam, T.M.C.; Tan, K.Y.; Tan, C.H. Thai Russell’s viper monospecific antivenom is immunoreactive and effective in neutralizing the venom of Daboia siamensis from Java, Indonesia. Toxicon 2019, 168, 95–97. [Google Scholar] [CrossRef]
- Tan, K.Y.; Tan, N.H.; Tan, C.H. Venom proteomics and antivenom neutralization for the Chinese eastern Russell’s viper, Daboia siamensis from Guangxi and Taiwan. Sci. Rep. 2018, 8, 8545. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).