Old World Vipers—A Review about Snake Venom Proteomics of Viperinae and Their Variations
Abstract
1. Introduction
2. Viperinae Venoms: A Proteomic Database
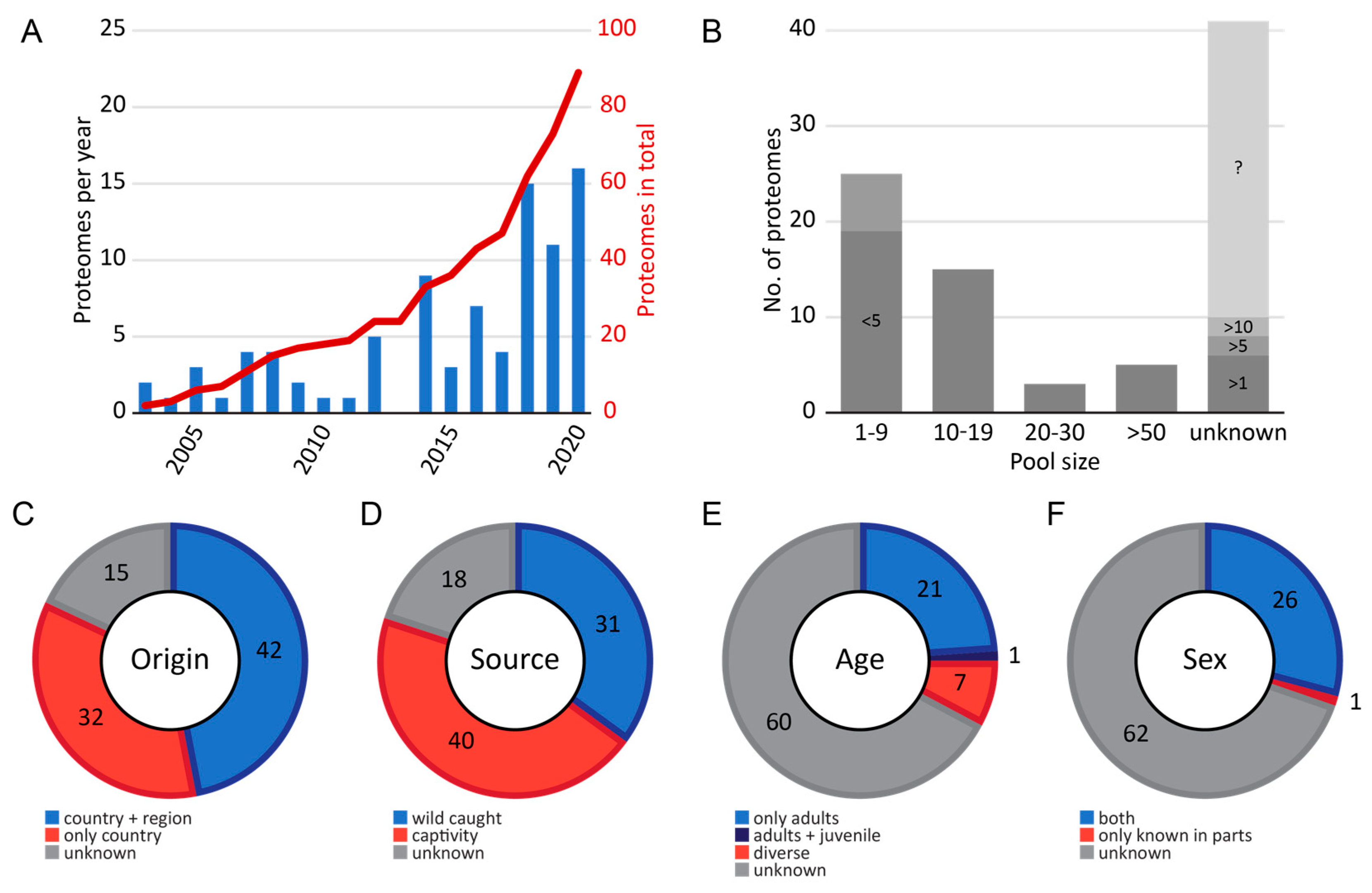
2.1. Meta Data of Investigated Snakes
2.2. Venom Proteome Data Accessibility
2.3. Identified Toxin Families
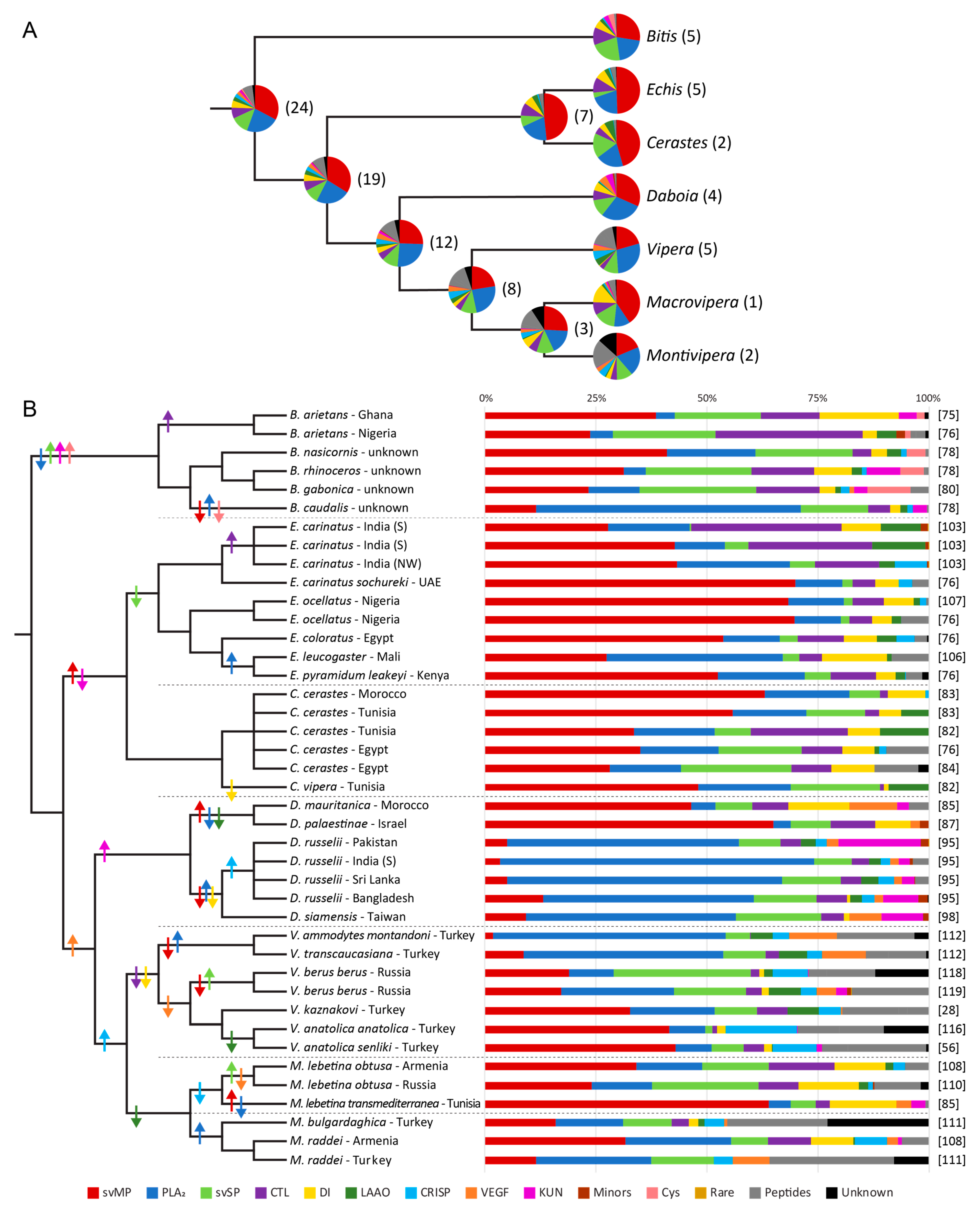
3. Venom Variations of Old World Vipers
3.1. The Bias of Quantification
3.2. Snake Venomics
3.3. Clade of African Adders
3.4. Clade of Echis and Cerastes
3.5. Clade of Eurasian Vipers
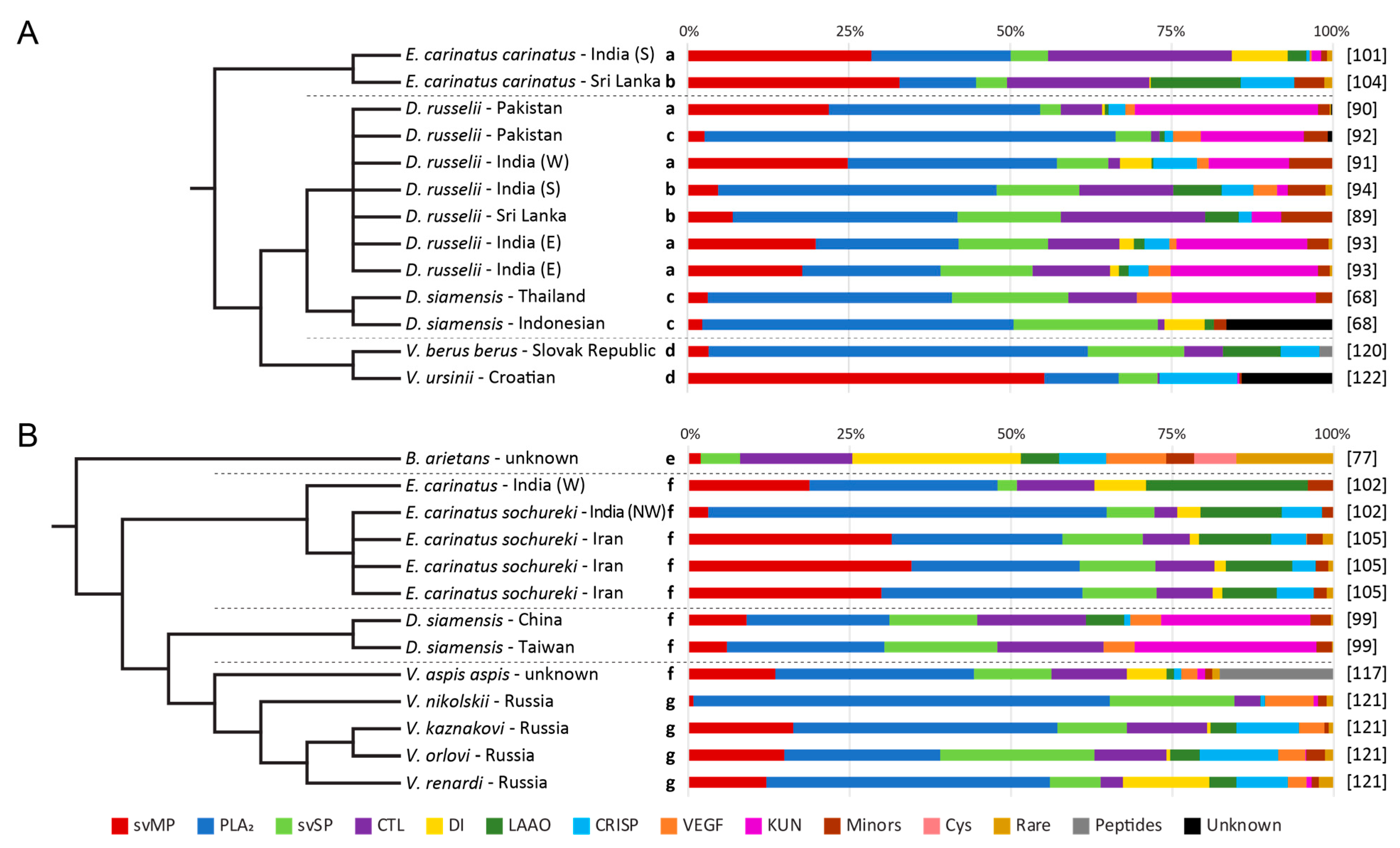
3.6. Other Quantification Workflows
3.7. Two-Step Quantifications
3.8. Whole Venom in-Solution Shotgun
3.9. Non-Quantified Venom Compositions
4. Outlook
5. Materials and Methods
5.1. Online Search and Selection Criteria
5.2. Taxonomic Status and Phylogentic Relationships
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alencar, L.R.V.; Quental, T.B.; Grazziotin, F.G.; Alfaro, M.L.; Martins, M.; Venzon, M.; Zaher, H. Diversification in vipers: Phylogenetic relationships, time of divergence and shifts in speciation rates. Mol. Phylogenet. Evol. 2016, 105, 50–62. [Google Scholar] [CrossRef]
- Figueroa, A.; McKelvy, A.D.; Grismer, L.L.; Bell, C.D.; Lailvaux, S.P. A species-level phylogeny of extant snakes with description of a new colubrid subfamily and genus. PLoS ONE 2016, 11, e0161070. [Google Scholar] [CrossRef] [PubMed]
- Šmíd, J.; Tolley, K.A. Calibrating the tree of vipers under the fossilized birth-death model. Sci. Rep. 2019, 9, 5510. [Google Scholar] [CrossRef] [PubMed]
- Wüster, W.; Peppin, L.; Pook, C.E.; Walker, D.E. A nesting of vipers: Phylogeny and historical biogeography of the Viperidae (Squamata: Serpentes). Mol. Phylogenet. Evol. 2008, 49, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Pook, C.E.; Joger, U.; Stümpel, N.; Wüster, W. When continents collide: Phylogeny, historical biogeography and systematics of the medically important viper genus Echis (Squamata: Serpentes: Viperidae). Mol. Phylogenet. Evol. 2009, 53, 792–807. [Google Scholar] [CrossRef]
- Zheng, Y.; Wiens, J.J. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 2016, 94, 537–547. [Google Scholar] [CrossRef]
- Mirtschin, P.J.; Dunstan, N.; Hough, B.; Hamilton, E.; Klein, S.; Lucas, J.; Millar, D.; Madaras, F.; Nias, T. Venom yields from Australian and some other species of snakes. Ecotoxicology 2006, 15, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Pucca, M.B.; Knudsen, C.; Oliveira, I.S.; Rimbault, C.; Cerni, F.A.; Wen, F.H.; Sachett, J.; Sartim, M.A.; Laustsen, A.H.; Monteiro, W.M. Current knowledge on snake dry bites. Toxins 2020, 12, 668. [Google Scholar] [CrossRef]
- Allon, N.; Kochva, E. The quantities of venom injected into prey of different size by Vipera palaestinae in a single bite. J. Exp. Zool. 1974, 188, 71–75. [Google Scholar] [CrossRef]
- Hayes, W.K.; Herbert, S.S.; Rehling, G.C.; Gennaro, J.F. Chapter 13. Factors that influence venom expenditure in viperids and other snake species during predatory and defensive contexts. In Biology of the Vipers; Schuett, G.W., Ed.; Eagle Mountain Pub: Eagle Mountain, UT, USA, 2002; pp. 207–333. ISBN 9780972015400. [Google Scholar]
- Morgenstern, D.; King, G.F. The venom optimization hypothesis revisited. Toxicon 2013, 63, 120–128. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Primers 2017, 3, 17063. [Google Scholar] [CrossRef]
- Williams, D.J.; Faiz, M.A.; Abela-Ridder, B.; Ainsworth, S.; Bulfone, T.C.; Nickerson, A.D.; Habib, A.G.; Junghanss, T.; Fan, H.W.; Turner, M.; et al. Strategy for a globally coordinated response to a priority neglected tropical disease: Snakebite envenoming. PLoS Negl. Trop. Dis. 2019, 13, e0007059. [Google Scholar] [CrossRef]
- Bolon, I.; Durso, A.M.; Botero Mesa, S.; Ray, N.; Alcoba, G.; Chappuis, F.; Ruiz de Castañeda, R. Identifying the snake: First scoping review on practices of communities and healthcare providers confronted with snakebite across the world. PLoS ONE 2020, 15, e0229989. [Google Scholar] [CrossRef]
- Albulescu, L.-O.; Xie, C.; Ainsworth, S.; Alsolaiss, J.; Crittenden, E.; Dawson, C.A.; Softley, R.; Bartlett, K.E.; Harrison, R.A.; Kool, J.; et al. A therapeutic combination of two small molecule toxin inhibitors provides broad preclinical efficacy against viper snakebite. Nat. Commun. 2020, 11, 6094. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, D.; Burns, D.S.; Wilson, D.; Warrell, D.A.; Lamb, L.E.M. Snakebites in Africa and Europe: A military perspective and update for contemporary operations. J. R. Army Med. Corps 2018, 164, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Paolino, G.; Di Nicola, M.R.; Pontara, A.; Didona, D.; Moliterni, E.; Mercuri, S.R.; Grano, M.; Borgianni, N.; Kumar, R.; Pampena, R. Vipera snakebite in Europe: A systematic review of a neglected disease. J. Eur. Acad. Dermatol. Venereol. 2020, 18, 485. [Google Scholar] [CrossRef]
- Amr, Z.S.; Abu Baker, M.A.; Warrell, D.A. Terrestrial venomous snakes and snakebites in the Arab countries of the Middle East. Toxicon 2020, 177, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Di Nicola, M.R.; Pontara, A.; Kass, G.E.N.; Kramer, N.I.; Avella, I.; Pampena, R.; Mercuri, S.R.; Dorne, J.L.C.M.; Paolino, G. Vipers of major clinical relevance in europe: Taxonomy, venom composition, toxicology and clinical management of human bites. Toxicology 2021, 152724. [Google Scholar] [CrossRef]
- Fry, B.G. (Ed.) Venomous Reptiles and Their Toxins. Evolution, Pathophysiology, and Biodiscovery; Oxford University Press: New York, NY, USA, 2015; ISBN 9780199309399. [Google Scholar]
- Casewell, N.R.; Jackson, T.N.W.; Laustsen, A.H.; Sunagar, K. Causes and Consequences of Snake Venom Variation. Trends Pharmacol. Sci. 2020, 41, 570–581. [Google Scholar] [CrossRef]
- Chippaux, J.-P.; Williams, V.; White, J. Snake venom variability: Methods of study, results and interpretation. Toxicon 1991, 29, 1279–1303. [Google Scholar] [CrossRef]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wüster, W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. Biol. Sci. 2009, 276, 2443–2449. [Google Scholar] [CrossRef] [PubMed]
- Davies, E.-L.; Arbuckle, K. Coevolution of snake venom toxic activities and diet: Evidence that ecological generalism favours toxicological diversity. Toxins 2019, 11, 711. [Google Scholar] [CrossRef] [PubMed]
- Daltry, J.C.; Wüster, W.; Thorpe, R.S. Diet and snake venom evolution. Nature 1996, 379, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Lyons, K.; Dugon, M.M.; Healy, K. Diet breadth mediates the prey specificity of venom potency in snakes. Toxins 2020, 12, 74. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, H.L.; Sanz, L.; Chiucchi, J.E.; Farrell, T.M.; Calvete, J.J. Proteomic analysis of ontogenetic and diet-related changes in venom composition of juvenile and adult Dusky Pigmy rattlesnakes (Sistrurus miliarius barbouri). J. Proteom. 2011, 74, 2169–2179. [Google Scholar] [CrossRef]
- Petras, D.; Hempel, B.-F.; Göçmen, B.; Karis, M.; Whiteley, G.; Wagstaff, S.C.; Heiss, P.; Casewell, N.R.; Nalbantsoy, A.; Süssmuth, R.D. Intact protein mass spectrometry reveals intraspecies variations in venom composition of a local population of Vipera kaznakovi in Northeastern Turkey. J. Proteom. 2019, 199, 31–50. [Google Scholar] [CrossRef]
- Durban, J.; Pérez, A.; Sanz, L.; Gómez, A.; Bonilla, F.; Rodríguez, S.; Chacón, D.; Sasa, M.; Angulo, Y.; Gutiérrez, J.M.; et al. Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Genom. 2013, 14, 234. [Google Scholar] [CrossRef] [PubMed]
- Zelanis, A.; Tashima, A.K.; Pinto, A.F.M.; Paes Leme, A.F.; Stuginski, D.R.; Furtado, M.F.; Sherman, N.E.; Ho, P.L.; Fox, J.W.; Serrano, S.M.T. Bothrops jararaca venom proteome rearrangement upon neonate to adult transition. Proteomics 2011, 11, 4218–4228. [Google Scholar] [CrossRef]
- Da Silva-Júnior, L.N.; Abreu, L.d.S.; Rodrigues, C.F.B.; Da Galizio, N.C.; Da Aguiar, W.S.; Serino-Silva, C.; Dos Santos, V.S.; Costa, I.A.; Oliveira, L.V.F.; Sant’Anna, S.S.; et al. Geographic variation of individual venom profile of Crotalus durissus snakes. J. Venom. Anim. Toxins Incl. Trop. Dis. 2020, 26, e20200016. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Rey-Suárez, P.; Fernández, J.; Sasa, M.; Pla, D.; Vargas, N.; Bénard-Valle, M.; Sanz, L.; Corrêa-Netto, C.; Núñez, V.; et al. Venoms of Micrurus coral snakes: Evolutionary trends in compositional patterns emerging from proteomic analyses. Toxicon 2016, 122, 7–25. [Google Scholar] [CrossRef]
- Huang, H.-W.; Liu, B.-S.; Chien, K.-Y.; Chiang, L.-C.; Huang, S.-Y.; Sung, W.-C.; Wu, W.-G. Cobra venom proteome and glycome determined from individual snakes of Naja atra reveal medically important dynamic range and systematic geographic variation. J. Proteom. 2015, 128, 92–104. [Google Scholar] [CrossRef]
- Amorim, F.G.; Costa, T.R.; Baiwir, D.; de Pauw, E.; Quinton, L.; Sampaio, S.V. Proteopeptidomic, Functional and immunoreactivity characterization of bothrops moojeni snake venom: Influence of snake gender on venom composition. Toxins 2018, 10, 177. [Google Scholar] [CrossRef]
- Menezes, M.C.; Furtado, M.F.; Travaglia-Cardoso, S.R.; Camargo, A.C.M.; Serrano, S.M.T. Sex-based individual variation of snake venom proteome among eighteen Bothrops jararaca siblings. Toxicon 2006, 47, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Amazonas, D.R.; Freitas-de-Sousa, L.A.; Orefice, D.P.; de Sousa, L.F.; Martinez, M.G.; Mourão, R.H.V.; Chalkidis, H.M.; Camargo, P.B.; Moura-da-Silva, A.M. Evidence for snake venom plasticity in a long-term study with individual captive Bothrops atrox. Toxins 2019, 11, 294. [Google Scholar] [CrossRef] [PubMed]
- McCleary, R.J.R.; Sridharan, S.; Dunstan, N.L.; Mirtschin, P.J.; Kini, R.M. Proteomic comparisons of venoms of long-term captive and recently wild-caught Eastern brown snakes (Pseudonaja textilis) indicate venom does not change due to captivity. J. Proteom. 2016, 144, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Zancolli, G.; Calvete, J.J.; Cardwell, M.D.; Greene, H.W.; Hayes, W.K.; Hegarty, M.J.; Herrmann, H.-W.; Holycross, A.T.; Lannutti, D.I.; Mulley, J.F.; et al. When one phenotype is not enough: Divergent evolutionary trajectories govern venom variation in a widespread rattlesnake species. Proc. Biol. Sci. 2019, 286, 20182735. [Google Scholar] [CrossRef]
- Kazandjian, T.D.; Petras, D.; Robinson, S.D.; van Thiel, J.; Greene, H.W.; Arbuckle, K.; Barlow, A.; Carter, D.A.; Wouters, R.M.; Whiteley, G.; et al. Convergent evolution of pain-inducing defensive venom components in spitting cobras. Science 2021, 371, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Arbuckle, K. From molecules to macroevolution: Venom as a model system for evolutionary biology across levels of life. Toxicon: X 2020, 6, 100034. [Google Scholar] [CrossRef]
- Barua, A.; Mikheyev, A.S. Many options, few solutions: Over 60 my snakes converged on a few optimal venom formulations. Mol. Biol. Evol. 2019, 36, 1964–1974. [Google Scholar] [CrossRef]
- Zancolli, G.; Casewell, N.R. Venom systems as models for studying the origin and regulation of evolutionary novelties. Mol. Biol. Evol. 2020, 37, 2777–2790. [Google Scholar] [CrossRef]
- Modahl, C.M.; Brahma, R.K.; Koh, C.Y.; Shioi, N.; Kini, R.M. omics technologies for profiling toxin diversity and evolution in snake venom: Impacts on the discovery of therapeutic and diagnostic agents. Annu. Rev. Anim. Biosci. 2020, 8, 91–116. [Google Scholar] [CrossRef]
- Kerkkamp, H.M.I.; Kini, R.M.; Pospelov, A.S.; Vonk, F.J.; Henkel, C.V.; Richardson, M.K. Snake genome sequencing: Results and future prospects. Toxins 2016, 8, 360. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M.T. Exploring snake venom proteomes: Multifaceted analyses for complex toxin mixtures. Proteomics 2008, 8, 909–920. [Google Scholar] [CrossRef]
- Calvete, J.J. Snake venomics—From low-resolution toxin-pattern recognition to toxin-resolved venom proteomes with absolute quantification. Expert Rev. Proteom. 2018, 15, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Calvete, J.J. Strategies in ‘snake venomics’ aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 26. [Google Scholar] [CrossRef]
- Mouchbahani-Constance, S.; Sharif-Naeini, R. Proteomic and transcriptomic techniques to decipher the molecular evolution of venoms. Toxins 2021, 13, 154. [Google Scholar] [CrossRef] [PubMed]
- Ghezellou, P.; Garikapati, V.; Kazemi, S.M.; Strupat, K.; Ghassempour, A.; Spengler, B. A perspective view of top-down proteomics in snake venom research. Rapid Commun. Mass Spectrom. 2019, 33, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Waridel, P.; Frank, A.; Thomas, H.; Surendranath, V.; Sunyaev, S.; Pevzner, P.; Shevchenko, A. Sequence similarity-driven proteomics in organisms with unknown genomes by LC-MS/MS and automated de novo sequencing. Proteomics 2007, 7, 2318–2329. [Google Scholar] [CrossRef]
- Abd El-Aziz, T.M.; Soares, A.G.; Stockand, J.D. Advances in venomics: Modern separation techniques and mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2020, 1160, 122352. [Google Scholar] [CrossRef]
- Walker, A.A.; Robinson, S.D.; Hamilton, B.F.; Undheim, E.A.B.; King, G.F. Deadly proteomes: A practical guide to proteotranscriptomics of animal venoms. Proteomics 2020, 20, e1900324. [Google Scholar] [CrossRef]
- Melani, R.D.; Nogueira, F.C.S.; Domont, G.B. It is time for top-down venomics. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 44. [Google Scholar] [CrossRef]
- Calvete, J.J.; Petras, D.; Calderón-Celis, F.; Lomonte, B.; Encinar, J.R.; Sanz-Medel, A. Protein-species quantitative venomics: Looking through a crystal ball. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 27. [Google Scholar] [CrossRef]
- Lemon, D.J.; Horvath, F.P.; Ford, A.A.; May, H.C.; Moffett, S.X.; Olivera, D.S.; Hwang, Y.Y. ICP-MS characterization of seven North American snake venoms. Toxicon 2020, 184, 62–67. [Google Scholar] [CrossRef]
- Hempel, B.-F.; Damm, M.; Mrinalini; Göçmen, B.; Karış, M.; Nalbantsoy, A.; Kini, R.M.; Süssmuth, R.D. Extended snake venomics by top-down in-source decay: Investigating the newly discovered anatolian meadow viper subspecies, vipera anatolica senliki. J. Proteome Res. 2020. [Google Scholar] [CrossRef]
- Zancolli, G.; Sanz, L.; Calvete, J.J.; Wüster, W. Venom on-a-chip: A fast and efficient method for comparative venomics. Toxins 2017, 9, 179. [Google Scholar] [CrossRef] [PubMed]
- Cristina, R.T.; Kocsis, R.; Tulcan, C.; Alexa, E.; Boldura, O.M.; Hulea, C.I.; Dumitrescu, E.; Radulov, I.; Muselin, F. Protein structure of the venom in nine species of snake: From bio-compounds to possible healing agents. Braz. J. Med. Biol. Res. 2020, 53, e9001. [Google Scholar] [CrossRef]
- Igci, N.; OZEL DEMİRALP, F.D. A Fourier transform infrared spectroscopic investigation of macrovipera lebetina lebetina and M. l. obtusa Crude Venoms. Eur. J. Biol. 2020. [Google Scholar] [CrossRef]
- Fasoli, E.; Sanz, L.; Wagstaff, S.; Harrison, R.A.; Righetti, P.G.; Calvete, J.J. Exploring the venom proteome of the African puff adder, Bitis arietans, using a combinatorial peptide ligand library approach at different pHs. J. Proteom. 2010, 73, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Junqueira-de-Azevedo, I.L.M.; Campos, P.F.; Ching, A.T.C.; Mackessy, S.P. Colubrid venom composition: An -omics perspective. Toxins 2016, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Isbister, G.K. A Review and Database of Snake Venom Proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [PubMed]
- Dam, S.H.; Friis, R.U.W.; Petersen, S.D.; Martos-Esteban, A.; Laustsen, A.H. Snake Venomics Display: An online toolbox for visualization of snake venomics data. Toxicon 2018, 152, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Kalita, B.; Mukherjee, A.K. Recent advances in snake venom proteomics research in India: A new horizon to decipher the geographical variation in venom proteome composition and exploration of candidate drug prototypes. J. Proteins. Proteom. 2019, 10, 149–164. [Google Scholar] [CrossRef]
- Rima, M.; Alavi Naini, S.M.; Karam, M.; Sadek, R.; Sabatier, J.-M.; Fajloun, Z. Vipers of the middle east: A rich source of bioactive molecules. Molecules 2018, 23, 2721. [Google Scholar] [CrossRef]
- Chanda, A.; Mukherjee, A.K. Mass spectrometric analysis to unravel the venom proteome composition of Indian snakes: Opening new avenues in clinical research. Expert Rev. Proteom. 2020, 1–13. [Google Scholar] [CrossRef]
- Siigur, J.; Aaspõllu, A.; Siigur, E. Biochemistry and pharmacology of proteins and peptides purified from the venoms of the snakes Macrovipera lebetina subspecies. Toxicon 2019, 158, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Lingam, T.M.C.; Tan, K.Y.; Tan, C.H. Proteomics and antivenom immunoprofiling of Russell’s viper (Daboia siamensis) venoms from Thailand and Indonesia. J. Venom. Anim. Toxins Incl. Trop. Dis. 2020, 26, e20190048. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, X.; König, E.; Zhou, M.; Wang, L.; Chen, T.; Shaw, C. Comparative Profiling of Three Atheris Snake Venoms: A. squamigera, A. nitschei and A. chlorechis. Protein J. 2018, 37, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.A.; Jackson, T.N.W.; Casewell, N.R.; Low, D.H.W.; Rossi, S.; Baumann, K.; Fathinia, B.; Visser, J.; Nouwens, A.; Hendrikx, I.; et al. Extreme venom variation in Middle Eastern vipers: A proteomics comparison of Eristicophis macmahonii, Pseudocerastes fieldi and Pseudocerastes persicus. J. Proteom. 2015, 116, 106–113. [Google Scholar] [CrossRef]
- Chippaux, J.-P. Snakebite envenomation turns again into a neglected tropical disease! J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 38. [Google Scholar] [CrossRef]
- WHO. Snakebite Envenoming—A Strategy for Prevention and Control; WHO: Geneva, Switzerland, 2019; ISBN 978 92 4 151564 1. [Google Scholar]
- Harrison, R.A.; Casewell, N.R.; Ainsworth, S.A.; Lalloo, D.G. The time is now: A call for action to translate recent momentum on tackling tropical snakebite into sustained benefit for victims. Trans. R. Soc. Trop. Med. Hyg. 2019, 113, 835–838. [Google Scholar] [CrossRef]
- Knudsen, C.; Ledsgaard, L.; Dehli, R.I.; Ahmadi, S.; Sørensen, C.V.; Laustsen, A.H. Engineering and design considerations for next-generation snakebite antivenoms. Toxicon 2019, 167, 67–75. [Google Scholar] [CrossRef]
- Juárez, P.; Wagstaff, S.C.; Oliver, J.; Sanz, L.; Harrison, R.A.; Calvete, J.J. Molecular cloning of disintegrin-like transcript BA-5A from a Bitis arietans venom gland cDNA library: A putative intermediate in the evolution of the long-chain disintegrin bitistatin. J. Mol. Evol. 2006, 63, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wagstaff, S.C.; Wüster, W.; Cook, D.A.N.; Bolton, F.M.S.; King, S.I.; Pla, D.; Sanz, L.; Calvete, J.J.; Harrison, R.A. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc. Natl. Acad. Sci. USA 2014, 111, 9205–9210. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.R.; Bubner, E.R.; Jovcevski, B.; Mittal, P.; Pukala, T.L. Interrogating the higher order structures of snake venom proteins using an integrated mass spectrometric approach. J. Proteom. 2020, 216, 103680. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Escolano, J.; Sanz, L. Snake venomics of Bitis species reveals large intragenus venom toxin composition variation: Application to taxonomy of congeneric taxa. J. Proteome Res. 2007, 6, 2732–2745. [Google Scholar] [CrossRef]
- Francischetti, I.M.B.; My-Pham, V.; Harrison, J.; Garfield, M.K.; Ribeiro, J.M.C. Bitis gabonica (Gaboon viper) snake venom gland: Toward a catalog for the full-length transcripts (cDNA) and proteins. Gene 2004, 337, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Marcinkiewicz, C.; Sanz, L. Snake venomics of Bitis gabonica gabonica. Protein family composition, subunit organization of venom toxins, and characterization of dimeric disintegrins bitisgabonin-1 and bitisgabonin-2. J. Proteome Res. 2007, 6, 326–336. [Google Scholar] [CrossRef]
- Coimbra, F.C.P.; Dobson, J.; Zdenek, C.N.; Op den Brouw, B.; Hamilton, B.; Debono, J.; Masci, P.; Frank, N.; Ge, L.; Kwok, H.F.; et al. Does size matter? Venom proteomic and functional comparison between night adder species (Viperidae: Causus) with short and long venom glands. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2018, 211, 7–14. [Google Scholar] [CrossRef]
- Bazaa, A.; Marrakchi, N.; El Ayeb, M.; Sanz, L.; Calvete, J.J. Snake venomics: Comparative analysis of the venom proteomes of the Tunisian snakes Cerastes cerastes, Cerastes vipera and Macrovipera lebetina. Proteomics 2005, 5, 4223–4235. [Google Scholar] [CrossRef]
- Fahmi, L.; Makran, B.; Pla, D.; Sanz, L.; Oukkache, N.; Lkhider, M.; Harrison, R.A.; Ghalim, N.; Calvete, J.J. Venomics and antivenomics profiles of North African Cerastes cerastes and C. vipera populations reveals a potentially important therapeutic weakness. J. Proteom. 2012, 75, 2442–2453. [Google Scholar] [CrossRef]
- Ozverel, C.S.; Damm, M.; Hempel, B.-F.; Göçmen, B.; Sroka, R.; Süssmuth, R.D.; Nalbantsoy, A. Investigating the cytotoxic effects of the venom proteome of two species of the Viperidae family (Cerastes cerastes and Cryptelytrops purpureomaculatus) from various habitats. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2019, 220, 20–30. [Google Scholar] [CrossRef]
- Makran, B.; Fahmi, L.; Pla, D.; Sanz, L.; Oukkache, N.; Lkhider, M.; Ghalim, N.; Calvete, J.J. Snake venomics of Macrovipera mauritanica from Morocco, and assessment of the para-specific immunoreactivity of an experimental monospecific and a commercial antivenoms. J. Proteom. 2012, 75, 2431–2441. [Google Scholar] [CrossRef] [PubMed]
- Chakir, S.; Daoudi, K.; Darkaoui, B.; Lafnoune, A.; Hmyene, A.; Oukkache, N. Screening of Active Biomolecules from the Venom of the Moroccan Viper Daboia mauritanica. EC Pharmacol. Toxicol. 2019, 7, 144–149. [Google Scholar]
- Momic, T.; Arlinghaus, F.T.; Arien-Zakay, H.; Katzhendler, J.; Eble, J.A.; Marcinkiewicz, C.; Lazarovici, P. Pharmacological aspects of Vipera xantina palestinae venom. Toxins 2011, 3, 1420–1432. [Google Scholar] [CrossRef]
- Sharma, M.; Das, D.; Iyer, J.K.; Kini, R.M.; Doley, R. Unveiling the complexities of Daboia russelii venom, a medically important snake of India, by tandem mass spectrometry. Toxicon 2015, 107, 266–281. [Google Scholar] [CrossRef] [PubMed]
- Tan, N.H.; Fung, S.Y.; Tan, K.Y.; Yap, M.K.K.; Gnanathasan, C.A.; Tan, C.H. Functional venomics of the Sri Lankan Russell’s viper (Daboia russelii) and its toxinological correlations. J. Proteom. 2015, 128, 403–423. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Kalita, B.; Mackessy, S.P. A proteomic analysis of Pakistan Daboia russelii russelii venom and assessment of potency of Indian polyvalent and monovalent antivenom. J. Proteom. 2016, 144, 73–86. [Google Scholar] [CrossRef]
- Kalita, B.; Patra, A.; Mukherjee, A.K. Unraveling the proteome composition and Immuno-profiling of western india russell’s viper venom for in-depth understanding of its pharmacological properties, clinical manifestations, and effective antivenom treatment. J. Proteome Res. 2017, 16, 583–598. [Google Scholar] [CrossRef]
- Faisal, T.; Tan, K.Y.; Sim, S.M.; Quraishi, N.; Tan, N.H.; Tan, C.H. Proteomics, functional characterization and antivenom neutralization of the venom of Pakistani Russell’s viper (Daboia russelii) from the wild. J. Proteom. 2018, 183, 1–13. [Google Scholar] [CrossRef]
- Kalita, B.; Patra, A.; Das, A.; Mukherjee, A.K. Proteomic Analysis and Immuno-Profiling of Eastern India Russell’s Viper (Daboia russelii) Venom: Correlation between RVV Composition and Clinical Manifestations Post RV Bite. J. Proteome Res. 2018, 17, 2819–2833. [Google Scholar] [CrossRef]
- Kalita, B.; Singh, S.; Patra, A.; Mukherjee, A.K. Quantitative proteomic analysis and antivenom study revealing that neurotoxic phospholipase A2 enzymes, the major toxin class of Russell’s viper venom from southern India, shows the least immuno-recognition and neutralization by commercial polyvalent antivenom. Int. J. Biol. Macromol. 2018, 118, 375–385. [Google Scholar] [CrossRef]
- Pla, D.; Sanz, L.; Quesada-Bernat, S.; Villalta, M.; Baal, J.; Chowdhury, M.A.W.; León, G.; Gutiérrez, J.M.; Kuch, U.; Calvete, J.J. Phylovenomics of Daboia russelii across the Indian subcontinent. Bioactivities and comparative in vivo neutralization and in vitro third-generation antivenomics of antivenoms against venoms from India, Bangladesh and Sri Lanka. J. Proteom. 2019, 207, 103443. [Google Scholar] [CrossRef] [PubMed]
- Nawarak, J.; Sinchaikul, S.; Wu, C.-Y.; Liau, M.-Y.; Phutrakul, S.; Chen, S.-T. Proteomics of snake venoms from Elapidae and Viperidae families by multidimensional chromatographic methods. Electrophoresis 2003, 24, 2838–2854. [Google Scholar] [CrossRef]
- Risch, M.; Georgieva, D.; von Bergen, M.; Jehmlich, N.; Genov, N.; Arni, R.K.; Betzel, C. Snake venomics of the Siamese Russell’s viper (Daboia russelli siamensis)—Relation to pharmacological activities. J. Proteom. 2009, 72, 256–269. [Google Scholar] [CrossRef]
- Sanz, L.; Quesada-Bernat, S.; Chen, P.Y.; Lee, C.D.; Chiang, J.R.; Calvete, J.J. Translational venomics: Third-generation antivenomics of anti-siamese russell’s viper, daboia siamensis, antivenom manufactured in Taiwan CDC’s vaccine center. Trop. Med. Infect. Dis. 2018, 3, 66. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.Y.; Tan, N.H.; Tan, C.H. Venom proteomics and antivenom neutralization for the Chinese eastern Russell’s viper, Daboia siamensis from Guangxi and Taiwan. Sci. Rep. 2018, 8, 8545. [Google Scholar] [CrossRef]
- Liu, C.-C.; Lin, C.-C.; Hsiao, Y.-C.; Wang, P.-J.; Yu, J.-S. Proteomic characterization of six Taiwanese snake venoms: Identification of species-specific proteins and development of a SISCAPA-MRM assay for cobra venom factors. J. Proteom. 2018, 187, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.; Kalita, B.; Chanda, A.; Mukherjee, A.K. Proteomics and antivenomics of Echis carinatus carinatus venom: Correlation with pharmacological properties and pathophysiology of envenomation. Sci. Rep. 2017, 7, 17119. [Google Scholar] [CrossRef] [PubMed]
- Senji Laxme, R.R.; Khochare, S.; de Souza, H.F.; Ahuja, B.; Suranse, V.; Martin, G.; Whitaker, R.; Sunagar, K. Beyond the ‘big four’: Venom profiling of the medically important yet neglected Indian snakes reveals disturbing antivenom deficiencies. PLoS Negl. Trop. Dis. 2019, 13, e0007899. [Google Scholar] [CrossRef]
- Bhatia, S.; Vasudevan, K. Comparative proteomics of geographically distinct saw-scaled viper (Echis carinatus) venoms from India. Toxicon X 2020, 7, 100048. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.; Mukherjee, A.K. Proteomic Analysis of Sri Lanka Echis carinatus Venom: Immunological Cross-Reactivity and Enzyme Neutralization Potency of Indian Polyantivenom. J. Proteome Res. 2020, 19, 3022–3032. [Google Scholar] [CrossRef] [PubMed]
- Ghezellou, P.; Albuquerque, W.; Garikapati, V.; Casewell, N.R.; Kazemi, S.M.; Ghassempour, A.; Spengler, B. Integrating top-down and bottom-up mass spectrometric strategies for proteomic profiling of iranian saw-scaled viper, echis carinatus sochureki, venom. J. Proteome Res. 2020. [Google Scholar] [CrossRef]
- Albulescu, L.-O.; Hale, M.S.; Ainsworth, S.; Alsolaiss, J.; Crittenden, E.; Calvete, J.J.; Evans, C.; Wilkinson, M.C.; Harrison, R.A.; Kool, J.; et al. Preclinical validation of a repurposed metal chelator as an early-intervention therapeutic for hemotoxic snakebite. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Wagstaff, S.C.; Sanz, L.; Juárez, P.; Harrison, R.A.; Calvete, J.J. Combined snake venomics and venom gland transcriptomic analysis of the ocellated carpet viper, Echis ocellatus. J. Proteom. 2009, 71, 609–623. [Google Scholar] [CrossRef]
- Sanz, L.; Ayvazyan, N.; Calvete, J.J. Snake venomics of the Armenian mountain vipers Macrovipera lebetina obtusa and Vipera raddei. J. Proteom. 2008, 71, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Igci, N.; Demiralp, D.O. A preliminary investigation into the venom proteome of Macrovipera lebetina obtusa (Dwigubsky, 1832) from Southeastern Anatolia by MALDI-TOF mass spectrometry and comparison of venom protein profiles with Macrovipera lebetina lebetina (Linnaeus, 1758) from Cyprus by 2D-PAGE. Arch. Toxicol. 2012, 86, 441–451. [Google Scholar] [CrossRef]
- Pla, D.; Quesada-Bernat, S.; Rodríguez, Y.; Sánchez, A.; Vargas, M.; Villalta, M.; Mesén, S.; Segura, Á.; Mustafin, D.O.; Fomina, Y.A.; et al. Dagestan blunt-nosed viper, Macrovipera lebetina obtusa (Dwigubsky, 1832), venom. Venomics, antivenomics, and neutralization assays of the lethal and toxic venom activities by anti-Macrovipera lebetina turanica and anti-Vipera berus berus antivenoms. Toxicon X 2020, 6, 100035. [Google Scholar] [CrossRef] [PubMed]
- Nalbantsoy, A.; Hempel, B.-F.; Petras, D.; Heiss, P.; Göçmen, B.; Iğci, N.; Yildiz, M.Z.; Süssmuth, R.D. Combined venom profiling and cytotoxicity screening of the Radde’s mountain viper (Montivipera raddei) and Mount Bulgar Viper (Montivipera bulgardaghica) with potent cytotoxicity against human A549 lung carcinoma cells. Toxicon 2017, 135, 71–83. [Google Scholar] [CrossRef]
- Hempel, B.-F.; Damm, M.; Göçmen, B.; Karis, M.; Oguz, M.A.; Nalbantsoy, A.; Süssmuth, R.D. Comparative Venomics of the Vipera ammodytes transcaucasiana and Vipera ammodytes montandoni from Turkey Provides Insights into Kinship. Toxins 2018, 10, 23. [Google Scholar] [CrossRef]
- Georgieva, D.; Risch, M.; Kardas, A.; Buck, F.; von Bergen, M.; Betzel, C. Comparative analysis of the venom proteomes of Vipera ammodytes ammodytes and Vipera ammodytes meridionalis. J. Proteome Res. 2008, 7, 866–886. [Google Scholar] [CrossRef]
- Leonardi, A.; Sajevic, T.; Pungerčar, J.; Križaj, I. comprehensive study of the proteome and transcriptome of the venom of the most venomous european viper: Discovery of a new subclass of ancestral snake venom metalloproteinase precursor-derived proteins. J. Proteome Res. 2019, 18, 2287–2309. [Google Scholar] [CrossRef]
- Gopcevic, K.; Karadzic, I.; Izrael-Zivkovic, L.; Medic, A.; Isakovic, A.; Popović, M.; Kekic, D.; Stanojkovic, T.; Hozic, A.; Cindric, M. Study of the venom proteome of Vipera ammodytes ammodytes (Linnaeus, 1758): A qualitative overview, biochemical and biological profiling. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 37, 100776. [Google Scholar] [CrossRef]
- Göçmen, B.; Heiss, P.; Petras, D.; Nalbantsoy, A.; Süssmuth, R.D. Mass spectrometry guided venom profiling and bioactivity screening of the Anatolian Meadow Viper, Vipera anatolica. Toxicon 2015, 107, 163–174. [Google Scholar] [CrossRef]
- Giribaldi, J.; Kazandjian, T.; Amorim, F.G.; Whiteley, G.; Wagstaff, S.C.; Cazals, G.; Enjalbal, C.; Quinton, L.; Casewell, N.R.; Dutertre, S. Venomics of the asp viper Vipera aspis aspis from France. J. Proteom. 2020, 103707. [Google Scholar] [CrossRef]
- Latinović, Z.; Leonardi, A.; Šribar, J.; Sajevic, T.; Žužek, M.C.; Frangež, R.; Halassy, B.; Trampuš-Bakija, A.; Pungerčar, J.; Križaj, I. Venomics of Vipera berus berus to explain differences in pathology elicited by Vipera ammodytes ammodytes envenomation: Therapeutic implications. J. Proteom. 2016, 146, 34–47. [Google Scholar] [CrossRef]
- Al-Shekhadat, R.I.; Lopushanskaya, K.S.; Segura, Á.; Gutiérrez, J.M.; Calvete, J.J.; Pla, D. Vipera berus berus Venom from Russia: Venomics, bioactivities and preclinical assessment of microgen antivenom. Toxins 2019, 11, 90. [Google Scholar] [CrossRef] [PubMed]
- Bocian, A.; Urbanik, M.; Hus, K.; Łyskowski, A.; Petrilla, V.; Andrejčáková, Z.; Petrillová, M.; Legath, J. Proteome and peptidome of vipera berus berus venom. Molecules 2016, 21, 1398. [Google Scholar] [CrossRef]
- Kovalchuk, S.I.; Ziganshin, R.H.; Starkov, V.G.; Tsetlin, V.I.; Utkin, Y.N. Quantitative proteomic analysis of venoms from russian vipers of pelias group: Phospholipases A2 are the main venom components. Toxins 2016, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Lang Balija, M.; Leonardi, A.; Brgles, M.; Sviben, D.; Kurtović, T.; Halassy, B.; Križaj, I. Biological activities and proteomic profile of the venom of Vipera ursinii ssp., a very rare karst viper from Croatia. Toxins 2020, 12, 187. [Google Scholar] [CrossRef] [PubMed]
- Diz, A.P.; Truebano, M.; Skibinski, D.O.F. The consequences of sample pooling in proteomics: An empirical study. Electrophoresis 2009, 30, 2967–2975. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Alpi, E.; Wang, R.; Hermjakob, H.; Vizcaíno, J.A. Making proteomics data accessible and reusable: Current state of proteomics databases and repositories. Proteomics 2015, 15, 930–949. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Csordas, A.; Sun, Z.; Jarnuczak, A.; Perez-Riverol, Y.; Ternent, T.; Campbell, D.S.; Bernal-Llinares, M.; Okuda, S.; Kawano, S.; et al. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017, 45, D1100–D1106. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Deutsch, E.W.; Wang, R.; Csordas, A.; Reisinger, F.; Ríos, D.; Dianes, J.A.; Sun, Z.; Farrah, T.; Bandeira, N.; et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Hargreaves, A.D.; Swain, M.T.; Hegarty, M.J.; Logan, D.W.; Mulley, J.F. Restriction and recruitment-gene duplication and the origin and evolution of snake venom toxins. Genome Biol. Evol. 2014, 6, 2088–2095. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Velasco, J.; Card, D.C.; Andrew, A.L.; Shaney, K.J.; Adams, R.H.; Schield, D.R.; Casewell, N.R.; Mackessy, S.P.; Castoe, T.A. Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom. Mol. Biol. Evol. 2015, 32, 173–183. [Google Scholar] [CrossRef]
- Silva, A.; Kuruppu, S.; Othman, I.; Goode, R.J.A.; Hodgson, W.C.; Isbister, G.K. Neurotoxicity in Sri Lankan Russell’s Viper (Daboia russelii) envenoming is primarily due to U1-viperitoxin-Dr1a, a pre-synaptic neurotoxin. Neurotox. Res. 2017, 31, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, G.; Duregotti, E.; Locatelli, C.A.; Giampreti, A.; Lonati, D.; Rossetto, O.; Pirazzini, M. Variability in venom composition of European viper subspecies limits the cross-effectiveness of antivenoms. Sci. Rep. 2018, 8, 9818. [Google Scholar] [CrossRef] [PubMed]
- Boldrini-França, J.; Cologna, C.T.; Pucca, M.B.; Bordon, K.d.C.F.; Amorim, F.G.; Anjolette, F.A.P.; Cordeiro, F.A.; Wiezel, G.A.; Cerni, F.A.; Pinheiro-Junior, E.L.; et al. Minor snake venom proteins: Structure, function and potential applications. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 824–838. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.R.; Resende, L.M.; Watanabe, R.K.; Carregari, V.C.; Huancahuire-Vega, S.; da S. Caldeira, C.A.; Coutinho-Neto, A.; Soares, A.M.; Vale, N.; de C. Gomes, P.A.; et al. Snake venom peptides and low mass proteins: Molecular tools and therapeutic agents. Curr. Med. Chem. 2017, 24, 3254–3282. [Google Scholar] [CrossRef] [PubMed]
- Villar-Briones, A.; Aird, S.D. Organic and peptidyl constituents of snake venoms: The picture is vastly more complex than we imagined. Toxins 2018, 10, 392. [Google Scholar] [CrossRef]
- Olaoba, O.T.; Karina Dos Santos, P.; Selistre-de-Araujo, H.S.; Ferreira de Souza, D.H. Snake Venom Metalloproteinases (SVMPs): A structure-function update. Toxicon X 2020, 7, 100052. [Google Scholar] [CrossRef]
- Fox, J.W.; Serrano, S.M.T. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008, 275, 3016–3030. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wagstaff, S.C.; Harrison, R.A.; Renjifo, C.; Wüster, W. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 2011, 28, 2637–2649. [Google Scholar] [CrossRef] [PubMed]
- Markland, F.S.; Swenson, S. Snake venom metalloproteinases. Toxicon 2013, 62, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, B.A.; Deconte, S.R.; de Moura, F.B.R.; Tomiosso, T.C.; Clissa, P.B.; Andrade, S.P.; Araújo, F.d.A. Inflammation, angiogenesis and fibrogenesis are differentially modulated by distinct domains of the snake venom metalloproteinase jararhagin. Int. J. Biol. Macromol. 2018, 119, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M. Excitement ahead: Structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon 2003, 42, 827–840. [Google Scholar] [CrossRef] [PubMed]
- Hiu, J.J.; Yap, M.K.K. Cytotoxicity of snake venom enzymatic toxins: Phospholipase A2 and l-amino acid oxidase. Biochem. Soc. Trans. 2020, 48, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.B.; Scott-Davey, T. Secreted phospholipases A2 of snake venoms: Effects on the peripheral neuromuscular system with comments on the role of phospholipases A2 in disorders of the CNS and their uses in industry. Toxins 2013, 5, 2533–2571. [Google Scholar] [CrossRef]
- Mora-Obando, D.; Fernández, J.; Montecucco, C.; Gutiérrez, J.M.; Lomonte, B. Synergism between basic Asp49 and Lys49 phospholipase A2 myotoxins of viperid snake venom in vitro and in vivo. PLoS ONE 2014, 9, e109846. [Google Scholar] [CrossRef]
- Jan, V.M.; Guillemin, I.; Robbe-Vincent, A.; Choumet, V. Phospholipase A2 diversity and polymorphism in European viper venoms: Paradoxical molecular evolution in Viperinae. Toxicon 2007, 50, 1140–1161. [Google Scholar] [CrossRef] [PubMed]
- Serrano, S.M.T. The long road of research on snake venom serine proteinases. Toxicon 2013, 62, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Masood, R.; Ali, I.; Ullah, K.; Ali, H.; Akbar, H.; Betzel, C. Thrombin-like enzymes from snake venom: Structural characterization and mechanism of action. Int. J. Biol. Macromol. 2018, 114, 788–811. [Google Scholar] [CrossRef] [PubMed]
- Castro, H.C.; Zingali, R.B.; Albuquerque, M.G.; Pujol-Luz, M.; Rodrigues, C.R. Snake venom thrombin-like enzymes: From reptilase to now. Cell. Mol. Life Sci. 2004, 61, 843–856. [Google Scholar] [CrossRef]
- Latinović, Z.; Leonardi, A.; Koh, C.Y.; Kini, R.M.; Trampuš Bakija, A.; Pungerčar, J.; Križaj, I. The procoagulant snake venom serine protease potentially having a dual, blood coagulation factor V and X-Activating activity. Toxins 2020, 12, 358. [Google Scholar] [CrossRef]
- Eble, J.A. Structurally Robust and Functionally Highly Versatile-C-Type Lectin (-Related) Proteins in Snake Venoms. Toxins 2019, 11, 136. [Google Scholar] [CrossRef]
- Morita, T. Structures and functions of snake venom CLPs (C-type lectin-like proteins) with anticoagulant-, procoagulant-, and platelet-modulating activities. Toxicon 2005, 45, 1099–1114. [Google Scholar] [CrossRef]
- Clemetson, K.J. Snaclecs (snake C-type lectins) that inhibit or activate platelets by binding to receptors. Toxicon 2010, 56, 1236–1246. [Google Scholar] [CrossRef]
- Calvete, J.J. Chapter 18: Brief history and molecular determinants of snake venom disintegrin evolution. In Toxins and Hemostasis; Kini, R.M., Clemetson, K.J., Markland, F.S., McLane, M.A., Morita, T., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 285–300. ISBN 978-90-481-9294-6. [Google Scholar]
- Rivas-Mercado, E.A.; Garza-Ocañas, L. Disintegrins obtained from snake venom and their pharmacological potential. Med. Univ. 2017, 19, 32–37. [Google Scholar] [CrossRef]
- Calvete, J.J.; Marcinkiewicz, C.; Monleón, D.; Esteve, V.; Celda, B.; Juárez, P.; Sanz, L. Snake venom disintegrins: Evolution of structure and function. Toxicon 2005, 45, 1063–1074. [Google Scholar] [CrossRef]
- Calvete, J.J.; Moreno-Murciano, M.P.; Theakston, R.D.G.; Kisiel, D.G.; Marcinkiewicz, C. Snake venom disintegrins: Novel dimeric disintegrins and structural diversification by disulphide bond engineering. Biochem. J. 2003, 372, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A. Structure-function studies and mechanism of action of snake venom L-Amino acid oxidases. Front. Pharmacol. 2020, 11, 110. [Google Scholar] [CrossRef] [PubMed]
- Izidoro, L.F.M.; Sobrinho, J.C.; Mendes, M.M.; Costa, T.R.; Grabner, A.N.; Rodrigues, V.M.; da Silva, S.L.; Zanchi, F.B.; Zuliani, J.P.; Fernandes, C.F.C.; et al. Snake venom L-amino acid oxidases: Trends in pharmacology and biochemistry. Biomed Res. Int. 2014, 2014, 196754. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W. A brief review of the scientific history of several lesser-known snake venom proteins: L-amino acid oxidases, hyaluronidases and phosphodiesterases. Toxicon 2013, 62, 75–82. [Google Scholar] [CrossRef]
- Tadokoro, T.; Modahl, C.M.; Maenaka, K.; Aoki-Shioi, N. Cysteine-rich secretory proteins (CRISPs) from venomous snakes: An overview of the functional diversity in A large and underappreciated superfamily. Toxins 2020, 12, 175. [Google Scholar] [CrossRef]
- Sunagar, K.; Johnson, W.E.; O’Brien, S.J.; Vasconcelos, V.; Antunes, A. Evolution of CRISPs associated with toxicoferan-reptilian venom and mammalian reproduction. Mol. Biol. Evol. 2012, 29, 1807–1822. [Google Scholar] [CrossRef]
- Ramazanova, A.S.; Starkov, V.G.; Osipov, A.V.; Ziganshin, R.H.; Filkin, S.Y.; Tsetlin, V.I.; Utkin, Y.N. Cysteine-rich venom proteins from the snakes of Viperinae subfamily—Molecular cloning and phylogenetic relationship. Toxicon 2009, 53, 162–168. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Matsunaga, Y.; Tokunaga, Y.; Obayashi, S.; Saito, M.; Morita, T. Snake venom vascular endothelial growth Factors (VEGF-Fs) exclusively vary their structures and functions among species. J. Biol. Chem. 2009, 284, 9885–9891. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, Y.; Yamazaki, Y.; Morita, T. Specific distribution of VEGF-F in Viperinae snake venoms: Isolation and characterization of a VGEF-F from the venom of Daboia russelli siamensis. Arch. Biochem. Biophys. 2005, 439, 241–247. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Takani, K.; Atoda, H.; Morita, T. Snake venom vascular endothelial growth factors (VEGFs) exhibit potent activity through their specific recognition of KDR (VEGF receptor 2). J. Biol. Chem. 2003, 278, 51985–51988. [Google Scholar] [CrossRef]
- Župunski, V.; Kordiš, D.; Gubenšek, F. Adaptive evolution in the snake venom Kunitz/BPTI protein family. FEBS Lett. 2003, 547, 131–136. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Dutta, S.; Kalita, B.; Jha, D.K.; Deb, P.; Mackessy, S.P. Structural and functional characterization of complex formation between two Kunitz-type serine protease inhibitors from Russell’s Viper venom. Biochimie 2016, 128–129, 138–147. [Google Scholar] [CrossRef]
- Mourão, C.B.F.; Schwartz, E.F. Protease inhibitors from marine venomous animals and their counterparts in terrestrial venomous animals. Mar. Drugs 2013, 11, 2069–2112. [Google Scholar] [CrossRef]
- Sunagar, K.; Fry, B.G.; Jackson, T.N.W.; Casewell, N.R.; Undheim, E.A.B.; Vidal, N.; Ali, S.A.; King, G.F.; Vasudevan, K.; Vasconcelos, V.; et al. Molecular evolution of vertebrate neurotrophins: Co-option of the highly conserved nerve growth factor gene into the advanced snake venom arsenalf. PLoS ONE 2013, 8, e81827. [Google Scholar] [CrossRef]
- Hogue-Angeletti, R.A.; Bradshaw, R.A. Chapter 8: Nerve growth factors in snake venoms. In Snake Venoms; Lee, C.-Y., Ed.; Springer: Berlin/Heidelberg, Germany, 1979; pp. 276–294. ISBN 978-3-642-66915-6. [Google Scholar]
- Trummal, K.; Tõnismägi, K.; Paalme, V.; Järvekülg, L.; Siigur, J.; Siigur, E. Molecular diversity of snake venom nerve growth factors. Toxicon 2011, 58, 363–368. [Google Scholar] [CrossRef]
- Dhananjaya, B.L.; D’Souza, C.J.M. The pharmacological role of nucleotidases in snake venoms. Cell Biochem. Funct. 2010, 28, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Sales, P.B.V.; Santoro, M.L. Nucleotidase and DNase activities in Brazilian snake venoms. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2008, 147, 85–95. [Google Scholar] [CrossRef]
- Uzair, B.; Khan, B.A.; Sharif, N.; Shabbir, F.; Menaa, F. Phosphodiesterases (PDEs) from Snake Venoms: Therapeutic Applications. Protein Pept. Lett. 2018, 25, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Ullah, K.; Ali, H.; Betzel, C.; Ur Rehman, S. The sequence and a three-dimensional structural analysis reveal substrate specificity among snake venom phosphodiesterases. Toxins 2019, 11, 625. [Google Scholar] [CrossRef] [PubMed]
- Girish, K.S.; Jagadeesha, D.K.; Rajeev, K.B.; Kemparaju, K. Snake venom hyaluronidase: An evidence for isoforms and extracellular matrix degradation. Mol. Cell. Biochem. 2002, 240, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Kemparaju, K.; Girish, K.S.; Nagaraju, S. Chapter 11: Hyaluronidases, a neglected class of glycosidases from snake venom. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; Taylor & Francis: Boca Raton, FL, USA, 2010; pp. 237–258. ISBN 978-0849391651. [Google Scholar]
- Ullah, A.; Masood, R. The sequence and three-dimensional structure characterization of snake venom phospholipases B. Front. Mol. Biosci. 2020, 7, 175. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, H. Chapter 8: Snake venom protease inhibitors: Enhanced identification, expanding biological function, and promising future. In Snake Venoms; Inagaki, H., Vogel, C.-W., Mukherjee, A.K., Rahmy, T.R., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 161–186. ISBN 978-94-007-6409-5. [Google Scholar]
- Mashiko, H.; Takahashi, H. Cysteine proteinase inhibitors in elapid and hydrophiid snake venoms. Toxicon 2002, 40, 1275–1281. [Google Scholar] [CrossRef]
- Evans, H.J.; Barrett, A.J. A cystatin-like cysteine proteinase inhibitor from venom of the African puff adder (Bitis arietans). Biochem. J. 1987, 246, 795–797. [Google Scholar] [CrossRef]
- Pawlak, J.; Manjunatha Kini, R. Snake venom glutaminyl cyclase. Toxicon 2006, 48, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-M.; Huang, K.-F.; Tsai, I.-H. Snake venom glutaminyl cyclases: Purification, cloning, kinetic study, recombinant expression, and comparison with the human enzyme. Toxicon 2014, 86, 40–50. [Google Scholar] [CrossRef]
- Vaiyapuri, S.; Wagstaff, S.C.; Watson, K.A.; Harrison, R.A.; Gibbins, J.M.; Hutchinson, E.G. Purification and functional characterisation of rhiminopeptidase A, a novel aminopeptidase from the venom of Bitis gabonica rhinoceros. PLoS Negl. Trop. Dis. 2010, 4, e796. [Google Scholar] [CrossRef]
- Faiz, M.A.; Falkous, G.; Harris, J.B.; Mantle, D. Comparison of protease and related enzyme activities in snake venoms. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1996, 113, 199–204. [Google Scholar] [CrossRef]
- Ogawa, Y.; Murayama, N.; Fujita, Y.; Yanoshita, R. Characterization and cDNA cloning of aminopeptidase A from the venom of Gloydius blomhoffi brevicaudus. Toxicon 2007, 49, 1172–1181. [Google Scholar] [CrossRef]
- Huang, K.-F.; Chiou, S.-H.; Ko, T.-P.; Wang, A.H.-J. Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues. Eur. J. Biochem. 2002, 269, 3047–3056. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, S.C.; Favreau, P.; Cheneval, O.; Laing, G.D.; Wilkinson, M.C.; Miller, R.L.; Stöcklin, R.; Harrison, R.A. Molecular characterisation of endogenous snake venom metalloproteinase inhibitors. Biochem. Biophys. Res. Commun. 2008, 365, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Sciani, J.M.; Pimenta, D.C. The modular nature of bradykinin-potentiating peptides isolated from snake venoms. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 45. [Google Scholar] [CrossRef]
- Péterfi, O.; Boda, F.; Szabó, Z.; Ferencz, E.; Bába, L. Hypotensive snake venom components-a mini-review. Molecules 2019, 24, 2778. [Google Scholar] [CrossRef]
- Babenko, V.V.; Ziganshin, R.H.; Weise, C.; Dyachenko, I.; Shaykhutdinova, E.; Murashev, A.N.; Zhmak, M.; Starkov, V.; Hoang, A.N.; Tsetlin, V.; et al. Novel bradykinin-potentiating peptides and three-finger toxins from viper venom: Combined NGS venom gland transcriptomics and quantitative venom proteomics of the Azemiops feae viper. Biomedicines 2020, 8, 249. [Google Scholar] [CrossRef] [PubMed]
- Vink, S.; Jin, A.H.; Poth, K.J.; Head, G.A.; Alewood, P.F. Natriuretic peptide drug leads from snake venom. Toxicon 2012, 59, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Mosbah, A.; Marrakchi, N.; Mansuelle, P.; Kouidhi, S.; Giralt, E.; El Ayeb, M.; Herbette, G.; Cherif, A.; Gigmes, D.; Darbon, H.; et al. Lebetin peptides, a new class of potent platelet aggregation inhibitors: Chemical synthesis, biological Activity and NMR spectroscopic study. Int. J. Pept. Res. Ther. 2020, 26, 21–31. [Google Scholar] [CrossRef]
- Bantscheff, M.; Schirle, M.; Sweetman, G.; Rick, J.; Kuster, B. Quantitative mass spectrometry in proteomics: A critical review. Anal. Bioanal. Chem. 2007, 389, 1017–1031. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, G.M.; Chalkley, R.J. The effect of using an inappropriate protein database for proteomic data analysis. PLoS ONE 2011, 6, e20873. [Google Scholar] [CrossRef]
- Zhu, W.; Smith, J.W.; Huang, C.-M. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010, 2010, 840518. [Google Scholar] [CrossRef]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Tashima, A.K.; Zelanis, A. Snake venom peptidomics. In Venom Genomics and Proteomics; Gopalakrishnakone, P., Calvete, J.J., Eds.; Springer: Dordrecht, The Netherlands, 2016; pp. 317–331. ISBN 978-94-007-6415-6. [Google Scholar]
- Hargreaves, A.D.; Swain, M.T.; Logan, D.W.; Mulley, J.F. Testing the Toxicofera: Comparative transcriptomics casts doubt on the single, early evolution of the reptile venom system. Toxicon 2014, 92, 140–156. [Google Scholar] [CrossRef] [PubMed]
- Barlow, A.; Wüster, W.; Kelly, C.M.R.; Branch, W.R.; Phelps, T.; Tolley, K.A. Ancient habitat shifts and organismal diversification are decoupled in the African viper genus Bitis (Serpentes: Viperidae). J. Biogeogr. 2019, 46, 1234–1248. [Google Scholar] [CrossRef]
- Laing, G.; Renjifo, J.; Ruiz, F.; Harrison, R.; Nasidi, A.; Gutierrez, J.-M.; Rowley, P.; Warrell, D.; Theakston, R. A new Pan African polyspecific antivenom developed in response to the antivenom crisis in Africa. Toxicon 2003, 42, 35–41. [Google Scholar] [CrossRef]
- Segura, A.; Villalta, M.; Herrera, M.; León, G.; Harrison, R.; Durfa, N.; Nasidi, A.; Calvete, J.J.; Theakston, R.D.G.; Warrell, D.A.; et al. Preclinical assessment of the efficacy of a new antivenom (EchiTAb-Plus-ICP) for the treatment of viper envenoming in sub-Saharan Africa. Toxicon 2010, 55, 369–374. [Google Scholar] [CrossRef]
- Youngman, N.J.; Debono, J.; Dobson, J.S.; Zdenek, C.N.; Harris, R.J.; Op den Brouw, B.; Coimbra, F.C.P.; Naude, A.; Coster, K.; Sundman, E.; et al. Venomous landmines: Clinical implications of extreme coagulotoxic diversification and differential neutralization by antivenom of venoms within the viperid snake genus bitis. Toxins 2019, 11, 422. [Google Scholar] [CrossRef]
- Martínez-Freiría, F. (Biogeographic History of Cerastes Vipers, CIBIO/InBIO, Research Center in Biodiversity and Genetic Resources of the University of Porto, Vairão, Portugal). Personal communication, 2020.
- Barros, M.I.O. Reconstructing the Evolutionary History of Desert-Adapted Cerastes Vipers in North Africa and the Arabian Peninsula. Master’s Thesis, University of Porto, Potro, Portugal, 2019. [Google Scholar]
- Freitas, I.; Ursenbacher, S.; Mebert, K.; Zinenko, O.; Schweiger, S.; Wüster, W.; Brito, J.C.; Crnobrnja-Isailović, J.; Halpern, B.; Fahd, S.; et al. Evaluating taxonomic inflation: Towards evidence-based species delimitation in Eurasian vipers (Serpentes: Viperinae). Amphib. Reptil. 2020, 1–27. [Google Scholar] [CrossRef]
- Kalita, B.; Mackessy, S.P.; Mukherjee, A.K. Proteomic analysis reveals geographic variation in venom composition of Russell’s Viper in the Indian subcontinent: Implications for clinical manifestations post-envenomation and antivenom treatment. Expert Rev. Proteom. 2018, 15, 837–849. [Google Scholar] [CrossRef]
- Chauhan, V.; Thakur, S. The North-South divide in snake bite envenomation in India. J. Emerg. Trauma Shock 2016, 9, 151–154. [Google Scholar] [CrossRef]
- Makarova, Y.V.; Kryukova, E.V.; Shelukhina, I.V.; Lebedev, D.S.; Andreeva, T.V.; Ryazantsev, D.Y.; Balandin, S.V.; Ovchinnikova, T.V.; Tsetlin, V.I.; Utkin, Y.N. The first recombinant viper three-finger toxins: Inhibition of muscle and neuronal nicotinic acetylcholine receptors. Dokl. Biochem. Biophys. 2018, 479, 127–130. [Google Scholar] [CrossRef]
- Maity, G.; Mandal, S.; Chatterjee, A.; Bhattacharyya, D. Purification and characterization of a low molecular weight multifunctional cytotoxic phospholipase A2 from Russell’s viper venom. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007, 845, 232–243. [Google Scholar] [CrossRef]
- Utkin, Y.N. Last decade update for three-finger toxins: Newly emerging structures and biological activities. World J. Biol. Chem. 2019, 10, 17–27. [Google Scholar] [CrossRef]
- Doley, R.; Pahari, S.; Mackessy, S.P.; Kini, R.M. Accelerated exchange of exon segments in Viperid three-finger toxin genes (Sistrurus catenatus edwardsii; Desert Massasauga). BMC Evol. Biol. 2008, 8, 196. [Google Scholar] [CrossRef]
- Pawlak, J.; Mackessy, S.P.; Fry, B.G.; Bhatia, M.; Mourier, G.; Fruchart-Gaillard, C.; Servent, D.; Ménez, R.; Stura, E.; Ménez, A.; et al. Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (Mangrove Catsnake) with bird-specific activity. J. Biol. Chem. 2006, 281, 29030–29041. [Google Scholar] [CrossRef]
- INEICH, I.; BONNET, X.; SHINE, R.; SHINE, T.; BRISCHOUX, F.; LEBRETON, M.; CHIRIO, L. What, if anything, is a ‘typical’ viper? Biological attributes of basal viperid snakes (genus Causus Wagler, 1830). Biol. J. Linn. Soc. 2006, 89, 575–588. [Google Scholar] [CrossRef]
- Tsai, M.C.; Lee, C.Y.; Bdolah, A. Mode of neuromuscular blocking action of a toxic phospholipase A2 from Pseudocerastes fieldi (Field’s horned viper) snake venom. Toxicon 1983, 21, 527–534. [Google Scholar] [CrossRef]
- Bdolah, A. Comparison of venoms from two subspecies of the false horned viper (Pseudocerastes persicus). Toxicon 1986, 24, 726–729. [Google Scholar] [CrossRef]
- Ainsworth, S.; Menzies, S.K.; Casewell, N.R.; Harrison, R.A. An analysis of preclinical efficacy testing of antivenoms for sub-Saharan Africa: Inadequate independent scrutiny and poor-quality reporting are barriers to improving snakebite treatment and management. PLoS Negl. Trop. Dis. 2020, 14, e0008579. [Google Scholar] [CrossRef] [PubMed]
- Alangode, A.; Rajan, K.; Nair, B.G. Snake antivenom: Challenges and alternate approaches. Biochem. Pharmacol. 2020, 181, 114135. [Google Scholar] [CrossRef]
- Jenkins, T.P.; Fryer, T.; Dehli, R.I.; Jürgensen, J.A.; Fuglsang-Madsen, A.; Føns, S.; Laustsen, A.H. Toxin neutralization using alternative binding proteins. Toxins 2019, 11, 53. [Google Scholar] [CrossRef]
- Xie, C.; Albulescu, L.-O.; Bittenbinder, M.A.; Somsen, G.W.; Vonk, F.J.; Casewell, N.R.; Kool, J. Neutralizing effects of small molecule inhibitors and metal chelators on coagulopathic viperinae snake venom toxins. Biomedicines 2020, 8, 297. [Google Scholar] [CrossRef]
- Escalante, T.; Franceschi, A.; Rucavado, A.; Gutiérrez, J.M. Effectiveness of batimastat, a synthetic inhibitor of matrix metalloproteinases, in neutralizing local tissue damage induced by BaP1, a hemorrhagic metalloproteinase from the venom of the snake Bothrops asper. Biochem. Pharmacol. 2000, 60, 269–274. [Google Scholar] [CrossRef]
- Lewin, M.; Samuel, S.; Merkel, J.; Bickler, P. Varespladib (LY315920) appears to be a potent, broad-spectrum, inhibitor of snake venom phospholipase A2 and a possible pre-referral treatment for envenomation. Toxins 2016, 8, 248. [Google Scholar] [CrossRef]
- Kini, R.M.; Sidhu, S.S.; Laustsen, A.H. Biosynthetic oligoclonal antivenom (BOA) for snakebite and next-generation treatments for snakebite victims. Toxins 2018, 10, 534. [Google Scholar] [CrossRef]
- Jackson, T.N.W.; Jouanne, H.; Vidal, N. Snake venom in context: Neglected clades and concepts. Front. Ecol. Evol. 2019, 7, 60. [Google Scholar] [CrossRef]
- Menegon, M.; Loader, S.P.; Marsden, S.J.; Branch, W.R.; Davenport, T.R.B.; Ursenbacher, S. The genus Atheris (Serpentes: Viperidae) in East Africa: Phylogeny and the role of rifting and climate in shaping the current pattern of species diversity. Mol. Phylogenet. Evol. 2014, 79, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, R.S.; Pook, C.E.; Malhotra, A. Phylogeography of the Russell’s viper (Daboia russelii) complex in relation to variation in the colour pattern and symptoms of envenoming. Herpetol. J. 2007, 17, 209–218. [Google Scholar]
| Majors | Secondaries | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genus Species | No. of Compositions | Examined Subspecies | Bottom-Up | Top-Down | Edman Degradation | Q—Snake Venomics | Q—Two-Step Workflows | Q—in-Solution Shotgun | No Quantification | ■svMP | ■PLA2 | ■svSP | ■CTL | ■DI | ■LAAO | ■CRISP | ■VEGF | ■KUN | ■Minor families | ■CYS | ■Rare Families | ■svMP-i | ■NP | ■Other Peptides | References |
| Atheris | 3 | ||||||||||||||||||||||||
| A. chlorechis | 1 | ■ | ● | 🗸 | 🗸 | - | - | 🗸 | - | - | - | - | - | - | - | - | - | 🗸 | [69] | ||||||
| A. nitschei | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | - | 🗸 | - | - | - | - | - | - | - | - | - | 🗸 | [69] | ||||||
| A. squamigera | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | - | 🗸 | - | - | - | - | - | - | - | - | - | 🗸 | [69] | ||||||
| Bitis | 9 | ||||||||||||||||||||||||
| B. arietans | 4 | ■ | ■ | ● | ● | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | [60,75,76,77] | |||
| B. caudalis | 1 | ■ | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | - | - | - | - | - | 🗸 | [78] | |||||
| B. gabonica | 2 | ■ | ■ | ● | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | - | - | 🗸 | 🗸 | [79,80] | ||||
| B. nasicornis | 1 | ■ | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | - | 🗸 | - | - | - | 🗸 | [78] | |||||
| B. rhinoceros | 1 | ■ | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | - | 🗸 | - | - | 🗸 | 🗸 | [78] | |||||
| Causus | 2 | ||||||||||||||||||||||||
| C. lichtensteinii | 1 | ■ | ● | 🗸 | - | 🗸 | - | - | 🗸 | 🗸 | - | - | - | - | - | - | - | - | [81] | ||||||
| C. rhombeatus | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | - | - | 🗸 | 🗸 | - | - | - | - | - | - | - | - | [81] | ||||||
| Cerastes | 6 | ||||||||||||||||||||||||
| C. cerastes | 5 | ■ | ■ | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | - | - | - | 🗸 | - | 🗸 | [76,82,83,84] | ||||
| C. vipera | 1 | ■ | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | - | - | - | - | 🗸 | - | - | [82] | |||||
| Daboia | 24 | ||||||||||||||||||||||||
| D. mauritanica | 2 | ■ | ■ | ● | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | 🗸 | 🗸 | - | - | - | - | 🗸 | - | [85,86] | ||||
| D. palaestinae | 1 | ■ | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | 🗸 | - | 🗸 | - | - | - | - | - | [87] | |||||
| D. russelii | 12 | ■ | ● | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | 🗸 | 🗸 | 🗸 | [88,89,90,91,92,93,94,95] | |||||
| D. siamensis | 9 | ■ | ■ | ● | ● | ● | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | - | - | - | [68,96,97,98,99,100] | ||
| Echis | 16 | ||||||||||||||||||||||||
| E. carinatus | 11 | (2) | ■ | ■ | ■ | ● | ● | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | 🗸 | 🗸 | 🗸 | [76,101,102,103,104,105] | |
| E. coloratus | 1 | ■ | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | - | - | - | 🗸 | - | - | [76] | |||||
| E. leucogaster | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | - | - | - | - | 🗸 | - | - | [106] | ||||||
| E. ocellatus | 2 | ■ | ■ | ● | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | - | - | - | 🗸 | - | 🗸 | [76,107] | ||||
| E. pyramidum | 1 | ■ | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | - | - | - | 🗸 | - | - | [76] | |||||
| Eristicophis | 1 | ||||||||||||||||||||||||
| E. macmahoni | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | - | - | - | 🗸 | - | [70] | ||||||
| Macrovipera | 5 | ||||||||||||||||||||||||
| M. lebetina | 5 | (2) | ■ | ■ | ● | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | 🗸 | 🗸 | - | [82,85,108,109,110] | |||
| Montivipera | 3 | ||||||||||||||||||||||||
| M. bulgardaghica | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | - | - | 🗸 | - | 🗸 | [111] | ||||||
| M. raddei | 2 | ■ | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | - | - | 🗸 | 🗸 | 🗸 | [108,111] | |||||
| Pseudocerastes | 2 | ||||||||||||||||||||||||
| P. fieldi | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | - | - | - | [70] | ||||||
| P. persicus | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | - | - | - | - | [70] | ||||||
| Vipera | 18 | ||||||||||||||||||||||||
| V. ammodytes | 5 | (3) | ■ | ■ | ■ | ● | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | 🗸 | 🗸 | 🗸 | [112,113,114,115] | ||
| V. anatolica | 2 | (2) | ■ | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | 🗸 | - | 🗸 | 🗸 | 🗸 | 🗸 | [56,116] | ||||
| V. aspis | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | 🗸 | - | 🗸 | [117] | ||||||
| V. berus | 3 | ■ | ■ | ● | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | 🗸 | 🗸 | 🗸 | [118,119,120] | ||||
| V. kaznakovi | 2 | ■ | ■ | ● | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | - | 🗸 | 🗸 | 🗸 | 🗸 | [28,121] | ||||
| V. nikolskii | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | - | 🗸 | - | [121] | ||||||
| V. orlovi | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | - | 🗸 | - | [121] | ||||||
| V. renardi | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | - | - | - | [121] | ||||||
| V. transcaucasiana | 1 | ■ | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | - | 🗸 | 🗸 | 🗸 | - | 🗸 | - | - | 🗸 | 🗸 | 🗸 | [112] | |||||
| V. ursinii | 1 | ■ | ● | 🗸 | 🗸 | 🗸 | 🗸 | - | - | 🗸 | - | 🗸 | 🗸 | - | 🗸 | - | - | - | [122] | ||||||
| Abbreviation | Snake Venom Toxin Family | Enzyme Class | Monomeric Size in kDa | No. of S-S | Observed in No. of Studies | References |
|---|---|---|---|---|---|---|
| Major toxin families | ||||||
| svMP | snake venom metalloproteinase | EC 3.4.24.- | 20–100 | 4–18 | 89 | [135,136,137,138,139] |
| PLA2 | phospholipase A2 | EC 3.1.1.4 | 13–15 | 6–8 | 87 | [140,141,142,143,144] |
| svSP | snake venom serine protease | EC 3.4.21.- | 22–67 | 6 | 86 | [145,146,147,148] |
| CTL incl. Snaclec | C-type lectin-related protein | - | 13–15 | 3 | 80 | [149,150,151] |
| Secondary toxin families | ||||||
| DI | disintegrin | - | 4–10 | 4–8 | 63 | [152,153,154,155] |
| LAAO | l-amino acid oxidase | EC 1.4.3.2 | 50–70 | 2 | 68 | [156,157,158] |
| CRISP | cysteine-rich secretory protein | - | 20–33 | 8 | 63 | [159,160,161] |
| VEGF | vascular endothelial growth factors F | - | 10–15 | 5 | 48 | [162,163,164] |
| KUN | Kunitz-type inhibitor | - | 6–7 | 3 | 42 | [20,165,166,167] |
| Minor toxin families | ||||||
| NGF | nerve growth factor | - | 12–37 | 3 | 41 | [168,169,170] |
| 5N | 5′-nucleotidase | EC 3.1.3.5 | 73–100 | 4 | 34 | [171,172] |
| PDE | phosphodiesterase | EC 3.1.4.1 | 90–140 | 16 | 33 | [173,174] |
| HYAL | hyaluronidase | EC 3.2.1.35 | 33–110 | 5 | 17 | [158,175,176] |
| PLB | phospholipase B | EC 3.1.1.5 | ~55 | 2 | 21 | [177] |
| CYS | cystatin | - | 12–15 | 2 | 8 | [178,179,180] |
| Rare families (selection) | ||||||
| QC | glutaminyl cyclotransferase | EC 2.3.2.5 | 33–40 | 1 | 17 | [181,182] |
| AP | Aminopeptidase | EC 3.4.11.- | 100–150 | * | 17 | [183,184,185] |
| Peptides | ||||||
| svMP-i | svMP-inhibitor | - | 0.3 | 0 | 22 | [134,186,187] |
| BPP | bradykinin potentiating peptide | - | 0.5–1.5 | 0 | 24 | [133,188,189,190] |
| other peptides | incl. further natriuretic peptides | - | 1–10 | - | 24 | [133,134,189,191,192] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damm, M.; Hempel, B.-F.; Süssmuth, R.D. Old World Vipers—A Review about Snake Venom Proteomics of Viperinae and Their Variations. Toxins 2021, 13, 427. https://doi.org/10.3390/toxins13060427
Damm M, Hempel B-F, Süssmuth RD. Old World Vipers—A Review about Snake Venom Proteomics of Viperinae and Their Variations. Toxins. 2021; 13(6):427. https://doi.org/10.3390/toxins13060427
Chicago/Turabian StyleDamm, Maik, Benjamin-Florian Hempel, and Roderich D. Süssmuth. 2021. "Old World Vipers—A Review about Snake Venom Proteomics of Viperinae and Their Variations" Toxins 13, no. 6: 427. https://doi.org/10.3390/toxins13060427
APA StyleDamm, M., Hempel, B.-F., & Süssmuth, R. D. (2021). Old World Vipers—A Review about Snake Venom Proteomics of Viperinae and Their Variations. Toxins, 13(6), 427. https://doi.org/10.3390/toxins13060427






