Abstract
Envenomation resulted from sea snake bite is a highly lethal health hazard in Southeast Asia. Although commonly caused by sea snakes of Hydrophiinae, each species is evolutionarily distinct and thus, unveiling the toxin gene diversity within individual species is important. Applying next-generation sequencing, this study investigated the venom-gland transcriptome of Hydrophis curtus (spine-bellied sea snake) from Penang, West Malaysia. The transcriptome was de novo assembled, followed by gene annotation and sequence analyses. Transcripts with toxin annotation were only 96 in number but highly expressed, constituting 48.18% of total FPKM in the overall transcriptome. Of the 21 toxin families, three-finger toxins (3FTX) were the most abundantly expressed and functionally diverse, followed by phospholipases A2. Lh_FTX001 (short neurotoxin) and Lh_FTX013 (long neurotoxin) were the most dominant 3FTXs expressed, consistent with the pathophysiology of envenomation. Lh_FTX001 and Lh_FTX013 were variable in amino acid compositions and predicted epitopes, while Lh_FTX001 showed high sequence similarity with the short neurotoxin from Hydrophis schistosus, supporting cross-neutralization effect of Sea Snake Antivenom. Other toxins of low gene expression, for example, snake venom metalloproteinases and L-amino acid oxidases not commonly studied in sea snake venom were also identified, enriching the knowledgebase of sea snake toxins for future study.
Keywords:
snakebite; envenomation; Lapemis hardwickii; Hydrophis hardwickii; three-finger toxin; alpha-neurotoxin Key Contribution:
The study reported the de novo venom-gland transcriptomics of H. curtus, a medically important sea snake species from the Peninsula of Malaysia and the diversity of venom genes in this species. The findings enriched the toxin database of sea snake and shed light on the medical importance of the venom.
1. Introduction
Snakebite envenomation is a World Health Organisation (WHO)-classified neglected tropical disease that heavily affects many impoverished populations in the tropics and subtropics [1]. Each year, it causes 81,000 to 138,000 deaths across the world and approximately three times as many permanent disabilities and psychological trauma in those who survive [2,3]. Snakebite cases are mostly reported from inland areas where agricultural activities are predominant, whereas sea snake bite remains an under-estimated, fatal occupational hazard to fishermen and coastal communities [4,5]. In recent years, the risk of sea snake bite increases due to various environmental and anthropogenic factors, for example, climate change (global warming) and economic practices such as sea snake hunting and trading, which are not uncommon in Asia [6,7].
Clinically, envenomation caused by sea snake bite is highly lethal, with a fatality rate between 3.2% and 30% [8]. The victims typically develop generalized neuromuscular paralysis, myotoxicity and complications such as acute kidney injury due to rhabdomyolysis [9,10]. The majority of studies on sea snake envenomation and pathophysiology were reported since early 1950s for the beaked sea snake (Hydrophis schistosus or Enhydrina schistosa prior to taxonomic revision [11]) in the western coast of Peninsular Malaya, ostensibly due to its common occurrence and frequent bites in the waters. More recent studies reported the venom proteomes of a few sea snake species, including the beaked sea (H. schistosus) and its congener species Hydrophis cyanocinctus, Hydrophis curtus and Hydrophis (Pelamis) platura [12,13,14,15,16], which represent the viviparous true sea snakes under the Hydrophiinae subfamily. The hydrophiids are relatively young radiations (approximately 3–5-million-year-old) comprising at least 16 genera, with the genera Aipysurus and Hydrophis being the two major clades [17,18]. Unlike Aipysurus which has a relatively stable taxonomic history and is mainly restricted in the Australo-Papuan region, the Hydrophis complex is a far more diverse monophyletic clade that consists of close to 50 species distributed in the waters of Indo-Malayan Archipelago, Indonesia, Australia and South China Sea. Each species of Hydrophis is evolutionarily distinct and thus, further genetic analysis is important to unveil the diversity and novelty of toxin genes within individual species.
In the current study, we investigated the venom gene profile of H. curtus, a medically and commercially important sea snake species in Southeast Asia through de novo venom-gland transcriptomics, applying next-generation sequencing technique. Hydrophis curtus (Synonym: Hydrophis hardwickii), commonly known as the spine-bellied sea snake, distributes widely from the Persian Gulf to the Indian coastline, Myanmar, Thailand, Straits of Malacca, Strait of Taiwan, South China Sea, the Philippines, Indonesia, Papua New Guinea and northern as well as eastern Australia [19]. In Southeast Asia, the increased human contact with sea snakes from various anthropogenic activities poses a threat to a wider community of people besides fishermen [20]. In this part of the world, sea snakes are a significant part of the global wildlife trade where they are captured, bred and harvested for live snake parts used in the production of accessories and for food (sea snake meat as an exotic delicacy), as well as for medicinal use (tonic soup, snake wine, gall bladder as traditional medicament) [7]. Admittedly, in the Gulf of Thailand, H. curtus is commonly implicated in the catch and trade industry of wild snakes and reported 80 tonnes of venomous sea snakes are harvested each year in the region. The economic benefit of sea snake trade to the fishers and traders clearly outweighs the risk of envenomation, while the snake population and the ecosystem are being jeopardized. To address the medical and ecological implications of sea snake toxins, the present study set to investigate the venom-gland transcriptomic profile of H. curtus, through which the toxin gene diversity of this species can be better understood and the existing venom database of sea snakes can be further enriched.
2. Results and Discussion
2.1. Sequencing and De Novo Transcriptome Assembly
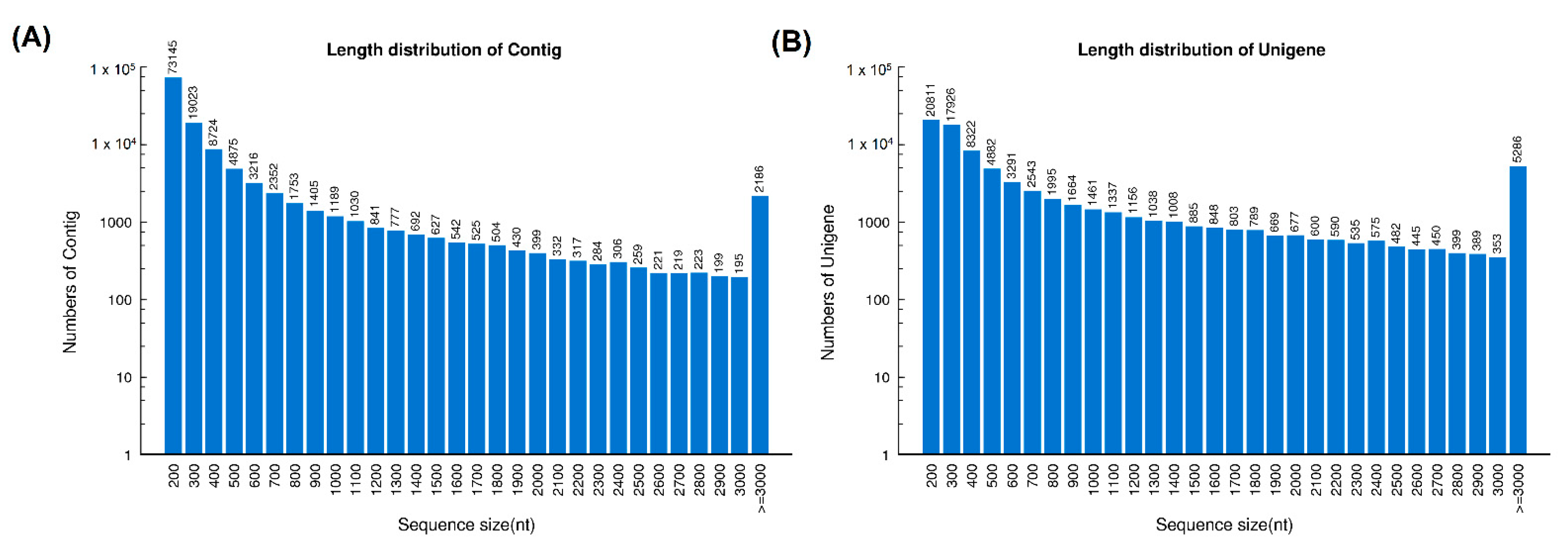
Sequencing of the cDNA libraries of H. curtus venom-gland tissue yielded a total of 54,140,326 clean reads. De novo assembly of the reads resulted in 126,790 contigs (N50 = 921), which were further clustered and streamlined into 82,209 unigenes (N50 = 2073). Of these, a total of 70,564 transcripts were identified at FPKM ≥ 1, a cut-off for gene expression of the current study (Table 1). The length distributions of contigs and unigenes from the de novo assembly of the venom-gland transcriptome were shown in Figure 1.

Table 1.
Overview of the output statistics. The sequencing and assembly quality of the venom gland transcriptome of Hydrophis curtus.
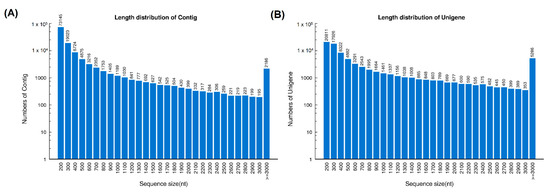
Figure 1.
Length distribution of contigs (A) and unigenes (B) from the de novo assembly of H. curtus venom-gland transcriptome.
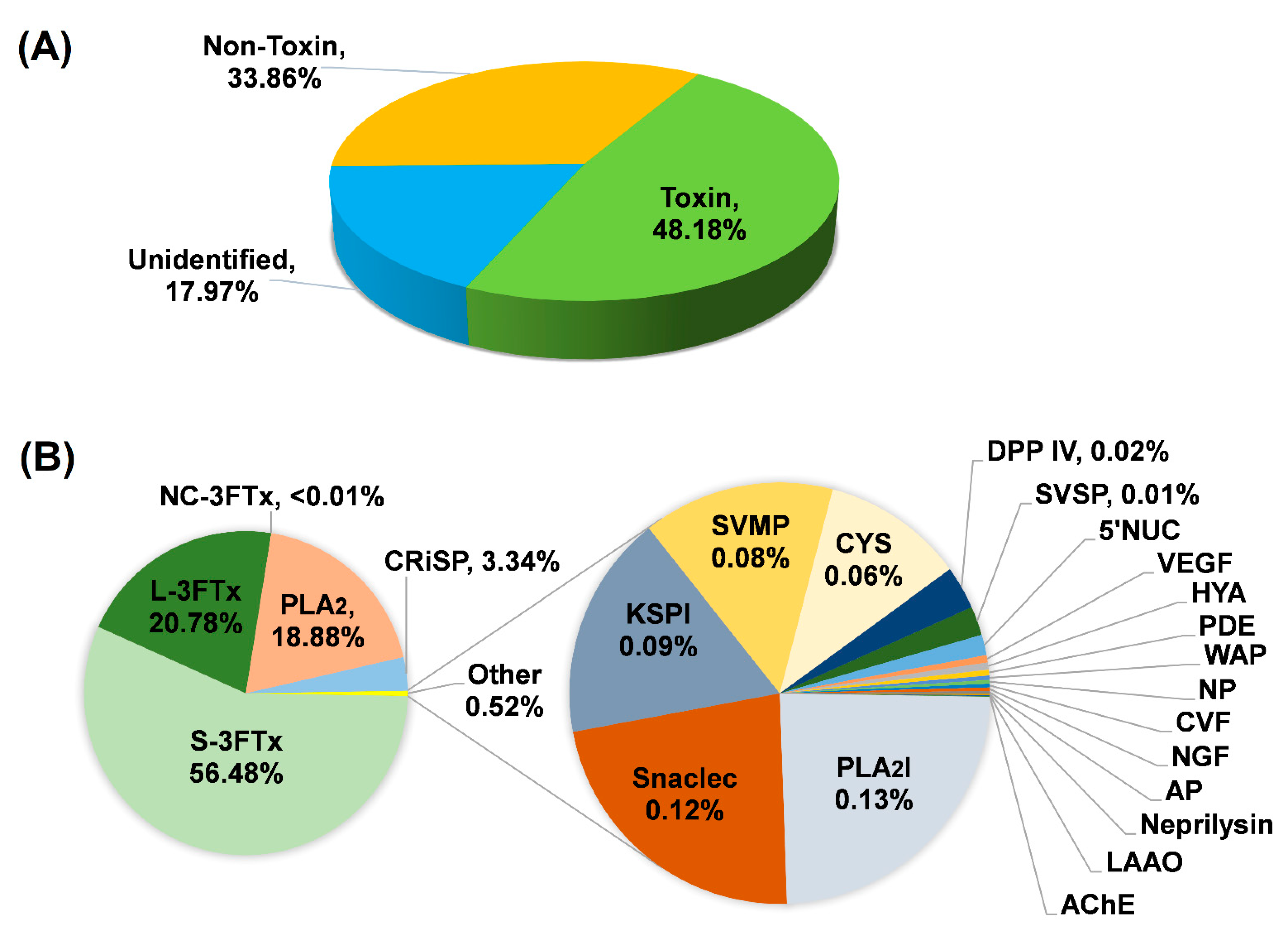
Based on BLASTx search, the transcripts were assigned into three categories: (a) “Toxin”; (b) “Non-toxin”; and (c) “Unidentified” (Figure 2A; Table 1). Transcripts in the “Toxin” category encoded known and putative snake toxins; these constituted the venom-gland transcriptome by a total FPKM of 48.18%. The remaining portion of the transcriptome was shared between “Non-toxin” transcripts, which represent cellular or house-keeping genes (33.86% of the total FPKM) and transcripts with no identifiable hits from the BLASTx search (17.97% of total FPKM). The venom-gland transcriptome of H. curtus was dominated (virtually 50%) by transcripts with toxin annotation, reflective of the specialized toxin-secreting function of the gland tissue. The high expression of “genes for toxins” is comparable to previous findings in other terrestrial elapid snakes, including cobras (Naja spp.) [21,22], king cobra (Ophiophagus hannah) [23] and Micrurus spp. (American coral snakes) [24,25], where toxin transcripts accounted for more than 40% of the venom-gland transcriptomes.
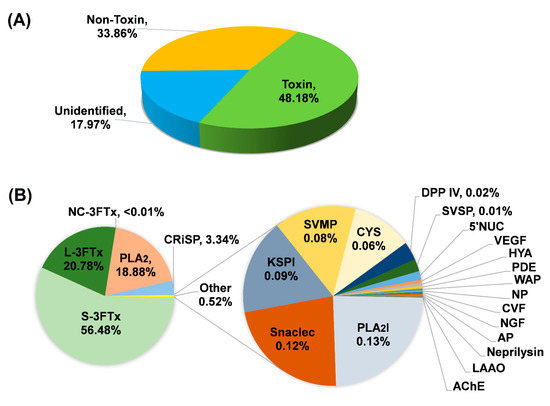
Figure 2.
De novo transcriptome of Hydrophis curtus venom glands: (A) Overview profile of all-transcript expression. (B) Profiling of toxin transcripts by gene families. Abbreviations: S-3FTx, short three-finger toxin; L-3FTx, long three-finger toxin; NC-3FTx, non-conventional three-finger toxin; PLA2, phospholipase A2; CRiSP, cysteine-rich secretory protein; PLA2I, phospholipase A2 inhibitor; Snaclec, snake venom C-type lectin/lectin-like protein; KSPI, Kunitz-type serine protease inhibitor; SVMP, snake venom metalloproteinase; CYS, cystatin; DPP IV, dipeptidylpeptidase IV; SVSP, snake venom serine protease; 5′NUC, 5′ nucleotidase; VEGF, vascular endothelial growth factor; HYA, hyaluronidase; PDE, phosphodiesterase; WAP, waprin; NP, natriuretic peptide; CVF, cobra venom factor; NGF, nerve growth factor; AP, aminopeptidase; LAAO, L-amino acid oxidase; and AChE, acetylcholinesterase. The expression of 5′NUC, VEGF, HYA, PDE, WAP, NP, CVF, NGF, AP, neprilysin, LAAO and AChE were each <0.01% of total toxin FPKM.
2.2. Toxin Gene Expression Profile
A total of 70,564 transcripts were expressed in the H. curtus venom-gland, while only 96 were classified under the “Toxin” category. The transcripts showed distinct, non-redundant sequences and were clustered by sequence similarity into 21 families of toxin genes (Figure 2B). Despite their extremely small number (96 out of 70,564 or 0.14% by total transcript count), these toxin transcripts were highly expressed, together contributing to an exceptionally high redundancy value of 4847.54 FPKM/transcript, which is in sharp contrast to the remaining genes with low expressions (non-toxin: 13.16 FPKM/transcript; unidentified: 3.89 FPKM/transcript, respectively) (Table 1). The high redundancy of toxin genes probably reflects multigene duplication in snake venom evolution, in which functional diversity of toxins with increasing prey-specificity is generated. By natural selection, this is essential for the evolving snakes to become more adapted to survive environmental perturbations and to occupy new niches for survival [26,27].
2.3. Profiling of Toxin Transcripts
The de novo transcriptome of H. curtus venom-gland also revealed high sequence similarity of all toxin transcripts to sequences from various elapid species (Table 2). Amongst the 96 toxin genes identified, 45 have full-length sequences with ≥90% amino acid coverage (Table 3). These proteins with full-length sequences included previously uncharacterized families of CRISP, CTL, KSPI, SVMP, cystatin, SVSP, 5NT, VEGF, hyaluronidase, waprin, CVF and neprilysin from sea snake venoms, in addition to the well-established 3FTx and PLA2 proteins. The de novo sequences of these toxins, which are unique to H. curtus are now available in the public repository database and provided in Supplementary Table S1 for deeper insights in the diversity of sea snake toxins.

Table 2.
Overview of toxin genes in venom-gland transcriptome of Malaysian Hydrophis curtus.

Table 3.
Full-length toxin transcripts derived from the venom-gland transcriptome of Malaysian Hydrophis curtus.
Of the 21 toxin gene families, 3FTxs are the most diversified and abundantly expressed (18 transcripts, 77.26% of total toxin FPKM), supporting that 3FTXs play a major role in the predatory function of H. curtus venom. More specifically, it was the alpha-neurotoxins that formed the bulk of 3FTx in the venom (~77% of total toxin FPKM) (Table 2). The diversely expressed 3FTx genes were further categorized into long-chain, short-chain and non-conventional groups (Table 2) and elaborated based on their functional attributes in the context of envenomation. PLA2 proteins constituted the second most abundantly expressed toxin genes (18.88% of total toxin FPKM), followed by CRISP (3.34%), PLA2 inhibitor (0.13%), CTL (0.12%), KSPI (0.09%), SVMP (0.087%), cystatin (0.06%), dipeptidyl peptidase (0.02%), SVSP (0.01%) and miscellaneous (5NT, VEGF, hyaluronidase, PDE, waprin, NP, CVF, NGF, aminopeptidase, neprilysin, LAAO and AChE, at <0.01% of total toxin FPKM, respectively). The transcriptomic profile showed a greater toxin diversity compared to the proteomic profile of H. curtus venom from the same geographical source (Penang) as reported previously [13], in which a few minor families (SVSP, NP, CTL, peptidases, neprilysin, AChE, waprin) were undetected at the protein level, implying that these minor proteins were present at a very low abundance in the venom but potentially serve ancillary function. From the evolutionary perspective, the transcriptomic finding indicates that the genes are conserved in the Hydrophis lineage, while their ecological significance awaits further elucidation.
2.4. Sequence Analysis and Phylogenetics of Three-Finger Toxins
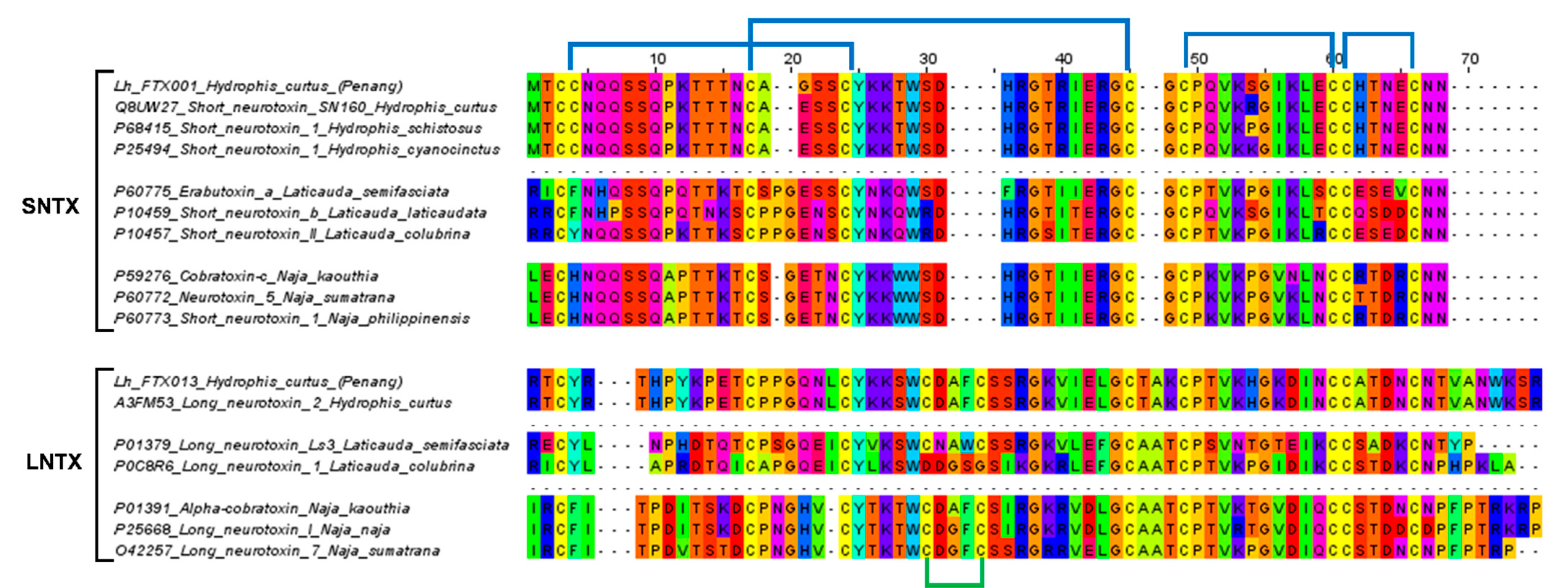
Three-finger toxins are non-enzymatic polypeptides containing 60–74 amino acid residues orientated in three beta-stranded loops, resembling three protruding fingers [28,29]. Based on the protein structure, we categorized the 19 3FTx transcripts in H. curtus venom-gland transcriptome into short-chain 3FTx (S-3FTx, with four disulfide bridges), long-chain 3FTx (L-3FTx, with an additional fifth disulfide bridge on the second loop) and non-conventional 3FTx (NC-3FTx, with an additional fifth disulfide bridge on the first loop) [29]. The majority of the transcripts that contributed to 56.48% of total toxin FPKM, were found to be S-3FTX (10 transcripts). The L-3FTX, comprising 7 transcripts, constituted 20.78% of total toxin FPKM, while there was only one NC-3FX transcript present at a negligible abundance (<0.01%) (Table 2).
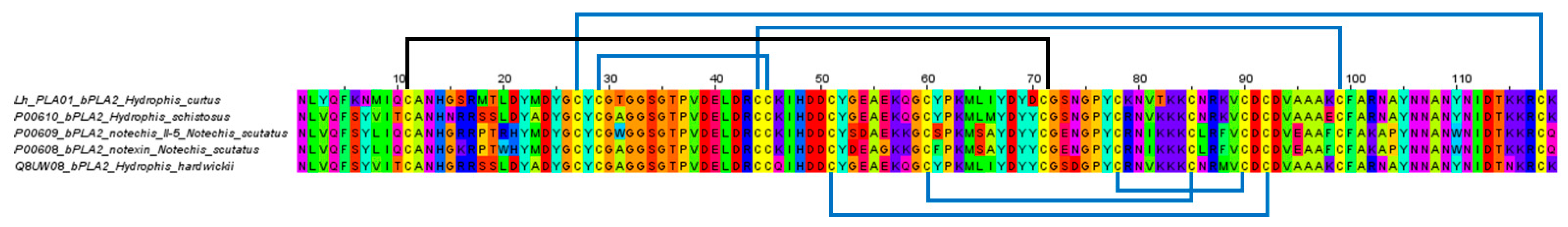
Within S-3FTX and L-3FTX subgroups, the short neurotoxin transcript, Lh_FTX01 and the long neurotoxin transcript, Lh_FTX13 were, respectively, the most abundantly expressed transcripts. Lh_FTX01 was most similar to the short neurotoxin SN160 (UniProt: Q8UW27), previously cloned from Lapemis hardwickii (Guangxi, China) [30]. Both Lh_FTX13 and SN160 (Q8UW27) encoded proteins consisting of 60 amino acid residues but with minor variation at two residual positions: Gly19 in Lh_FTX01 was substituted by Glu19 in SN160 and Ser46 in Lh_FTX_01 was substituted by Arg46 in SN160, as shown in Figure 3. The variation observed could be due to genetic differences between distant geographical populations, as the present specimen was from the northern waters of Malacca Straits (coastal Penang Island) while the previous specimen was sourced from Beihai, Guangxi, Southern China. In comparison, high homology was observed amongst the short neurotoxin (SNTX) sequences of congeneric sea snakes (Hydrophis spp.), sea kraits (Laticauda spp.) and Asiatic cobras (Naja spp.), all of which are polypeptides of 60 amino acid residues reinforced by 4 disulfide bridges (8 highly conserved cysteine residues) (Figure 3). The sequence of Lh_FTX13, on the other hand, matched identically to the long neurotoxin 2 (UniProt: A3FM53), which was cloned from the same Chinese specimen. Lh_FTX013 showed conserved cysteine residues and disulfide bridges as with the long neurotoxin sequences of other comparing elapid species, although long neurotoxin (LNTX) sequences, in general, were more variable in amino acid composition. Notably, the LNTX of L. colubrina (P0C8R6) has only four disulfide bridges instead of five [31]. Comparing to Lh_FTX013 and other related LNTX sequences, P0C8R6 lacks the additional fifth disulfide bridge with amino acid mutation at residue-26 (C→D) and residue-30 (C→G), although the mutation did not appear to compromise its neurotoxicity and lethality [32].
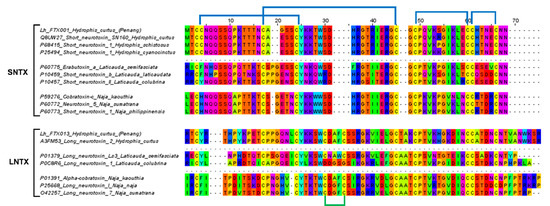
Figure 3.
Multiple sequence alignment of the major short neurotoxins (SNTX) and long neurotoxins (LNTX) from Hydrophis curtus (Penang) and related representative species of sea snakes, sea kraits and cobras. All SNTX shared highly conserved cysteine residues and four disulphide bridges that reinforce the three-finger structure of the molecule. LNTX has an additional fifth disulphide bond in the second loop, except Ls3 from Laticauda colubrina.
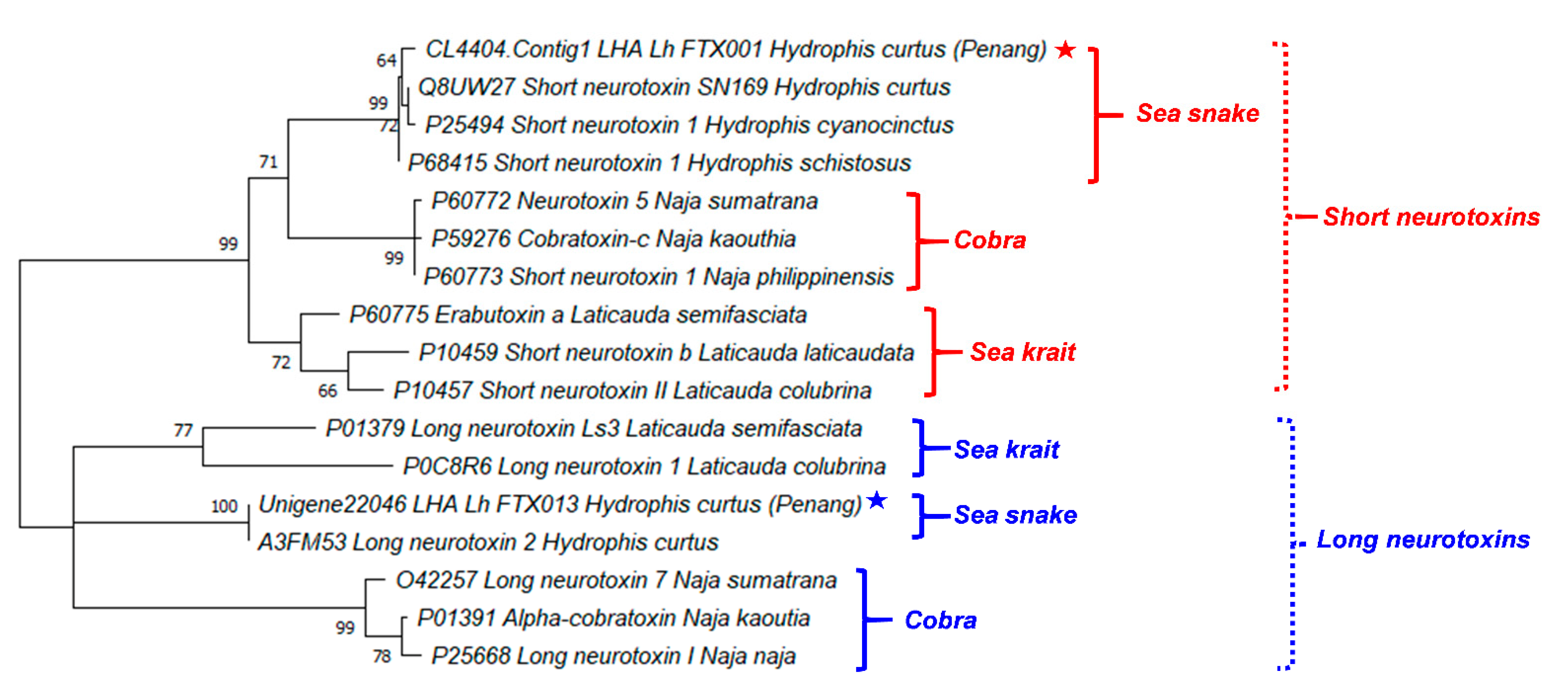
Figure 4 illustrates the phylogenetic tree of the major neurotoxins from H. curtus (Penang) and representative species of sea snakes, sea kraits and cobras. The SNTX and LNTX groups formed two distinct paraphyletic clades. The SNTX of sea snakes (Hydrophis spp.) and cobras (Naja spp.) appeared to be related to a recent ancestral protein that shared a common node with SNTX from the sea kraits (Laticauda spp.). Within the sea snake SNTX, Lh_FTX001 from H. curtus (Penang) is closely related to the more basal SNTX of H. schistosus and the further derived SN160 (H. curtus, Beihai) and Short Neurotoxin 1 (H. cyanocinctus) but the branch lengths were short and this implied little genetic differences. On the other hand, the LNTX sequences of Lh_FTX013 (Penang) and A3FM53 from H. curtus were identical, while there is no LNTX sequence of H. schistosus available for comparison.
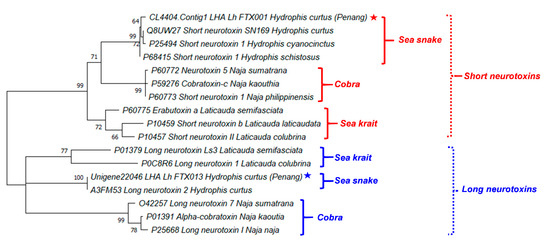
Figure 4.
A phylogenetic tree of short and long neurotoxins from representative species of sea snakes, sea kraits and cobras. Stars indicated the major short and long neurotoxins derived from de novo venom-gland transcriptome of Hydrophis curtus, Penang. Tree was constructed with PAM model of Dayhoff and bootstrapping was performed with 1000 replicates on MEGA Version X [35]. Numbers indicate branch support values. Red/blue stars indicate the specimen studied in this work.
The close phylogenetic relationship among the SNTX and LNTX of sea snakes, sea kraits and cobras support the wide cross-reactivity of Sea Snake Antivenom (SSAV) [33,34], which is the only specific antivenom indicated for the treatment of sea snake envenomation. SSAV is raised against the beaked sea snake (H. schistosus, Penang) specifically but studies have extensively demonstrated that it could effectively cross-neutralize the toxicity of most other marine elapids of various Hydrophis spp. (including H. platurus and its most important neurotoxin), sea kraits (Laticauda spp.) and related principal toxicity [13,14,32,33]. The extensive cross-reactivity of SSAV is indicative of substantially conserved antigenicity in the SNTX and LNTX, respectively. The antigenicity of SNTX and LNTX, however, may possibly vary in view of the more variable amino acid compositions and the further relatedness between the two toxin groups.
2.5. Clinical Relevance and Antigencity of Three-Finger Toxins
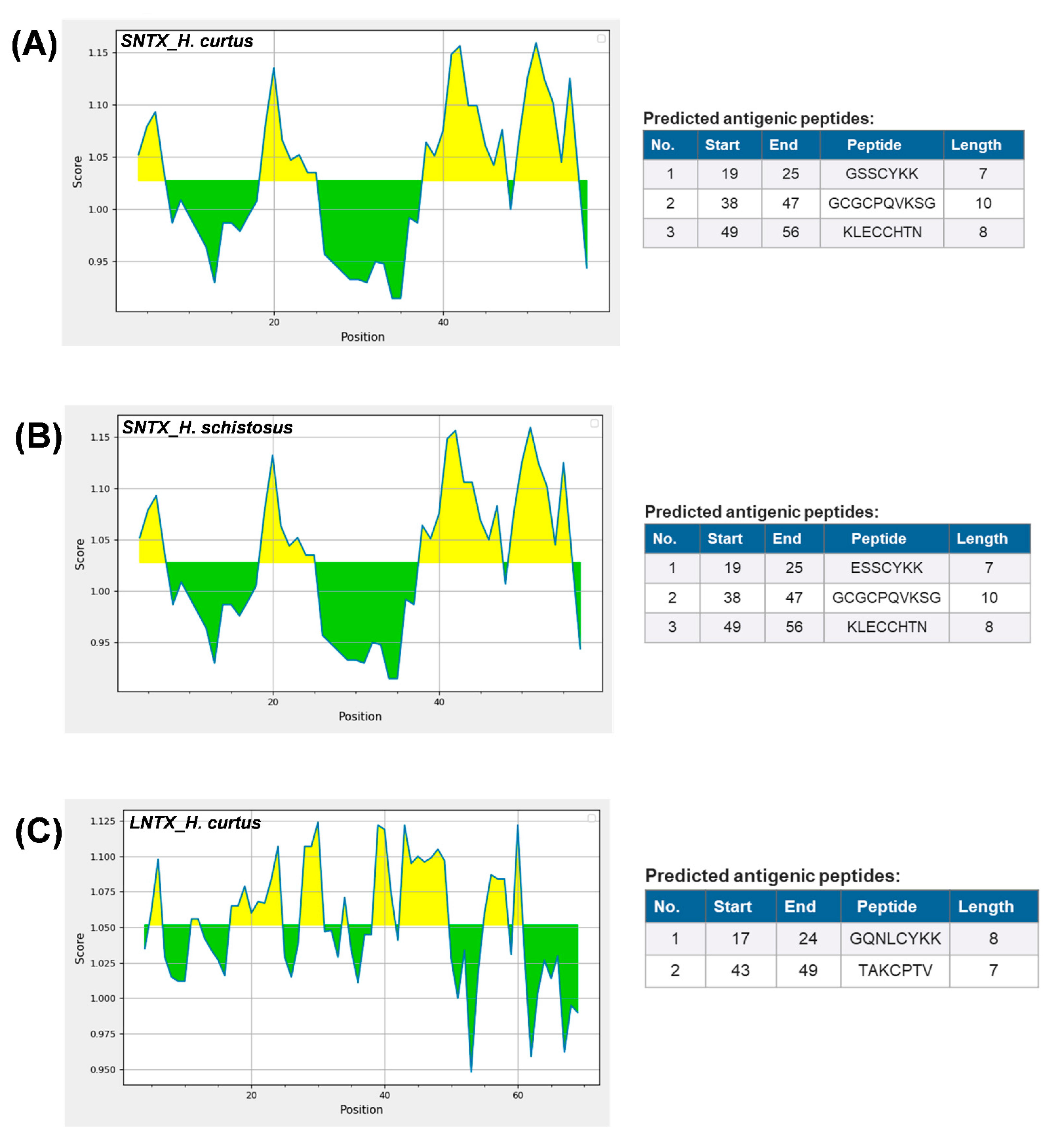
The dominant 3FTX expressed, Lh-FTX13 and Lh_FTX01, were corresponding to the major SNTX and LNTX reported in the venom proteome of H. curtus [13]. Both the SNTX and LNTX of H. curtus are highly lethal (LD50 = 0.10 µg/g and 0.24 µg/g, respectively) and contributing to the neurotoxicity and lethality of the venom (LD50 = 0.20 µg/g). In the current transcriptomic study, Lh_FTX001 (SNTX) has a higher relative abundance compared to Lh_FTX013 (LNTX), in agreement with the proteome reported in which SNTX was more abundantly present than LNTX in the venom. The SNTX-predominating venom phenotype is common in several other sea snake species besides H. curtus (Penang), including the congeneric H. schistosus [36], H. platura [14], H. cyanocinctus [16] and the paraphyletic Aispyrus laevus [37]. The ecological role of SNTX and LNTX in the venom is associated with predatory function, whereby the venom composition is streamlined to incapacitate the fast-moving teleost-based prey (fishes). In envenomation, these are the toxins that block post-junctional nicotinic receptors, resulting in neuromuscular paralysis, respiratory failure and death [38]. Ergo, the treatment outcome of envenomation is principally determined by the antivenom efficacy in neutralizing the principal toxins of the venom. It has been shown that SNTX are more reversible than LNTX in the binding of nicotinic receptors (nAChR), notwithstanding the fact that they are less effectively neutralized by antivenoms [32,34,39,40]. From the immunological perspective, it is possible that LNTX and SNTX vary in their antigenicity, hence the discrepancy in immunorecognition and efficacy of antivenom. Figure 5 shows the predicted antigenicity of alpha-neurotoxin proteins from H. curtus (SNTX and LNTX), H. schistosus (SNTX) and L. colubrina (LNTX). SNTX of H. curtus and H. schistosus have, respectively, 3 prominent epitopes with antigenicity scores beyond 1.10. All three antigenic peptide segments of the two Hydrophis sea snakes comprise residues across 19–25, 38–47 and 49–56, with each antigenic pair sharing highly conserved amino acid residues. The epitope prediction suggested that SNTX of H. schistosus, the species whose venom is used in raising Sea Snake Antivenom, is antigenic to produce antibodies that should be equally effective in cross-neutralizing the SNTX of H. curtus. This is in line with the reported neutralization potency of Sea Snake Antivenom against the neurotoxins of H. schistosus and H. curtus at 0.35 mg/mL and 0.34 mg/mL, respectively. On the other hand, the LNTX of H. curtus exhibited two epitopes (residues 17–24 and 43–49), while the LNTX sequence of H. schistosus is not available from the database for comparison. We predicted that the LNTX of H. schistosus should share similar epitopes with the LNTX of H. curtus, since the Sea Snake Antivenom could effectively cross-neutralize the H. curtus LNTX (potency = 0.78 mg/mL), albeit less potent than it was against that of H. schistosus (potency = 1.38 mg/mL) [13,34]. Interestingly, the SNTX and LNTX do not seem to share much common epitopes, implying limited synergistic cross-reactivity that can be resulted from one antibody toward both toxins. Furthermore, the neutralization potency of antivenom against SNTX is generally lower than LNTX, despite the presence of prominent epitopes in the SNTX protein. Hence, antivenom manufacturers should ensure that the product contains adequate antibodies that are sufficiently immunoreactive toward both types of neurotoxins, so that the reversal of neurotoxicity caused by either SNTX or LNTX can be effective. The production of antivenom toward specific toxin targets can be improved through recent innovations of recombinant technologies [41,42] and re-formulation of specific toxin-targeting antivenom [43,44] to achieve higher potency against the different toxins.
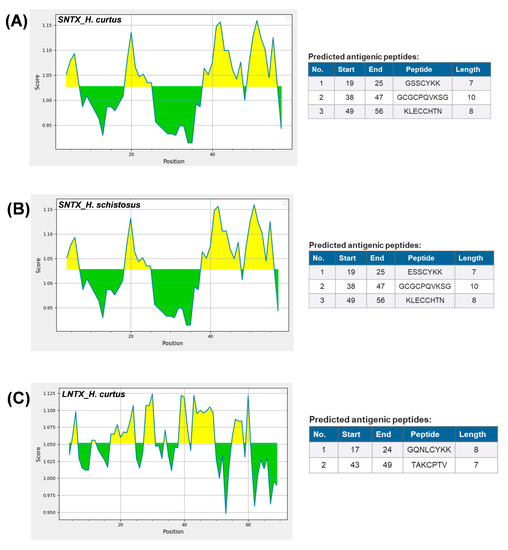
Figure 5.
Predicted antigenicity of alpha-neurotoxin proteins from Hydrophis curtus and Hydrophis schistosus with Kolaskar and Tongaonkar method [45]. (A) Short neurotoxin, H. curtus, accession: Lh_FTX001. (B) Short neurotoxin, H. schistosus, accession: P68415. (C) Long neurotoxin, H. curtus, Lh_FTX013. Yellow areas corresponded to antigenicity score above threshold (1.024) proposed to be part of B-cell epitope.
2.6. Phospholipases A2
Phospholipase A2 transcripts represented the second most abundantly expressed toxin genes in H. curtus venom-gland transcriptome. The major transcript coding for PLA2, that is, Lh_PLA01 contains a full sequence of 118 amino acid residues and was annotated to the basic PLA2 73 (Q8UW30) from Hardwick’s sea snake (of unknown locale, possibly from southern China), based on 92% sequence similarity. Lh_PLA01 belongs to Group IA PLA2 and is a D49 subtype of snake venom PLA2. It has a conserved Ca2+ binding loop that lies between residues 25 and 33 (consensus sequence: Y25-G-C-Y/F-C-G-X-G-G33) and His48 as well as Asp49 which are critical for enzymatic activity [46] (Figure 6). High sequence similarity was also observed when comparing Lh_PLA01 with the basic PLA2 of H. schistosus (P00610) (Figure 6), a highly lethal myotoxin that causes systemic myotoxicity and renal failure secondary to rhabdomyolysis [47]. Unlike the myotoxic PLA2 of H. schistosus, the major enzymatic PLA2 of H. curtus was found to be non-lethal in mice [13,36]. The finding implied that Lh_PLA01 has a variable sequence that probably does not contribute to toxicity, or, it requires the presence of subunit to form PLA2 complex in order to produce toxic activity. Lind and Eaker [48] pointed out that in toxic elapid PLA2s that act in monomeric form, such as the myotoxin from H. schcistosus, notexin and notechis-II5 (both are neurotoxic systemically while myotoxic locally) from Notechis scutatus, have a unique Lys-Lys-Lys sequence at positions 82-84 (Figure 6) not shared by beta-bungarotoxin PLA2 chain and most other non-myotoxic PLA2 variants. This sequence thus could be important for basic PLA2 to exert myotoxic and/or neurotoxic activity in monomeric form. Lh-PLA01 lacks this feature: the positively charged Lys82 was substituted with the neutral Thr82 (Figure 6) and the mutation probably has modified the characteristic cationic site crucial for the myotoxic activity of monomeric PLA2 [46]. More extensive sequence comparison in conjunction with chemical modification studies should clarify the phenomenon.
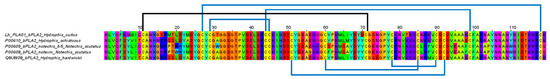
Figure 6.
Multiple sequence alignment of the major phospholipase A2 (PLA2, Lh_PLA01) of Hydrophis curtus (Penang) and related sequences. The PLA2 shared highly conserved cysteine residues and seven disulfide bridges. Blue lines: conservative disulfide bonds; black lines: additional disulfide bond for Group IA PLA2.
In the present study, the transcript expression level of PLA2 (~20% of total toxin FPKM) was lower than the protein abundance of PLA2 reported in proteomics (50–70% of the total venom proteins) [12,13]. The discrepancy could be due to the fact that the mRNAs of various toxins were synthesized at different rates over days and weeks, while the venom-gland tissue was harvested at a certain time point, typically a few days after venom milking. Moreover, it is reasonable to think that the diverse mRNAs had varying half-lives and were subjected to complex regulation processes like post-transcriptional and post-translational modifications [49] which further modulated the maturation of the proteins in the final venom product. The lack of correlation between venom gene expression levels and protein abundances has also been observed in several previous studies [22,23,24,25,50], presumably due to the reason(s) above.
3. Conclusions
The venom-gland transcriptome of H. curtus from the Peninsula of Malaysia was de novo assembled, unveiling the diversity of venom genes in this species. Three-finger toxins constituted the major genes expressed in the venom glands, with SNTX and LNTX being the most abundant, consistent with their role as the principal toxins implicated in the pathophysiology of snakebite envenomation. The findings enriched the toxin knowledgebase of sea snakes and shed light on the medical importance of the venom.
4. Materials and Methods
4.1. Preparation of Snake Venom-Gland Tissue
The sea snake, H. curtus was an adult specimen from the northern waters of Penang Island west of Peninsular Malaysia. The venom was milked four days prior to venom gland tissue collection to promote transcription [51]. The venom glands were collected following euthanasia and sectioned into dimensions of 5 × 5 mm. The sectioned tissue was immersed in RNAlater® solution (Ambion, TX, USA) at 4 °C overnight and stored at −80 °C until further use. The study was carried out in line with protocols approved by the Institutional Animal Use and Care Committee (IACUC) of University of Malaya, Malaysia (Approval code: #2013-11-12/PHAR/R/TCH).
4.2. RNA Extraction and Purification
The venom-gland tissue was homogenized in a 1 mL glass homogenizer with TRIzol solution (Invitrogen, Carlsbad, CA, USA). This was followed by the isolation using chloroform and treated with RNA-free DNAase I (Thermo Fisher Scientific, Waltham, MA, USA), to separate cellular debris and residual DNA. The isolated RNA was then purified via isopropyl alcohol ethanol precipitation. Polyadenylated mRNA was subsequently purified with oligo(dT) magnetic beads (Illumina TruSeq Stranded mRNA) (Illumina, San Diego, CA, USA) as per manufacturer’s instructions. The quality of the purified total RNA was assessed using Agilent 2100 Bioanalyzer (RNA 6000 NanoKit) (Agilent Technologies, Waldbronn, Germany).
Enriched poly(A)+ mRNA isolated from the total venom-gland RNA was used for cDNA construction. The isolated mRNA was fragmented into short fragments, which acted as templates for cDNA synthesis [52]. Random hexamer-primer (N6) was used to synthesis the first-stranded cDNA, followed by second-strand cDNA synthesis with the double-stranded cDNA as input materials, using second strand buffers, dNTPs, RNase H and DNA polymerase I. From these cDNA, a paired-end library was synthesized using the Genomic Sample Prep kit (Illumina, San Diego, CA, USA), according to the manufacturer’s instructions. The cDNA fragments generated were purified with QIAquick PCR extraction kit (Qiagen, Valencia, CA, USA) and dissolved in elution buffer for end repair and the addition of poly(A) to aid in the subsequent ligation of Illumina adaptors that contain a single thymine (T) base overhang at the 3′ ends. Following the ligation, these cDNA fragments were amplified via polymerase chain reaction (PCR) electrophoresed on a 1.5–2% TAE (Tris base, acetic acid and EDTA) agarose gel. From the gel, suitable fragments (200–700 bp) were selected as templates for subsequent PCR amplification. Sequencing of the amplified samples library was achieved in a single lane on the Illumina HiSeq™ 2000 platform (Illumina, San Diego, CA, USA)) with 100-base-pair, paired-end reads.
4.3. Filtration of Raw Sequenced Reads
Sequenced data generated from Illumina HiSeq™ 2000 platform were transformed by base calling into sequence data, called the raw reads and stored in a FASTQ format. Prior to transcriptome assembly, raw reads were filtered to generate clean reads as part of the quality control process in the pre-analysis stage [53]. This involved the removal of (i) adaptors; (ii) reads with >5% of unknown nucleotides or (iii) low-quality reads with >20% of low-quality bases (determined as base quality < 10).
4.4. De Novo Transcriptome Assembly
The de novo transcriptome assembly was performed using a short-reads assembly program, Trinity (version 2.0.6) [54,55]. Three independent software modules, that is, Inchworm, Chrysalis and Butterfly, comprised the Trinity program were sequentially applied to process the large volumes of RNA-seq reads. In brief, this was based on the algorithm of de Bruijn graphs construction, which began by aligning k-mers (k = 25) and reads with a certain length of overlap were joined to form linear contigs. The reads were mapped back onto contigs and by referring to paired-end reads, contigs from the same transcript, as well as the distances between them were determined. The contigs were then partitioned into clusters, each of which carried a complete set of de Bruijn graphs (representing the transcriptional complexity at a given gene or locus). The graphs were independently processed to obtained full-length transcripts for alternatively spliced isoforms and to tease apart transcripts that corresponded to paralogous genes. The clean read Q20 percentage, a point of reference for quality control assessment was obtained as a benchmark for successful de novo assembly of the transcriptome.
4.5. Clustering and Functional Annotation of Transcripts
The transcript sequences generated through Trinity were called Unigenes. Unigenes from the transcriptome assembly were further processed for sequence splicing and redundancy removal with TGI clustering tools (TGICL, version 2.1) to acquire non-redundant (NR) transcripts at the longest possible length [56]. The transcripts were then subjected to family clustering, which resulted in two classes of transcripts: (a) clusters, with a prefix CL and the cluster ID behind as contig; (b) singletons, whose ID was simply left with a prefix of Unigene. In each cluster, there were several transcripts with sequence similarities among them being >70%; while the singletons ‘Unigenes’ lack overlapping with other fragments at the given stringency. The value 70% was used to categorize the assembled sequences based on similarity; sequences similar to each other (may or may not be homologous as having >90% similarity) were grouped under a cluster comprising various contigs.
Following this, transcript Unigenes were then aligned with BLASTx to protein database in priority order to NCBI non-redundant database (NR), with a cut-off value of E < 10−5. Proteins with the highest ranks in the BLASTx results were referred to determine the coding region sequences of Unigenes, followed by translation into amino acid sequences (using standard codon table). Hence, both nucleotide sequences (5′ to 3′) and amino acid sequences of the Unigene-coding regions was acquired. To remove redundancy from each cluster, the longest sequence in each cluster was chosen as the transcript, meanwhile, the length of scaffold was extended based on overlapping sequences using Phrap assembler (release 23.0) (http://www.phrap.org). The distributions of the length of contigs, scaffolds and Unigenes were calculated and the N50 length (assembly quality indicator) was set at N50 > 500 for assembly success.
4.6. Quantifying Transcript Abundance
Clean reads were aligned to Unigene using Bowtie2 [57]. The transcript abundances were calculated using RNA-seq with expectation maximization (RSEM) tool [58].
Fragments per kilobase of exon model per million reads mapped (FPKM) were used to determine the transcript abundance for the identified genes [59]. FPKM is the summation of normalized read counts based on gene length and the total number of mapped reads. The data was obtained using RSEM tool in conjunction with Trinity based on a computational formula:
FPKM is the expression of gene A; B is the number of fragments/reads which are aligned to gene A; N is the total number of fragments/reads that are aligned to all genes; C is the base number in the coding sequence of gene A.
4.7. Categorization of Transcripts
The de novo assembled transcripts were subjected to BLASTx search to obtain the closest resembling sequences from the NR protein database for further classification based on functional annotations. The transcripts (Unigenes) were then sifted to remove those with an FPKM value of less than 1, followed by categorization into three groups: “toxins,” “non-toxins” and “unidentified” [21,23]. “Toxin” transcripts were recruited by toxin-related keyword searches against the annotated transcripts. “Non-toxin” and “unidentified” groups contain transcripts of cellular proteins or house-keeping genes and transcripts that could not be identified, respectively. The redundancy of gene expression was determined by dividing the total FPKM of each group by the total number of transcripts in the respective group of transcripts [21]. In the toxin group, the amino acid sequences were used to further validate the toxin identity through BLASTp suite (Basic Local Alignment Search Tool-Protein) in the UniProt (Universal Protein Resource Knowledgebase) database platform. The transcripts were searched against Serpentes database (taxid: 8570) and validated based on the lowest E-score value with the highest percentage of sequence similarity (updated as of 29 June 2020).
4.8. Multiple Sequence Alignment
Multiple sequence alignment was conducted using Jalview software (version 2.11.1.0) [60] and MUSCLE (Multiple Sequence Comparison by Log-Expectation) [61] program. Sequences of related species used in multiple sequence alignment were retrieved from UniProtKB depository (accessed date: 14 September 2020) (http://www.uniprot.org/). The selection was based on their relevance to the toxins in comparison to elucidate the similarity and variation as well as conserved regions of the sequences.
4.9. Phylogenetic Analysis
Sequences of long and short neurotoxins annotated for H. curtus of Penang (this work) and representative species of sea snakes, sea kraits and cobras (retrieved from Universal Protein Knowledgebase, UniProtKB, http://www.uniprot.org/) (accessed date: 14 September 2020) were used to construct the phylogenetic tree. The tree was constructed with Molecular Evolutionary Genetics Analysis (MEGA) Version X [35] applying the Dayhoff (PAM) substitution model (+G) [62]. Bootstrap test (1000 replicates) was computed for the confidence limits of the constructed phylogenetic tree [63].
4.10. Scale-Based B-Cell Epitope Prediction
The antigenic determinants (epitopes) of toxins were predicted using a scale-based B-cell epitope prediction software applying Kolaskar and Tongaonkar antigenicity prediction algorithm (http://tools.immuneepitope.org) (accessed on 27 November 2020) [45]. Default parameters and window size 7 were used in the analysis for predicting potentially antigenic regions of amino acids in the sequences
4.11. Supporting Data
Sequencing data from the de novo venom-gland transcriptomics of H. curtus was deposited in National Centre for Biotechnology Information (NCBI) Sequence Read Archive (https://submit.ncbi.nlm.nih.gov/subs/sra/) (submitted on 29 December 2020) under SRA accession: PRJNA688573.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6651/13/2/127/s1, Table S1: Venom-gland transcriptomics of Hydrophis curtus (Penang): Toxin annotation and sequences.
Author Contributions
Conceptualization, C.H.T.; Data curation, K.Y.T.; Formal analysis, C.H.T. and K.Y.T.; Funding acquisition, C.H.T.; Investigation, C.H.T. and K.Y.T.; Methodology, C.H.T.; Project administration, C.H.T.; Resources, C.H.T.; Software, K.Y.T.; Validation, K.Y.T.; Writing–original draft, C.H.T.; Writing–review & editing, C.H.T. and K.Y.T. All authors have read and agreed to the published version of the manuscript.
Funding
Institut Pengurusan dan Pemantauan Penyelidikan, Universiti Malaya: ST011-2020.
Data Availability Statement
Sequencing data from the de novo venom-gland transcriptomics of H. curtus was deposited in National Centre for Biotechnology Information (NCBI) Sequence Read Archive (https://submit.ncbi.nlm.nih.gov/subs/sra/) under SRA accession: PRJNA688573.
Acknowledgments
The authors thank Jin Wei Tan, Tzu Shan Ng and Kok Onn Kwong for technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Primers 2017, 3, 17063. [Google Scholar]
- World Health Organization. Guidelines for the Management of Snake-Bites; Regional Office for South-East Asia: New Delhi, India, 2016. [Google Scholar]
- Feola, A.; Marella, G.L.; Carfora, A.; Della Pietra, B.; Zangani, P.; Campobasso, C.P. Snakebite Envenoming a Challenging Diagnosis for the Forensic Pathologist: A Systematic Review. Toxins 2020, 12, 699. [Google Scholar] [CrossRef]
- Jamaiah, I.; Rohela, M.; Ng, T.K.; Ch’ng, K.B.; Teh, Y.S.; Nurulhuda, A.L.; Suhaili, N. Retrospective prevalence of snakebites from Hospital Kuala Lumpur (HKL) (1999–2003). Southeast Asian J. Trop. Med. Public Health 2006, 37, 200–205. [Google Scholar]
- Reid, H.A. Sea-snake bite research. Trans. R. Soc. Trop. Med. Hyg. 1956, 50, 517–538. [Google Scholar] [CrossRef]
- Alirol, E.; Sharma, S.K.; Bawaskar, H.S.; Kuch, U.; Chappuis, F. Snake Bite in South Asia: A Review. PLoS Negl. Trop. Dis. 2010, 4, e603. [Google Scholar] [CrossRef]
- Cao, N.; Tao, N.T.; Moore, A.; Montoya, A.; Rasmussen, A.R.; Broad, K.; Voris, H.K.; Takacs, Z. Sea Snake Harvest in the Gulf of Thailand. Conserv. Biol. 2014, 28, 1677–1687. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, S.K.; Tibballs, J. Australian Animal Toxins: The Creatures, Their Toxins and Care of the Poisoned Patient; Oxford University Press: New York, NY, USA, 2001. [Google Scholar]
- Marsden, A.T.; Reid, H.A. Pathology of sea-snake poisoning. BMJ 1961, 1, 1290–1293. [Google Scholar] [CrossRef]
- Kularatne, S.A.; Hettiarachchi, R.; Dalpathadu, J.; Mendis, A.S.; Appuhamy, P.D.; Zoysa, H.D.; Maduwage, K.; Weerasinghe, V.S.; de Silva, A. Enhydrina schistosa (Elapidae: Hydrophiinae) the most dangerous sea snake in Sri Lanka: Three case studies of severe envenoming. Toxicon 2014, 77, 78–86. [Google Scholar] [CrossRef]
- Sanders, K.L.; Lee, M.S.; Mumpuni; Bertozzi, T.; Rasmussen, A.R. Multilocus phylogeny and recent rapid radiation of the viviparous sea snakes (Elapidae: Hydrophiinae). Mol. Phylogenet. Evol. 2013, 66, 575–591. [Google Scholar] [CrossRef]
- Neale, V.; Sotillo, J.; Seymour, J.E.; Wilson, D. The Venom of the Spine-Bellied Sea Snake (Hydrophis curtus): Proteome, Toxin Diversity and Intraspecific Variation. Int. J. Mol. Sci. 2017, 18, 2695. [Google Scholar] [CrossRef]
- Tan, C.H.; Tan, K.Y.; Ng, T.S.; Sim, S.M.; Tan, N.H. Venom Proteome of Spine-Bellied Sea Snake (Hydrophis curtus) from Penang, Malaysia: Toxicity Correlation, Immunoprofiling and Cross-Neutralization by Sea Snake Antivenom. Toxins 2019, 11, 3. [Google Scholar] [CrossRef]
- Lomonte, B.; Pla, D.; Sasa, M.; Tsai, W.C.; Solorzano, A.; Urena-Diaz, J.M.; Fernandez-Montes, M.L.; Mora-Obando, D.; Sanz, L.; Gutierrez, J.M.; et al. Two color morphs of the pelagic yellow-bellied sea snake, Pelamis platura, from different locations of Costa Rica: Snake venomics, toxicity, and neutralization by antivenom. J. Proteom. 2014, 103, 137–152. [Google Scholar] [CrossRef]
- Tan, C.H.; Tan, N.H.; Tan, K.Y.; Kwong, K.O. Antivenom cross-neutralization of the venoms of Hydrophis schistosus and Hydrophis curtus, two common sea snakes in Malaysian waters. Toxins 2015, 7, 572–581. [Google Scholar] [CrossRef]
- Calvete, J.J.; Ghezellou, P.; Paiva, O.; Matainaho, T.; Ghassempour, A.; Goudarzi, H.; Kraus, F.; Sanz, L.; Williams, D.J. Snake venomics of two poorly known Hydrophiinae: Comparative proteomics of the venoms of terrestrial Toxicocalamus longissimus and marine Hydrophis cyanocinctus. J. Proteom. 2012, 75, 4091–4101. [Google Scholar] [CrossRef]
- Sanders, K.L.; Lee, M.S. Uncoupling ecological innovation and speciation in sea snakes (Elapidae, Hydrophiinae, Hydrophiini). J. Evol. Biol. 2010, 23, 2685–2693. [Google Scholar] [CrossRef]
- Sanders, K.L.; Lee, M.S.; Leys, R.; Foster, R.; Keogh, J.S. Molecular phylogeny and divergence dates for Australasian elapids and sea snakes (hydrophiinae): Evidence from seven genes for rapid evolutionary radiations. J. Evol. Biol. 2008, 21, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Uetz, P.; Freed, P.; Hošek, J.E. The Reptile Database. Available online: http://www.reptile-database.org (accessed on 15 November 2020).
- Reid, H.A. Epidemiology of sea-snake bites. J. Trop. Med. Hyg. 1975, 78, 106–113. [Google Scholar]
- Chong, H.P.; Tan, K.Y.; Tan, N.H.; Tan, C.H. Exploring the Diversity and Novelty of Toxin Genes in Naja sumatrana, the Equatorial Spitting Cobra from Malaysia through De Novo Venom-Gland Transcriptomics. Toxins 2019, 11, 104. [Google Scholar] [CrossRef]
- Tan, K.Y.; Tan, C.H.; Chanhome, L.; Tan, N.H. Comparative venom gland transcriptomics of Naja kaouthia (monocled cobra) from Malaysia and Thailand: Elucidating geographical venom variation and insights into sequence novelty. PeerJ 2017, 5, e3142. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, K.Y.; Fung, S.Y.; Tan, N.H. Venom-gland transcriptome and venom proteome of the Malaysian king cobra (Ophiophagus hannah). BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Correa-Netto, C.; Junqueira-de-Azevedo Ide, L.; Silva, D.A.; Ho, P.L.; Leitao-de-Araujo, M.; Alves, M.L.; Sanz, L.; Foguel, D.; Zingali, R.B.; Calvete, J.J. Snake venomics and venom gland transcriptomic analysis of Brazilian coral snakes, Micrurus altirostris and M. corallinus. J. Proteom. 2011, 74, 1795–1809. [Google Scholar] [CrossRef]
- Margres, M.J.; Aronow, K.; Loyacano, J.; Rokyta, D.R. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genom. 2013, 14, 531. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, A.D.; Swain, M.T.; Hegarty, M.J.; Logan, D.W.; Mulley, J.F. Restriction and recruitment-gene duplication and the origin and evolution of snake venom toxins. Genome Biol. Evol. 2014, 6, 2088–2095. [Google Scholar] [CrossRef]
- Kordis, D.; Gubensek, F. Adaptive evolution of animal toxin multigene families. Gene 2000, 261, 43–52. [Google Scholar] [CrossRef]
- Tsetlin, V. Snake venom alpha-neurotoxins and other ‘three-finger’ proteins. Eur. J. Biochem. 1999, 264, 281–286. [Google Scholar] [CrossRef]
- Kini, R.M.; Doley, R. Structure, function and evolution of three-finger toxins: Mini proteins with multiple targets. Toxicon 2010, 56, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.F.; Peng, L.S.; Wu, W.Y.; Wei, J.W.; Yang, H.; Yang, Y.Z.; Xu, A.L. Identification and Funtional Characterization of Three Postsynaptic Short-chain Neurotoxins from Hydrophiinae, Lapemis hardwickii Gray. Acta Biochim. Biophys. Sin. 2001, 33, 457–462. [Google Scholar] [PubMed]
- Kim, H.S.; Tamiya, N. Amino acid sequences of two novel long-chain neurotoxins from the venom of the sea snake Laticauda colubrina. Biochem. J. 1982, 207, 215–223. [Google Scholar] [CrossRef]
- Tan, C.H.; Wong, K.Y.; Tan, K.Y.; Tan, N.H. Venom proteome of the yellow-lipped sea krait, Laticauda colubrina from Bali: Insights into subvenomic diversity, venom antigenicity and cross-neutralization by antivenom. J. Proteom. 2017, 166, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Chetty, N.; Du, A.; Hodgson, W.C.; Winkel, K.; Fry, B.G. The in vitro neuromuscular activity of Indo-Pacific sea-snake venoms: Efficacy of two commercially available antivenoms. Toxicon 2004, 44, 193–200. [Google Scholar] [CrossRef]
- Tan, K.Y.; Tan, C.H.; Fung, S.Y.; Tan, N.H. Neutralization of the Principal Toxins from the Venoms of Thai Naja kaouthia and Malaysian Hydrophis schistosus: Insights into Toxin-Specific Neutralization by Two Different Antivenoms. Toxins 2016, 8, 86. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tan, C.H.; Tan, K.Y.; Lim, S.E.; Tan, N.H. Venomics of the beaked sea snake, Hydrophis schistosus: A minimalist toxin arsenal and its cross-neutralization by heterologous antivenoms. J. Proteom. 2015, 126, 121–130. [Google Scholar] [CrossRef]
- Laustsen, A.H.; Gutierrez, J.M.; Rasmussen, A.R.; Engmark, M.; Gravlund, P.; Sanders, K.L.; Lohse, B.; Lomonte, B. Danger in the reef: Proteome, toxicity, and neutralization of the venom of the olive sea snake, Aipysurus laevis. Toxicon 2015, 107, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Ranawaka, U.K.; Lalloo, D.G.; de Silva, H.J. Neurotoxicity in snakebite--the limits of our knowledge. PLoS Negl. Trop. Dis. 2013, 7, e2302. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.; Cristofori-Armstrong, B.; Rash, L.D.; Hodgson, W.C.; Isbister, G.K. Defining the role of post-synaptic alpha-neurotoxins in paralysis due to snake envenoming in humans. Cell. Mol. Life Sci. 2018, 75, 4465–4478. [Google Scholar] [CrossRef]
- Wong, K.Y.; Tan, C.H.; Tan, N.H. Venom and Purified Toxins of the Spectacled Cobra (Naja naja) from Pakistan: Insights into Toxicity and Antivenom Neutralization. Am. J. Trop. Med. Hyg. 2016, 94, 1392–1399. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, C.; Laustsen, A.H. Recent Advances in Next Generation Snakebite Antivenoms. Trop. Med. Infect. Dis. 2018, 3, 42. [Google Scholar] [CrossRef]
- de la Rosa, G.; Olvera, F.; Archundia, I.G.; Lomonte, B.; Alagon, A.; Corzo, G. Horse immunization with short-chain consensus alpha-neurotoxin generates antibodies against broad spectrum of elapid venomous species. Nat. Commun. 2019, 10, 3642. [Google Scholar] [CrossRef]
- Ratanabanangkoon, K.; Tan, K.Y.; Pruksaphon, K.; Klinpayom, C.; Gutierrez, J.M.; Quraishi, N.H.; Tan, C.H. A pan-specific antiserum produced by a novel immunization strategy shows a high spectrum of neutralization against neurotoxic snake venoms. Sci. Rep. 2020, 10, 11261. [Google Scholar] [CrossRef] [PubMed]
- Ratanabanangkoon, K.; Tan, K.Y.; Eursakun, S.; Tan, C.H.; Simsiriwong, P.; Pamornsakda, T.; Wiriyarat, W.; Klinpayom, C.; Tan, N.H. A Simple and Novel Strategy for the Production of a Pan-specific Antiserum against Elapid Snakes of Asia. PLoS Negl. Trop. Dis. 2016, 10, e0004565. [Google Scholar] [CrossRef] [PubMed]
- Kolaskar, A.S.; Tongaonkar, P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990, 276, 172–174. [Google Scholar] [CrossRef]
- Doley, R.; Zhou, X.; Kini, R.M. Snake Venom Phospholipase A2 Enzymes. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press Taylor & Francis Group: Boca Raton, FL, USA, 2009. [Google Scholar]
- Fohlman, J.; Eaker, D. Isolation and characterization of a lethal myotoxic phospholipase A from the venom of the common sea snake Enhydrina schistosa causing myoglobinuria in mice. Toxicon 1977, 15, 385–393. [Google Scholar] [CrossRef]
- Lind, P.; Eaker, D. Amino acid sequence of a lethal myotoxic phospholipase A2 from the venom of the common sea snake (Enhydrina schistosa). Toxicon 1981, 19, 11–24. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Rodrigues, R.S.; Boldrini-Franca, J.; Fonseca, F.P.; de la Torre, P.; Henrique-Silva, F.; Sanz, L.; Calvete, J.J.; Rodrigues, V.M. Combined snake venomics and venom gland transcriptomic analysis of Bothropoides pauloensis. J. Proteom. 2012, 75, 2707–2720. [Google Scholar] [CrossRef]
- Rotenberg, D.; Bamberger, E.S.; Kochva, E. Studies on ribonucleic acid synthesis in the venom glands of Vipera palaestinae (Ophidia, Reptilia). Biochem. J. 1971, 121, 609–612. [Google Scholar] [CrossRef][Green Version]
- Wery, M.; Descrimes, M.; Thermes, C.; Gautheret, D.; Morillon, A. Zinc-mediated RNA fragmentation allows robust transcript reassembly upon whole transcriptome RNA-Seq. Methods 2013, 63, 25–31. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 1–19. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.G.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Dayhoff, M.O.; Schwartz, R.M.; Orcutt, B.C.; Dayhoff, M.O. A Model of Evolutionary Change in Proteins. In Atlas of Protein Sequence and Structure; National Biomedical Research Foundation: Washington, DC, USA, 1978; Volume 5, pp. 345–352. [Google Scholar]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).