N-Terminal Tagging with GFP Enhances Selectivity of Agitoxin 2 to Kv1.3-Channel Binding Site
Abstract
:1. Introduction
2. Results
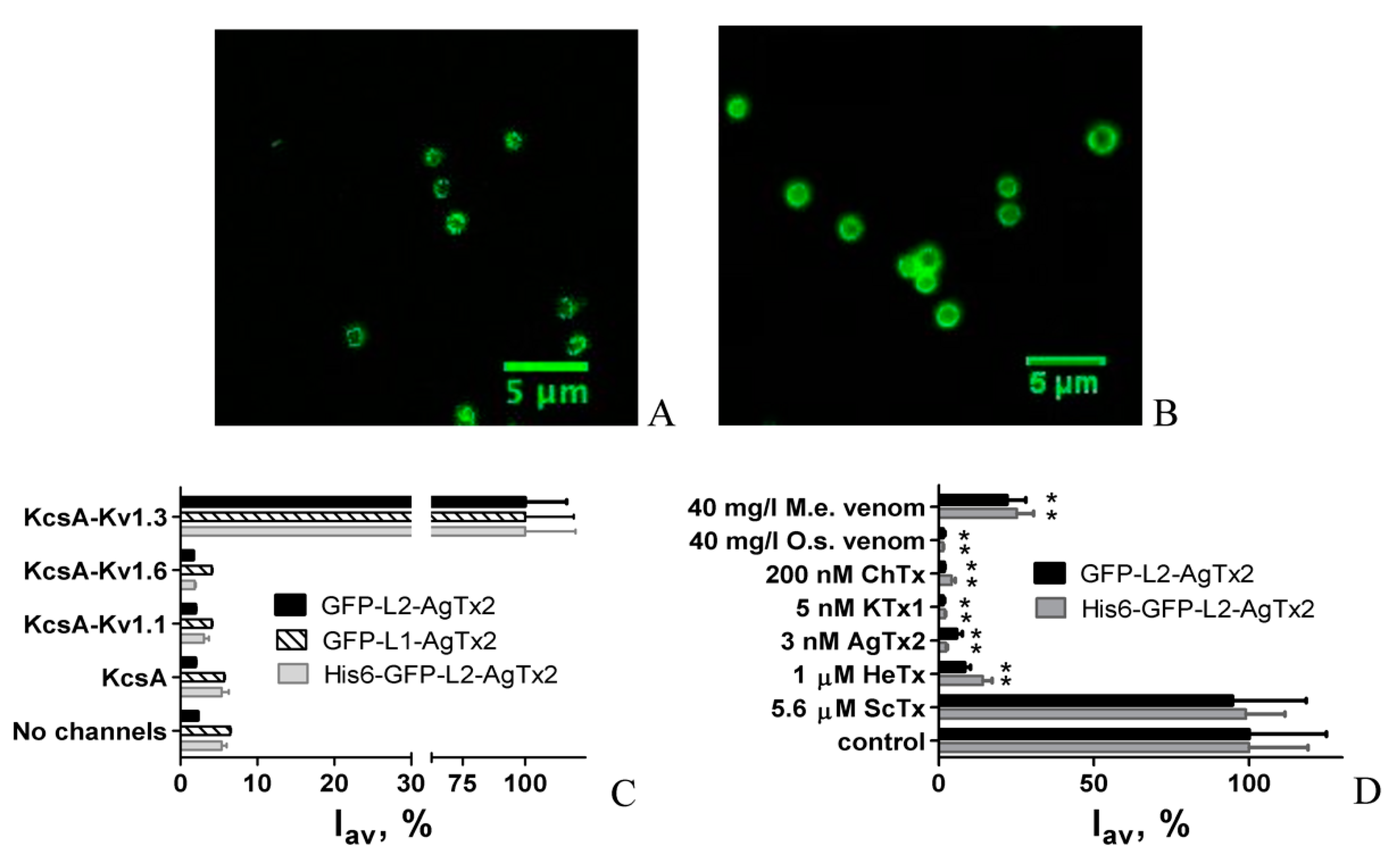
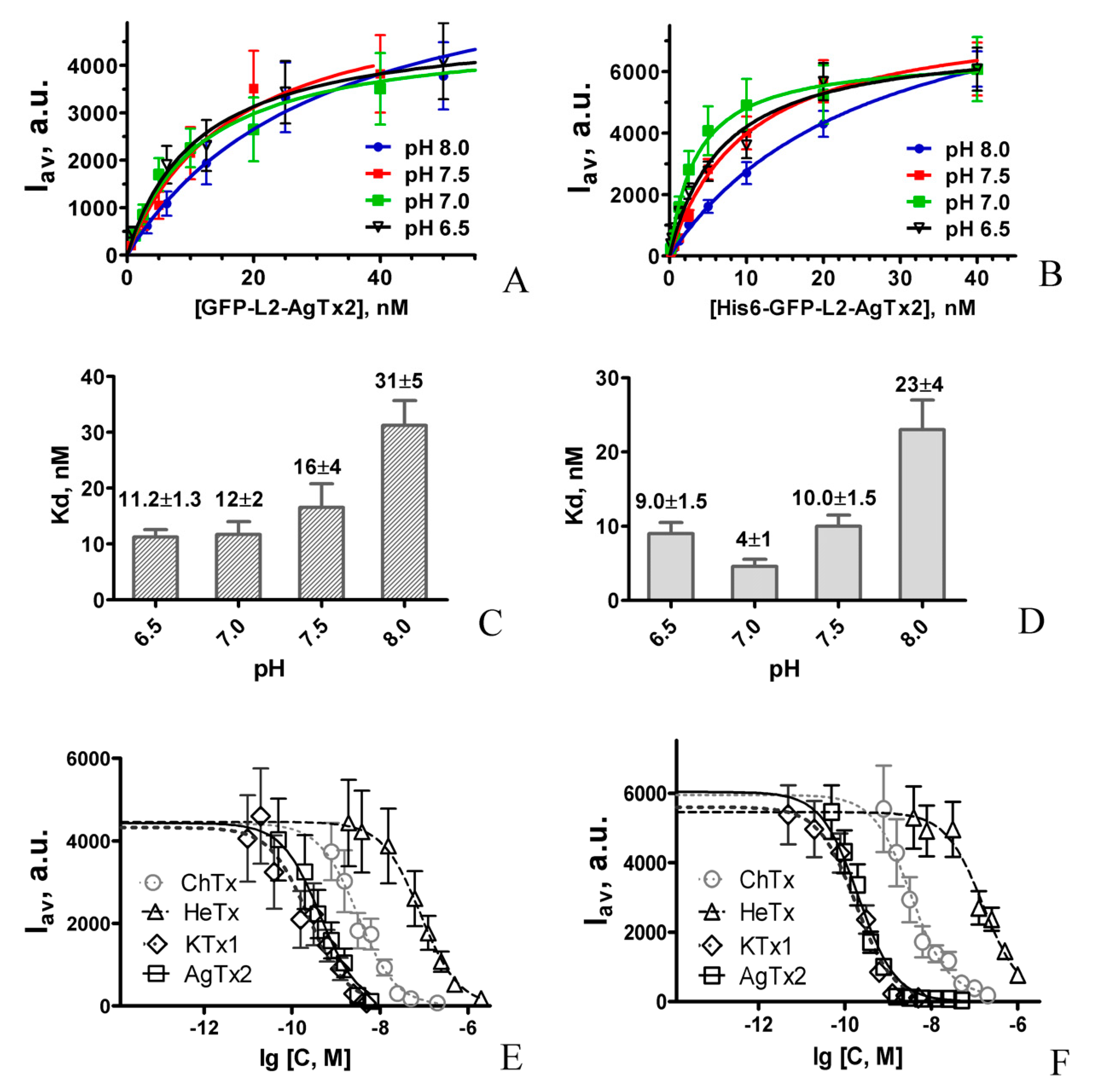
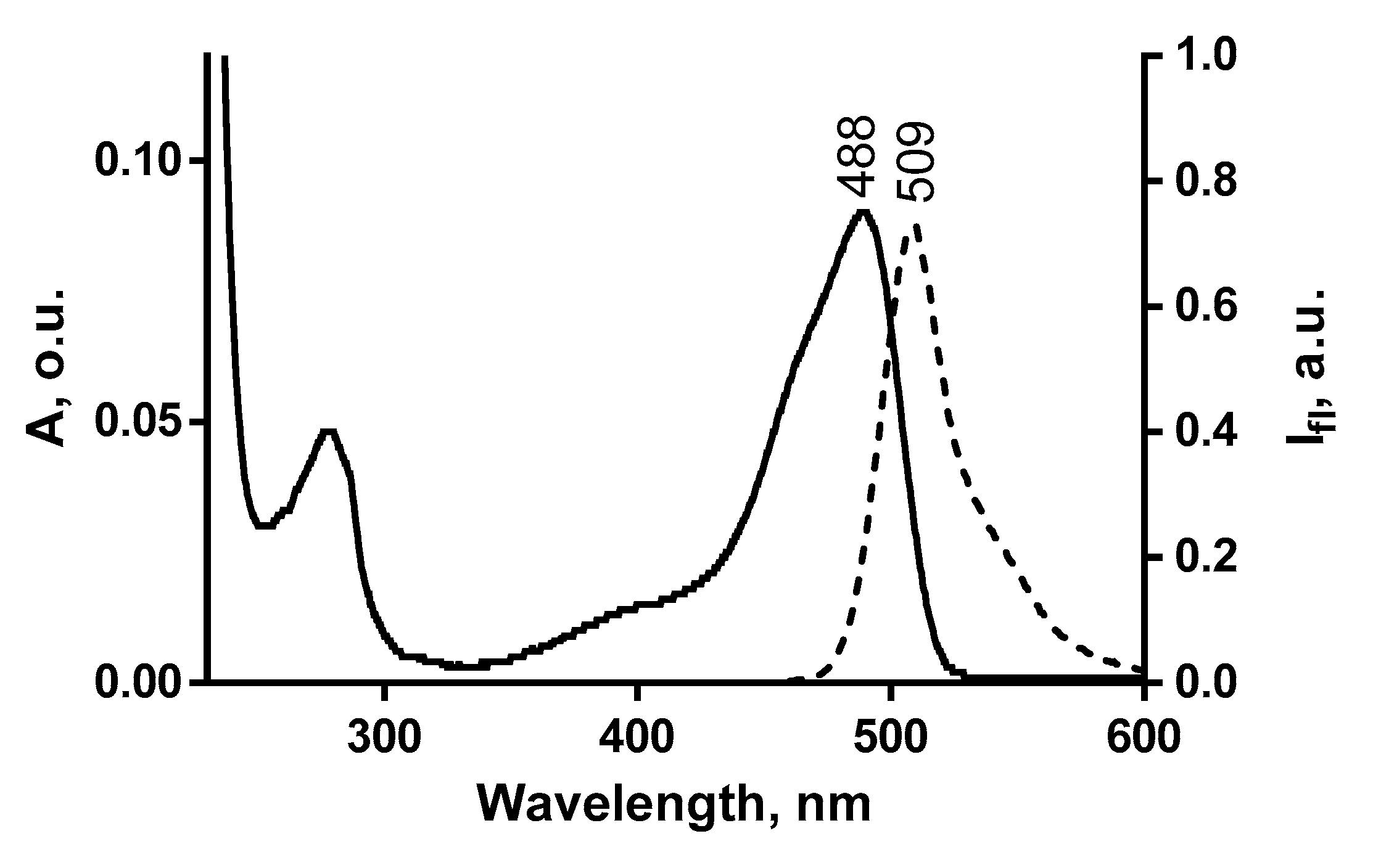
2.1. Design and Properties of GFP-L2-AgTx2
2.2. Study of Structural Factors Defining GFP-L2-AgTx2 Selectivity
2.3. Properties of His6-GFP-L2-AgTx2 Ligand
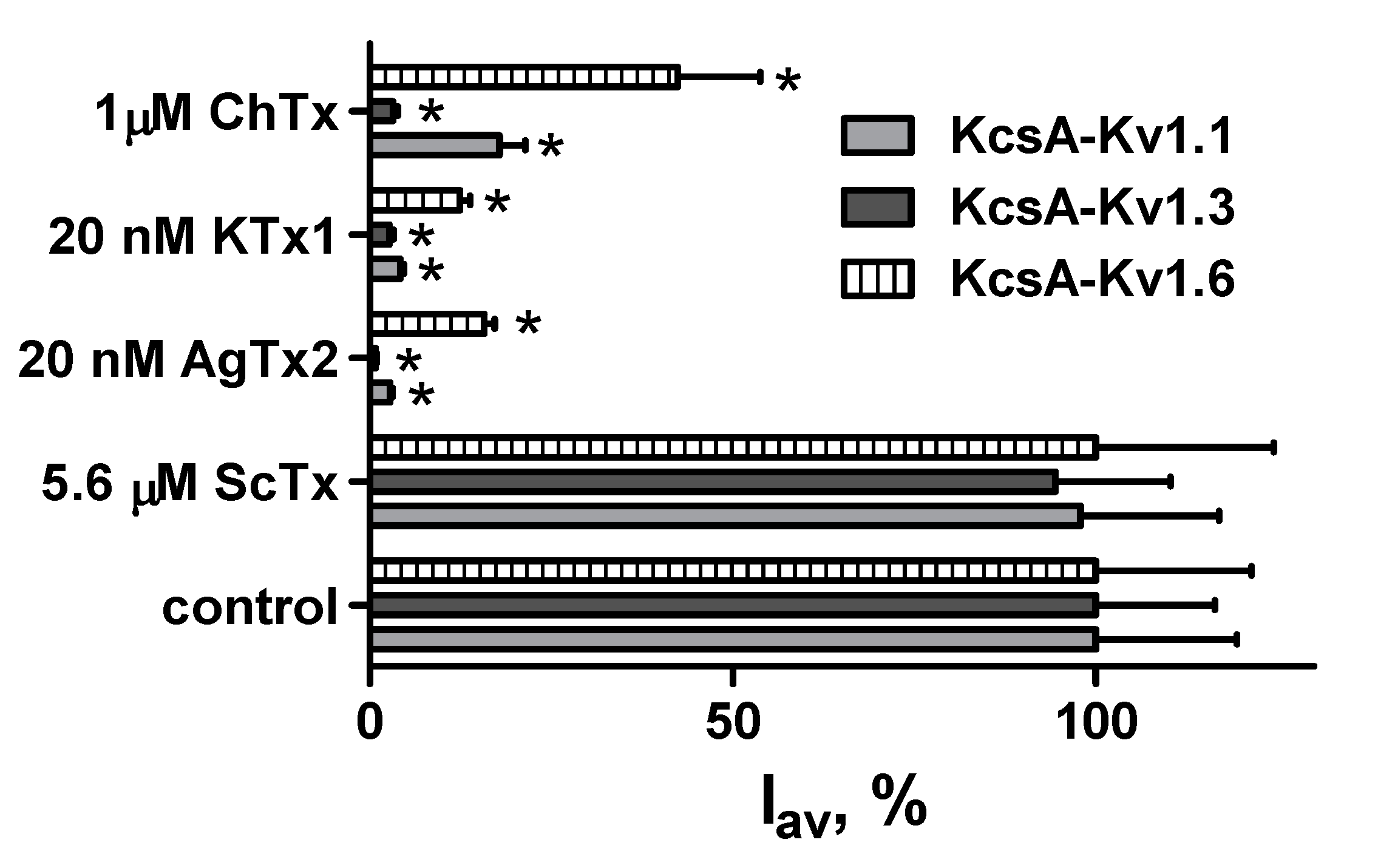
2.4. GFP-Tagged AgTx2 as a Component of the KcsA-Kv1.3-Based Bioengineering System
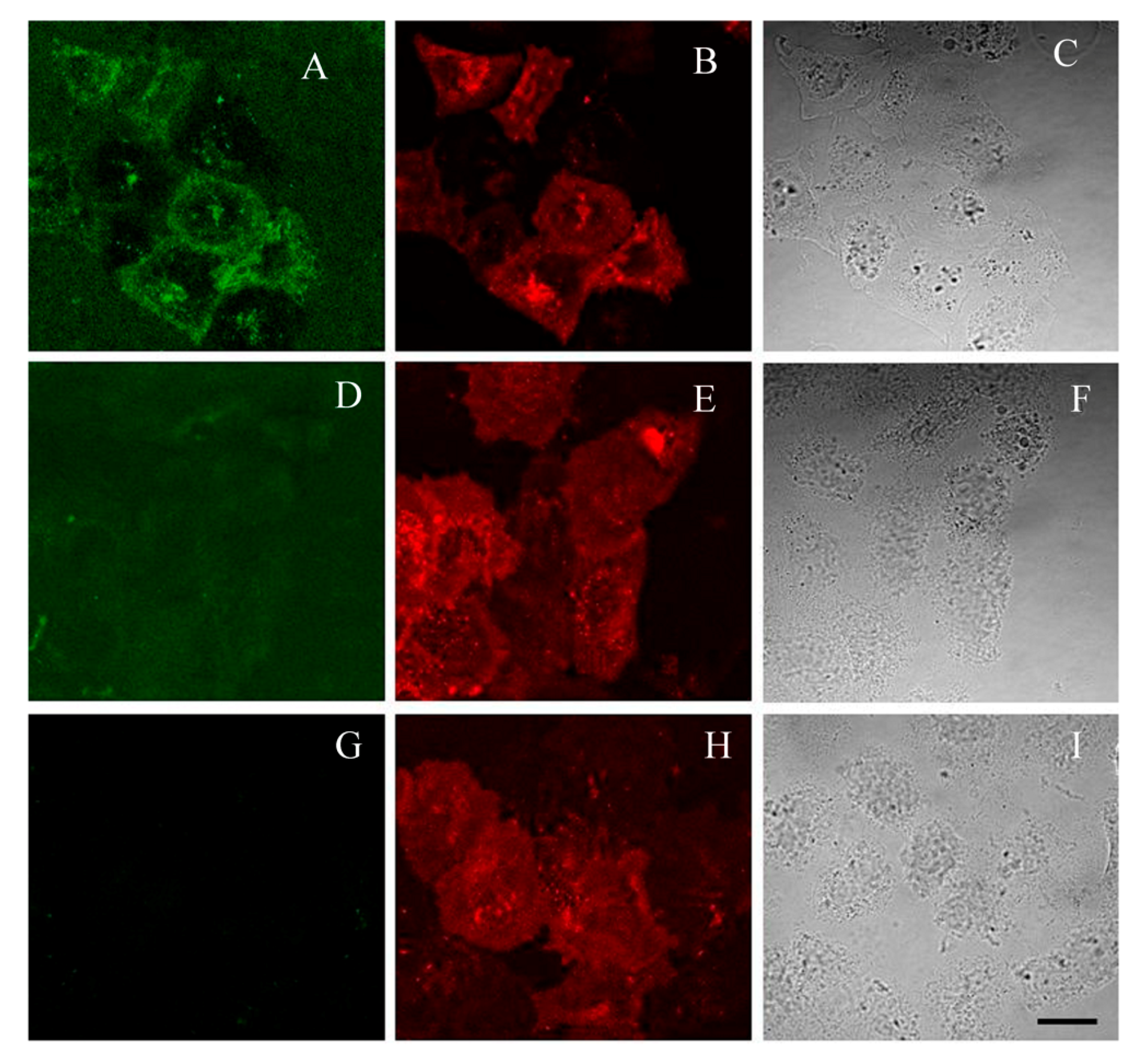
2.5. GFP-Tagged AgTx2 as a Fluorescent Probe of Kv1.3 on the Membrane of HEK293 Cells
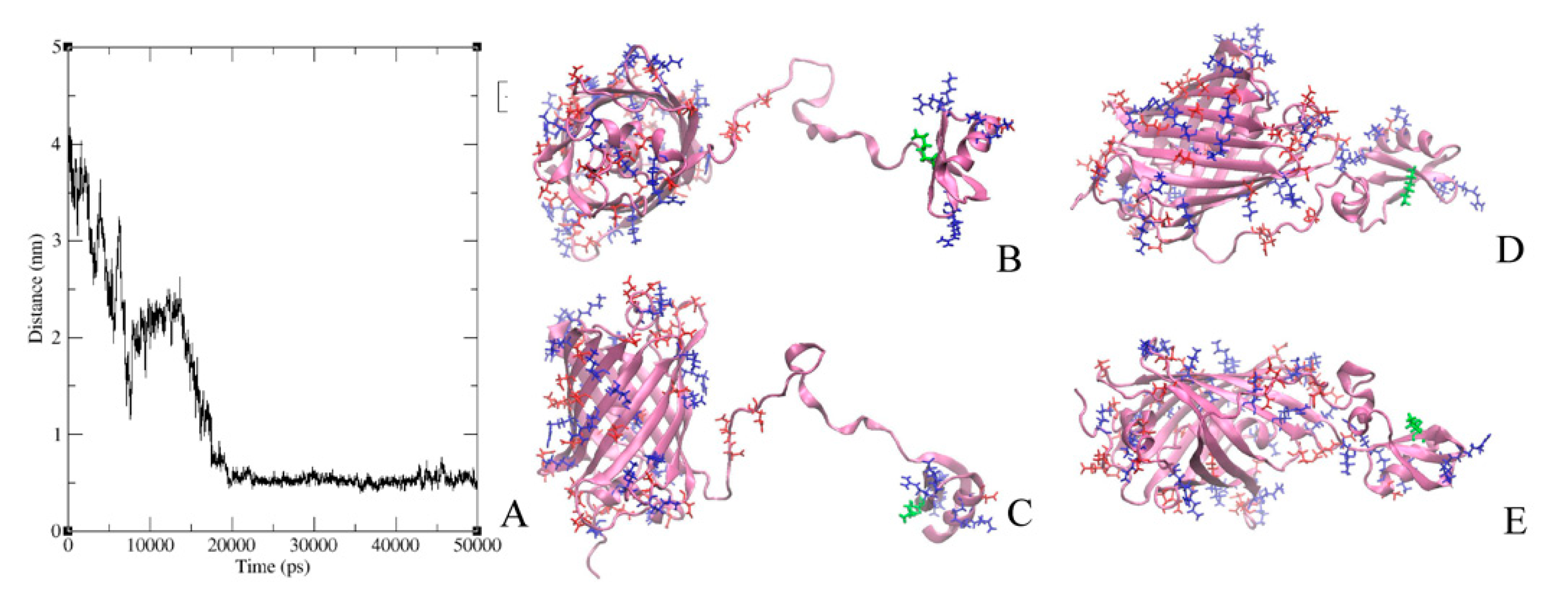
2.6. Modeling of GFP-L2-AgTx2 and AgTx2-L3-GFP Chimeric Proteins
3. Discussion
4. Conclusions
5. Materials and Methods
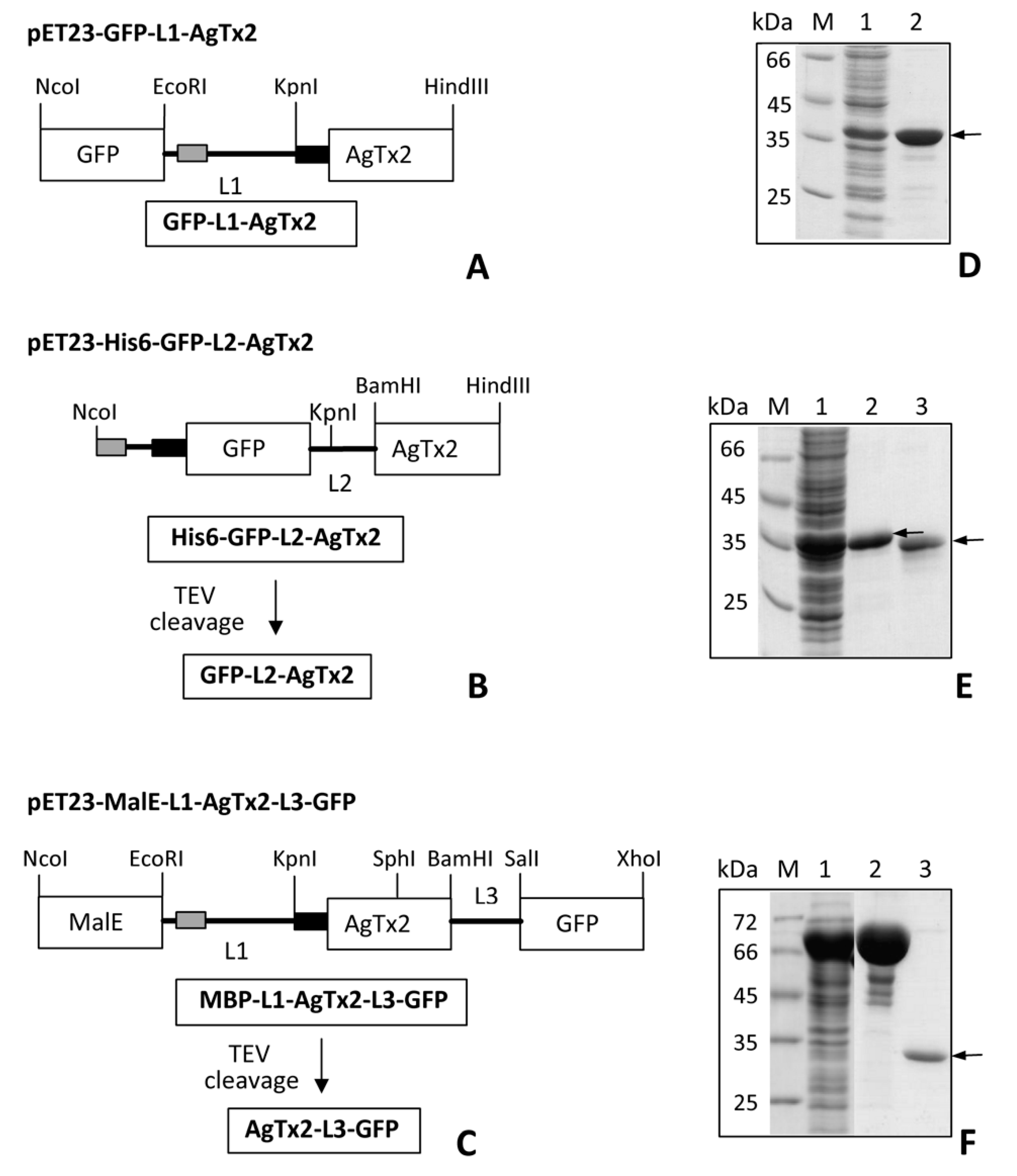
5.1. Gene Design, Synthesis and Cloning
5.2. Expression and Purification of GFP-Tagged AgTx2
5.3. Preparation of GFP-L2-AgTx2 and AgTx2-L3-GFP
5.4. Recombinant Toxins
5.5. Venoms
5.6. Preparation of Spheroplasts and Binding Protocol
5.7. Construction of mKate2-KCNA3-del Gene
5.8. Experiments with Cells
5.9. Microscopy
5.10. Optical Spectroscopy
5.11. Computer Modeling of Chimeric Proteins GFP-L2-AgTx2 and AgTx2-L3-GFP
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mobli, M.; Undheim, E.A.B.; Rash, L.D. Modulation of ion channels by cysteine-rich peptides. Adv. Pharmacol. 2017, 79, 199–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norton, R.S. Enhancing the therapeutic potential of peptide toxins. Expert Opin. Drug Discov. 2017, 12, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S. Animal toxins for channelopathy treatment. Neuropharmacology 2018, 132, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Cao, Z.; Li, W.; Sabatier, J.M.; Wu, Y. Treating autoimmune disorders with venom-derived peptides. Expert Opin. Biol. Ther. 2017, 17, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- de Souza, J.M.; Goncalves, B.D.C.; Gomez, M.V.; Vieira, L.B.; Ribeiro, F.M. Animal Toxins as Therapeutic Tools to Treat Neurodegenerative Diseases. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Lewis, R.J. Use of venom peptides to probe ion channel structure and function. J. Biol. Chem. 2010, 285, 13315–13320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batool, S.; Noureen, N.; Kamal, M.A. Structure activity relationship of venom toxins targeting potassium channels. Curr. Drug Metab. 2017, 19, 714–720. [Google Scholar] [CrossRef]
- Kuzmenkov, A.I.; Vassilevski, A.A. Labelled animal toxins as selective molecular markers of ion channels: Applications in neurobiology and beyond. Neurosci. Lett. 2018, 679, 15–23. [Google Scholar] [CrossRef]
- Beeton, C.; Wulff, H.; Singh, S.; Botsko, S.; Crossley, G.; Gutman, G.A.; Cahalan, M.D.; Pennington, M.; Chandy, K.G. A novel fluorescent toxin to detect and investigate Kv1.3 channel up-regulation in chronically activated T lymphocytes. J. Biol. Chem. 2003, 278, 9928–9937. [Google Scholar] [CrossRef] [Green Version]
- Pragl, B.; Koschak, A.; Trieb, M.; Obermair, G.; Kaufmann, W.A.; Gerster, U.; Blanc, E.; Hahn, C.; Prinz, H.; Schütz, G.; et al. Synthesis, characterization, and application of Cy-Dye- and Alexa-Dye-Labeled Hongotoxin 1 Analogues. The first high affinity fluorescence probes for Voltage-Gated K + channels. Bioconjug. Chem. 2002, 13, 416–425. [Google Scholar] [CrossRef]
- Nekrasova, O.V.; Ignatova, A.A.; Nazarova, A.I.; Feofanov, A.V.; Korolkova, Y.V.; Boldyreva, E.F.; Tagvei, A.I.; Grishin, E.V.; Arseniev, A.S.; Kirpichnikov, M.P. Recombinant Kv channels at the membrane of escherichia coli bind specifically agitoxin2. J. Neuroimmune Pharmacol. 2009, 4, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenkov, A.I.; Nekrasova, O.V.; Kudryashova, K.S.; Peigneur, S.; Tytgat, J.; Stepanov, A.V.; Kirpichnikov, M.P.; Grishin, E.V.; Feofanov, A.V.; Vassilevski, A.A. Fluorescent protein-scorpion toxin chimera is a convenient molecular tool for studies of potassium channels. Sci. Rep. 2016, 6, 33314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, M.L.; Garcia-Calvo, M.; Hidalgo, P.; Lee, A.; MacKinnon, R. Purification and Characterization of Three Inhibitors of Voltage-Dependent K+ Channels from Leiurus quinquestriatus var. hebraeus Venom. Biochemistry 1994, 33, 6834–6839. [Google Scholar] [CrossRef]
- Takacs, Z.; Toups, M.; Kollewe, A.; Johnson, E.; Cuello, L.G.; Driessens, G.; Biancalana, M.; Koide, A.; Ponte, C.G.; Perozo, E.; et al. A designer ligand specific for Kv1.3 channels from a scorpion neurotoxin-based library. Proc. Natl. Acad. Sci. USA 2009, 106, 22211–22216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudryashova, K.S.; Nekrasova, O.V.; Kuzmenkov, A.I.; Vassilevski, A.A.; Ignatova, A.A.; Korolkova, Y.V.; Grishin, E.V.; Kirpichnikov, M.P.; Feofanov, A. V Fluorescent system based on bacterial expression of hybrid KcsA channels designed for Kv1.3 ligand screening and study. Anal. Bioanal. Chem. 2013, 405, 2379–2389. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenkov, A.I.; Vassilevski, A.A.; Kudryashova, K.S.; Nekrasova, O.V.; Peigneur, S.; Tytgat, J.; Feofanov, A.V.; Kirpichnikov, M.P.; Grishin, E.V. Variability of potassium channel blockers in Mesobuthus eupeus scorpion venom with focus on Kv1.1: An integrated transcriptomic and proteomic study. J. Biol. Chem. 2015, 290, 12195–12209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nekrasova, O.V.; Volyntseva, A.D.; Kudryashova, K.S.; Novoseletsky, V.N.; Lyapina, E.A.; Illarionova, A.V.; Yakimov, S.A.; Korolkova, Y.V.; Shaitan, K.V.; Kirpichnikov, M.P.; et al. Complexes of peptide blockers with Kv1.6 pore domain: Molecular modeling and studies with KcsA-Kv1.6 channel. J. Neuroimmune Pharmacol. 2017, 12, 260–276. [Google Scholar] [CrossRef]
- Hoang, A.N.; Vo, H.D.M.; Vo, N.P.; Kudryashova, K.S.; Nekrasova, O.V.; Feofanov, A.V.; Kirpichnikov, M.P.; Andreeva, T.V.; Serebryakova, M.V.; Tsetlin, V.I.; et al. Vietnamese Heterometrus laoticus scorpion venom: Evidence for analgesic and anti-inflammatory activity and isolation of new polypeptide toxin acting on Kv1.3 potassium channel. Toxicon 2014, 77, 40–48. [Google Scholar] [CrossRef]
- Legros, C.; Pollmann, V.; Knaus, H.G.; Farrell, A.M.; Darbon, H.; Bougis, P.E.; Martin-Eauclaire, M.F.; Pongs, O. Generating a high affinity scorpion toxin receptor in KcsA-Kv1.3 chimeric potassium channels. J. Biol. Chem. 2000, 275, 16918–16924. [Google Scholar] [CrossRef] [Green Version]
- Orlov, N.; Ignatova, A.; Nekrasova, O.; Kirpichnikov, M.; Feofanov, A. Design of far-red fluorescent Kv1.3 channel for membrane expression in eukaryotic cells and its interactions with hongotoxin1. Microsc. Microanal. 2020, 1–2. [Google Scholar] [CrossRef]
- Voros, O.; Szilagyi, O.; Balajthy, A.; Somodi, S.; Panyi, G.; Hajdu, P. The C-terminal HRET sequence of Kv1.3 regulates gating rather than targeting of Kv1.3 to the plasma membrane. Sci. Rep. 2018, 8, 5937. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Chen, Y. Progress in Research of KV1.1 and KV1.3 Channels as Therapeutic Targets. Curr. Top. Med. Chem. 2016, 16, 1877–1885. [Google Scholar] [CrossRef] [PubMed]
- Volyntseva, A.D.; Novoseletsky, V.N.; Shaitan, K.V.; Feofanov, A.V. Molecular modeling of interactions of agitoxin 2 with Kv1.3 voltage-gated potassium channel. Moscow Univ. Biol. Sci. Bull. 2017, 72, 25–29. [Google Scholar] [CrossRef]
- Suzuki, N.; Hiraki, M.; Yamada, Y.; Matsugaki, N.; Igarashi, N.; Kato, R.; Dikic, I.; Drew, D.; Iwata, S.; Wakatsuki, S.; et al. Crystallization of small proteins assisted by green fluorescent protein. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Akerboom, J.; Rivera, J.D.V.; Guilbe, M.M.R.; Malavé, E.C.A.; Hernandez, H.H.; Tian, L.; Hires, S.A.; Marvin, J.S.; Looger, L.L.; Schreiter, E.R. Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design. J. Biol. Chem. 2009, 284, 6455–6464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrer-Miralles, N.; Corchero, J.L.; Kumar, P.; Cedano, J.A.; Gupta, K.C.; Villaverde, A.; Vazquez, E. Biological activities of histidine-rich peptides; merging biotechnology and nanomedicine. Microb. Cell Fact. 2011, 10, 101. [Google Scholar] [CrossRef] [Green Version]
- Nekrasova, O.; Kudryashova, K.; Fradkov, A.; Yakimov, S.; Savelieva, M.; Kirpichnikov, M.; Feofanov, A. Straightforward approach to produce recombinant scorpion toxins—Pore blockers of potassium channels. J. Biotechnol. 2017, 241, 127–135. [Google Scholar] [CrossRef]
- Nekrasova, O.; Yakimov, S.; Kirpichnikov, M.; Feofanov, A. Recombinant scorpion toxins: Focus on four-disulfide peptide blockers of Kv1-channels. Bioengineered 2018, 9, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Kubota, T.; Correa, A.M.; Bezanilla, F. Mechanism of functional interaction between potassium channel Kv1.3 and sodium channel NavBeta1 subunit. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; De Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2016, 14, 71–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]

| Kap (pM) | ||||
|---|---|---|---|---|
| Fluorescent Ligand | AgTx2 | KTx1 | ChTx | HeTx |
| GFP-L2-AgTx2 | 230 ± 50 | 100 ± 40 | 1500 ± 300 | 39,000 ± 4000 |
| His6-GFP-L2-AgTx2 | 70 ± 4 | 71 ± 7 | 900 ± 170 | 45,000 ± 8000 |
| Notation | Nucleotide Sequence * |
|---|---|
| GFP-f1 | 5′-TTCTTCCATGGTGAGCAAGGGCGAGGAGCTGTT-3′ |
| GFP-r1 | 5′-TTCTTGAATTCTTGTACAGCTCGTCCATGCCGAGA-3′ |
| GFP-f2 | 5′-CACAGCAGTGGCGAAAACCTGTACTTTCAGGGTATGGTGAG CAAGGGCGAG-3′ |
| GFP-r2 | 5′-TTCTTGGTACCTCCCGAACCTCCCGAGCCTCCCTC GTAC AGCTCGTCCATGCCGA-3′ |
| GFP-f3 | 5′-CCTCCTCCATGGGCAGCTCTCATCACCATCACCATCACAGCAGT GGCGAAAAC-3′ |
| Ag-1f | 5′-TTCTAGGTACCGGTGGCGCGGGAGGCGCGGGAGGTACGG GTGGATCC GGCGTTCCGA-3′ |
| Ag-2f | 5′-GGTTCTCCACAGTGTATCAAACCGTGCAAAGATGCAGGCATGC GCTTTGGCAAATG-3′ |
| Ag-1r | 5′-TGATACACTGTGGAGAACCCGTGCAGCTCACGTTGATCGG AACGCCGGAT-3′ |
| Ag-2r | 5′-TTCTTCAAGCTTACTTCGGCGTGCAGTGACACTTACGATT CATGCATTTGCCAAAGCGCA-3′ |
| GFP-f4 | 5′-CGGAGGTAGTGGAGGCAGCGGAGGCACCGGTGGCGCAG GCGGTGCAACGTCGACCATGGTGAGCAAGGGCGAG-3′ |
| GFP-r3 | 5′-TTCTTCTCGAGTTACTTGTACAGCTCGTCCATGCCGAGAG-3′ |
| GFP-f5 | 5′-TGCAGGCATGCGCTTTGGCAAATGCATGAATCGTAAGTGTC ACTACGCCGAAGGGATCCGGAGGTAGTGGAGGCA-3′ |
| KCNA-del-f1 | 5′-TTCTCAGATCTATGACCGTGGTGCCCGGGGACCA-3′ |
| KCNA-r1 | 5′-TTCTCAAGCTTAAACATCGGTGAATATCTTTTTGATGTTGA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nekrasova, O.V.; Primak, A.L.; Ignatova, A.A.; Novoseletsky, V.N.; Geras’kina, O.V.; Kudryashova, K.S.; Yakimov, S.A.; Kirpichnikov, M.P.; Arseniev, A.S.; Feofanov, A.V. N-Terminal Tagging with GFP Enhances Selectivity of Agitoxin 2 to Kv1.3-Channel Binding Site. Toxins 2020, 12, 802. https://doi.org/10.3390/toxins12120802
Nekrasova OV, Primak AL, Ignatova AA, Novoseletsky VN, Geras’kina OV, Kudryashova KS, Yakimov SA, Kirpichnikov MP, Arseniev AS, Feofanov AV. N-Terminal Tagging with GFP Enhances Selectivity of Agitoxin 2 to Kv1.3-Channel Binding Site. Toxins. 2020; 12(12):802. https://doi.org/10.3390/toxins12120802
Chicago/Turabian StyleNekrasova, Oksana V., Alexandra L. Primak, Anastasia A. Ignatova, Valery N. Novoseletsky, Olga V. Geras’kina, Ksenia S. Kudryashova, Sergey A. Yakimov, Mikhail P. Kirpichnikov, Alexander S. Arseniev, and Alexey V. Feofanov. 2020. "N-Terminal Tagging with GFP Enhances Selectivity of Agitoxin 2 to Kv1.3-Channel Binding Site" Toxins 12, no. 12: 802. https://doi.org/10.3390/toxins12120802
APA StyleNekrasova, O. V., Primak, A. L., Ignatova, A. A., Novoseletsky, V. N., Geras’kina, O. V., Kudryashova, K. S., Yakimov, S. A., Kirpichnikov, M. P., Arseniev, A. S., & Feofanov, A. V. (2020). N-Terminal Tagging with GFP Enhances Selectivity of Agitoxin 2 to Kv1.3-Channel Binding Site. Toxins, 12(12), 802. https://doi.org/10.3390/toxins12120802






