Several New Putative Bacterial ADP-Ribosyltransferase Toxins Are Revealed from In Silico Data Mining, Including the Novel Toxin Vorin, Encoded by the Fire Blight Pathogen Erwinia amylovora
Abstract
1. Introduction
2. Results and Discussion
2.1. Bioinformatics Genome Mining and Candidate Mono-ADP-Ribosyltransferase (mART) Sequence Filtering
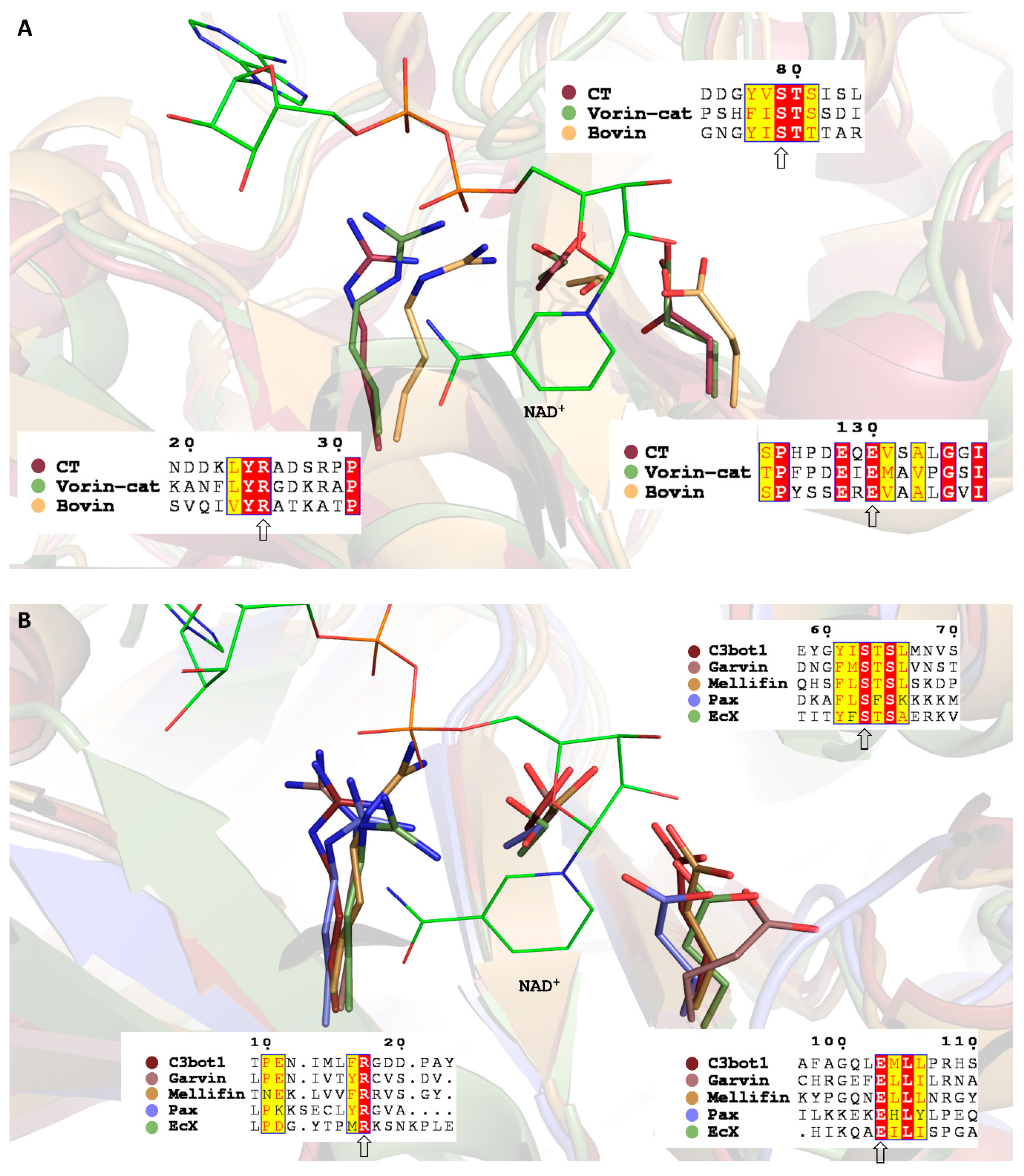
2.2. Bovin
2.3. EcX
2.4. Mellifin
2.5. Pax
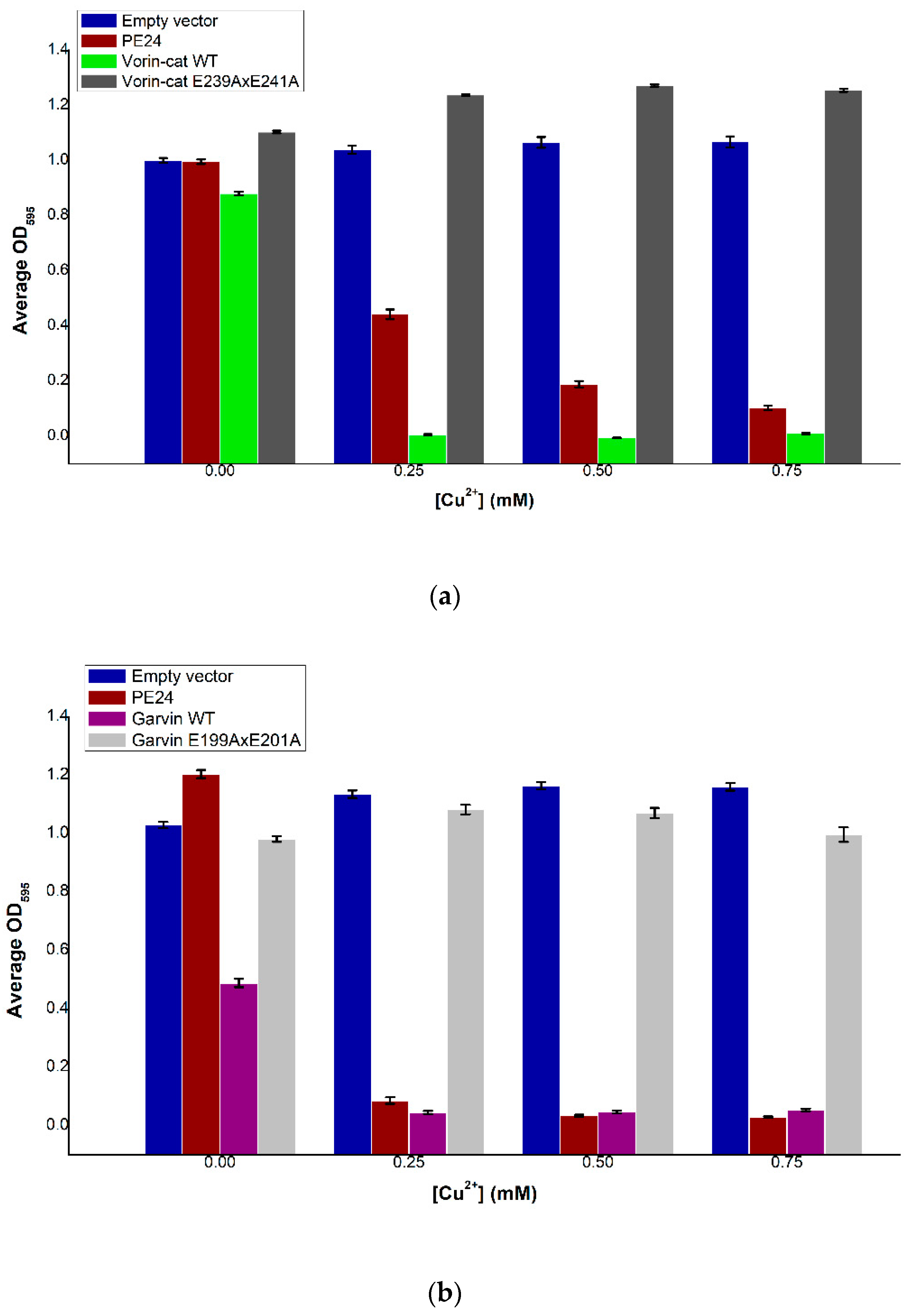
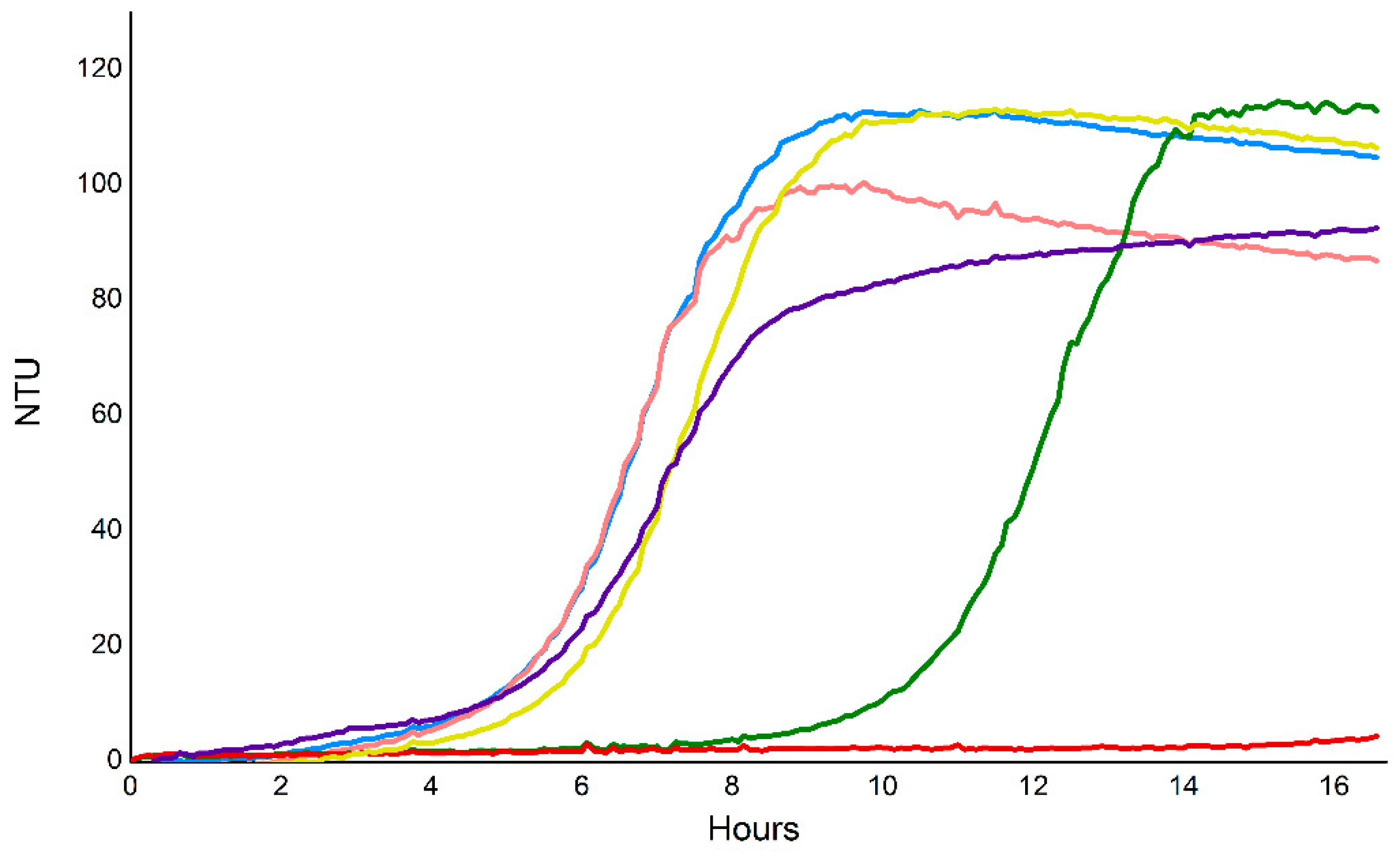
2.6. Garvin
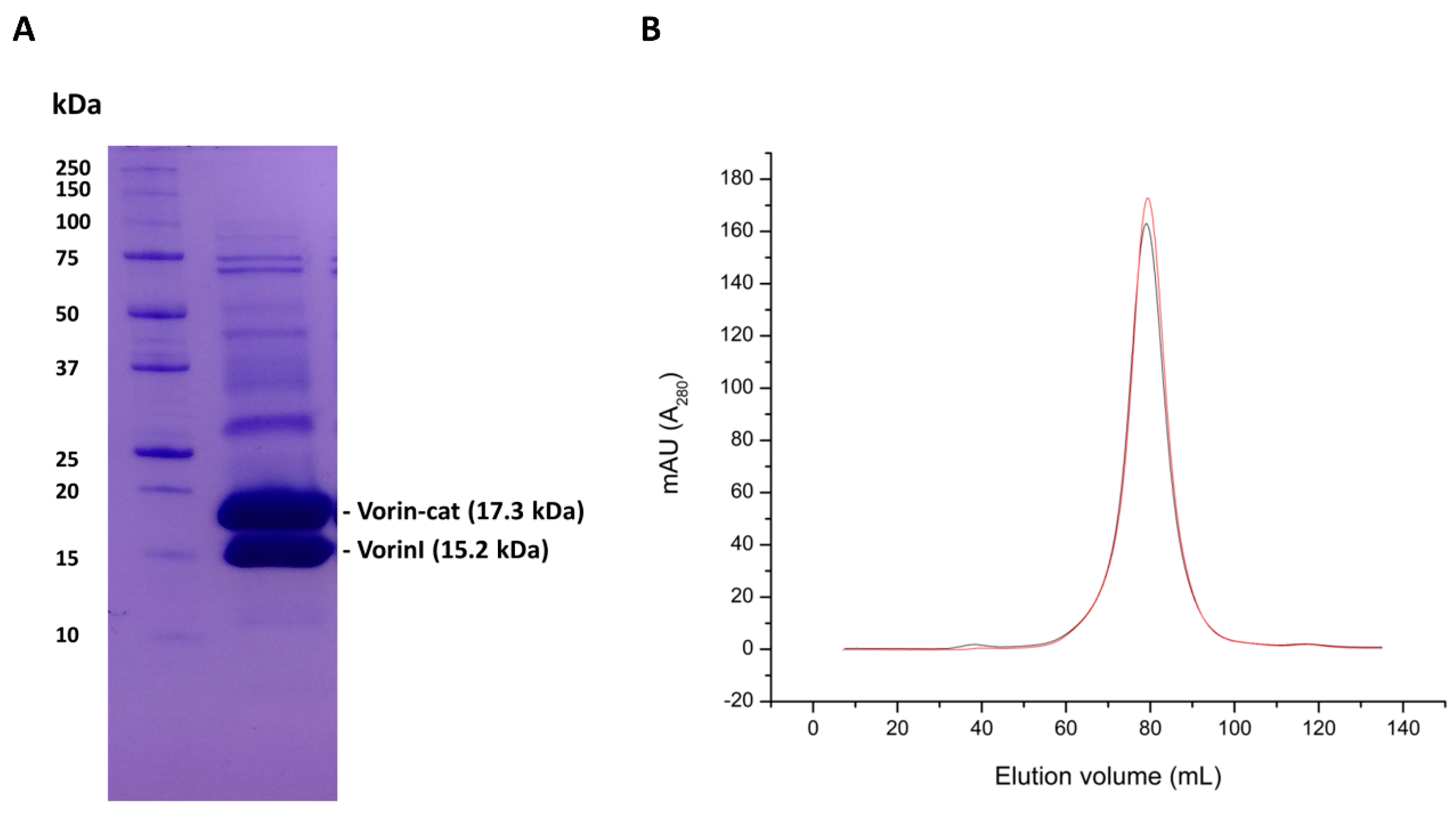
2.7. Vorin
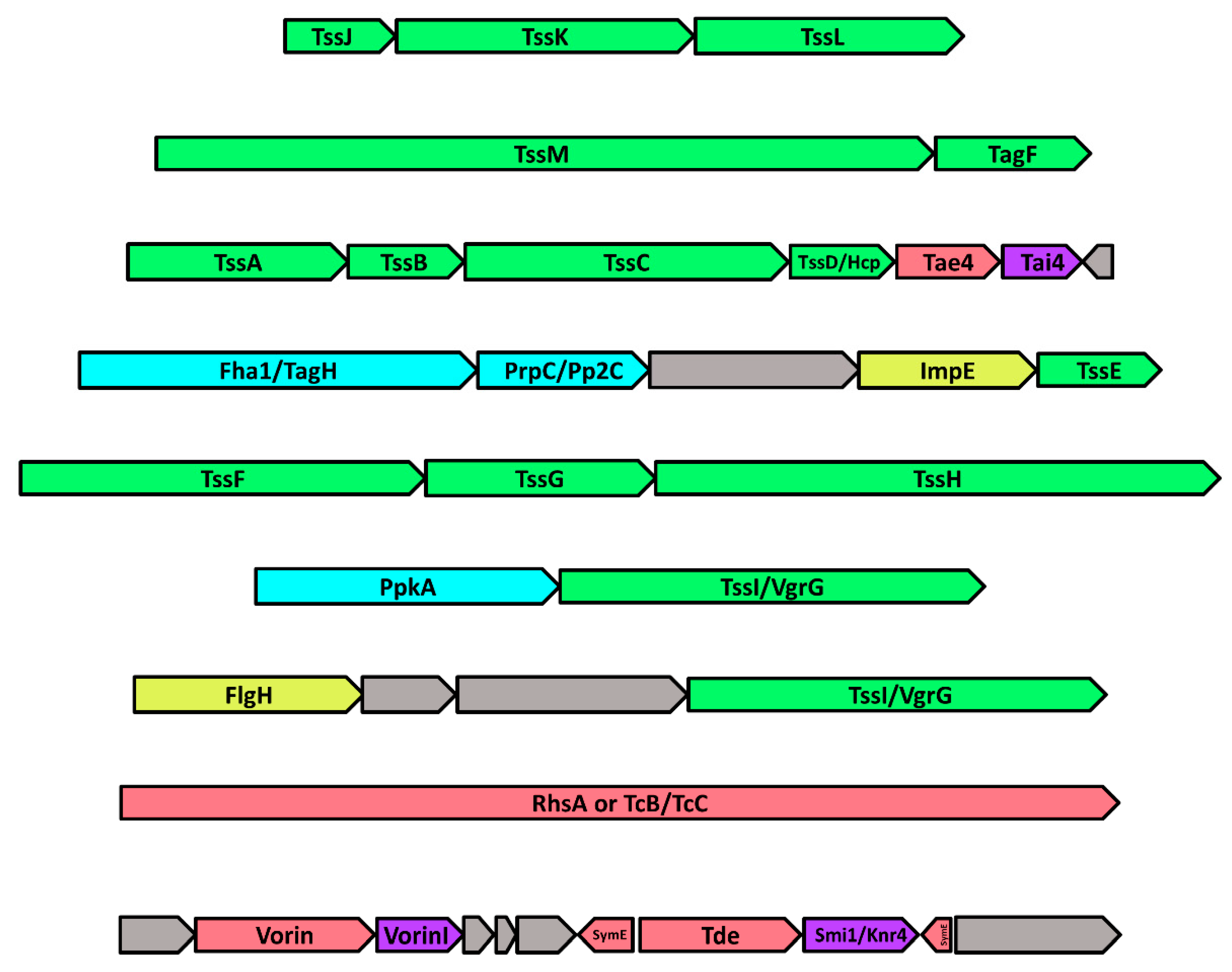
2.8. The Genomic Neighbourhood of Vorin: Type-VI Secretion and Toxin-Antitoxin Modules
2.9. Vorin and Garvin Yeast Growth Deficiency Assays: Validation of Bioinformatics Predictions
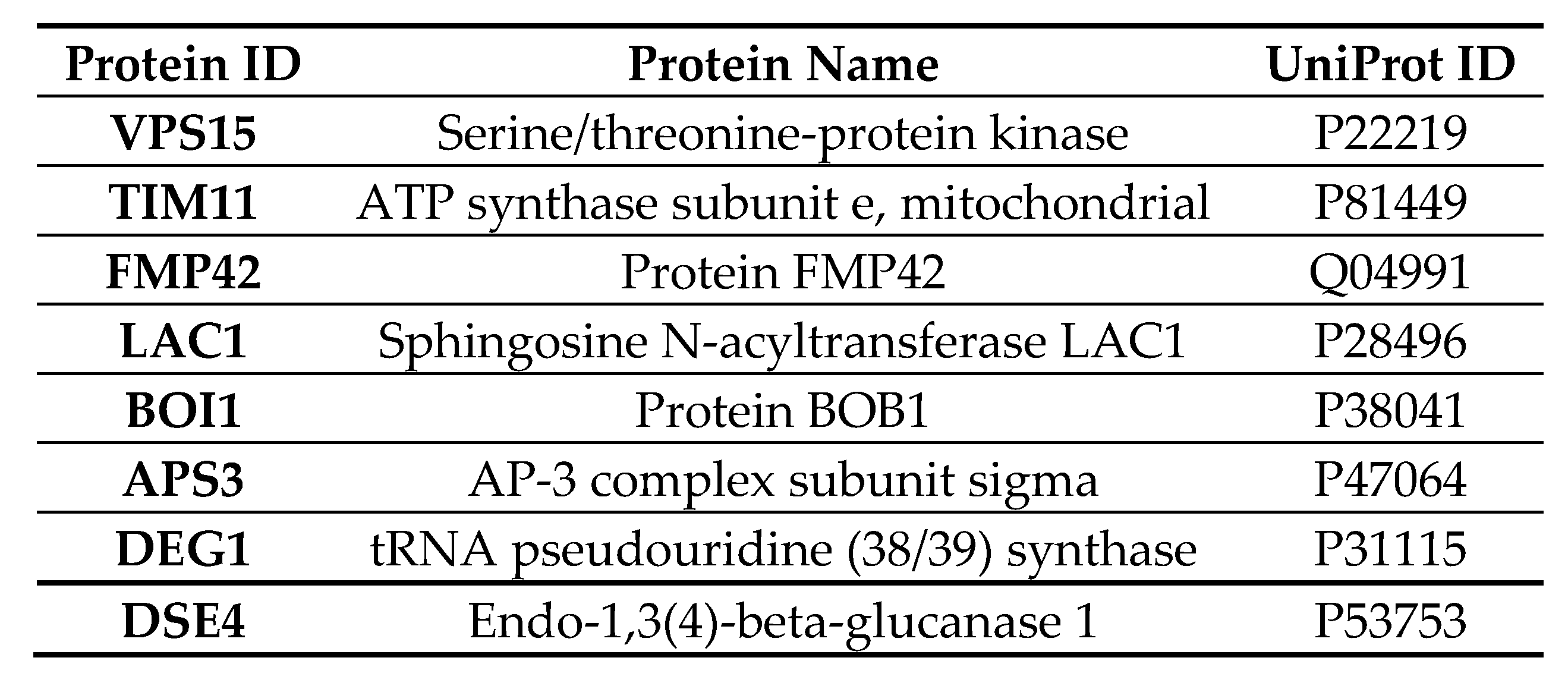
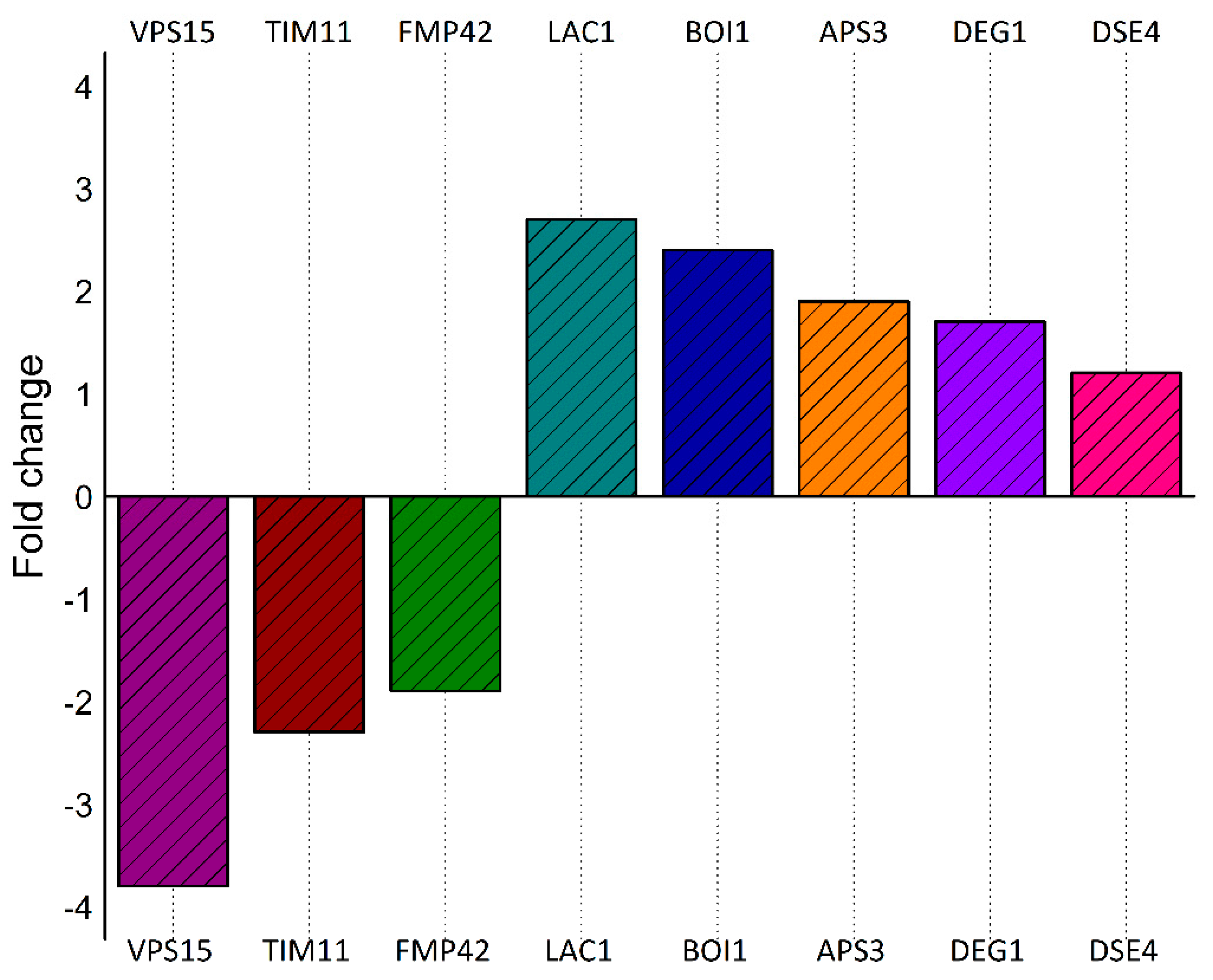
2.10. Quantitative Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS) of Cells Expressing Vorin-Cat: A Possible Role in the Suppression of Autophagy
2.11. Expression of Vorin-Cat Requires Co-Expression with Its Cognate Immunity Protein VorinI
3. Conclusions
4. Materials and Methods
4.1. Database Search for Candidate mART Toxin Sequences and Sequence Processing
4.2. Bacterial Genomic DNA
4.3. Transformation of Saccharomyces Cerevisiae and Yeast Growth Deficiency Assay
4.4. Site-Directed Mutants of Vorin-Cat and Garvin
4.5. Preparation of Yeast Cell Protein for LC-MS/MS
4.6. LC-MS/MS Data Analysis
4.7. Transformation, Expression and Purification of Vorin-Cat and VorinI
4.8. E. coli Cell Viability Assays
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Holbourn, K.P.; Shone, C.C.; Acharya, K.R. A family of killer toxins: Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 2006, 273, 4579–4593. [Google Scholar] [CrossRef]
- Corda, D.; Di Girolamo, M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003, 22, 1953–1958. [Google Scholar] [CrossRef] [PubMed]
- Simon, N.C.; Aktories, K.; Barbieri, J.T. Novel bacterial ADP-ribosylating toxins: Structure and function. Nat. Rev. Microbiol. 2014, 12, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Liu, C.; Shan, L.; He, P. Protein ADP-Ribosylation Takes Control in Plant–Bacterium Interactions. PLoS Pathog. 2016, 12, e1005941. [Google Scholar] [CrossRef] [PubMed]
- Lyons, B.; Ravulapalli, R.; Lanoue, J.; Lugo, M.R.; Dutta, D.; Carlin, S.; Merrill, A.R. Scabin, a novel DNA-acting ADP-ribosyltransferase from Streptomyces scabies. J. Biol. Chem. 2016, 291, 11198–11215. [Google Scholar] [CrossRef] [PubMed]
- Ebeling, J.; Funfhaus, A.; Knispel, H.; Krska, D.; Ravulapalli, R.; Heney, K.A.; Lugo, M.R.; Merrill, A.R.; Genersch, E. Characterization of the toxin Plx2A, a RhoA-targeting ADP-ribosyltransferase produced by the honey bee pathogen Paenibacillus larvae. Environ. Microbiol. 2017, 19, 5100–5116. [Google Scholar] [CrossRef] [PubMed]
- Jank, T.; Aktories, K. Strain-alleviation model of ADP-ribosylation. Proc. Natl. Acad. Sci. USA 2013, 110, 4163–4164. [Google Scholar] [CrossRef]
- Tsurumura, T.; Tsumori, Y.; Qiu, H.; Oda, M.; Sakurai, J.; Nagahama, M.; Tsuge, H. Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex. Proc. Natl. Acad. Sci. USA 2013, 110, 4267–4272. [Google Scholar] [CrossRef]
- Mateyak, M.K.; Kinzy, T.G. ADP-ribosylation of translation elongation factor 2 by diphtheria toxin in yeast inhibits translation and cell separation. J. Biol. Chem. 2013, 288, 24647–24655. [Google Scholar] [CrossRef]
- Jørgensen, R.; Merrill, A.R.; Andersen, G.R. The life and death of translation elongation factor 2. Biochem. Soc. Trans. 2006, 34, 1–6. [Google Scholar] [CrossRef]
- Gupta, P.K.; Liu, S.; Batavia, M.P.; Leppla, S.H. The diphthamide modification on elongation factor-2 renders mammalian cells resistant to ricin. Cell. Microbiol. 2008, 10, 1687–1694. [Google Scholar] [CrossRef] [PubMed]
- Torgersen, M.L.; Skretting, G.; van Deurs, B.; Sandvig, K. Internalization of cholera toxin by different endocytic mechanisms. J. Cell Sci. 2001, 114, 3737–3747. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.G.; Scott, D.L.; Westbrook, M.L.; Nance, S.; Spangler, B.D.; Shipley, G.G.; Westbrook, E.M. The three-dimensional crystal structure of cholera toxin. J. Mol. Biol. 1995, 251, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Aktories, K.; Lang, A.E.; Schwan, C.; Mannherz, H.G. Actin as target for modification by bacterial protein toxins. FEBS J. 2011, 278, 4526–4543. [Google Scholar] [CrossRef]
- Margarit, S.M.; Davidson, W.; Frego, L.; Stebbins, C.E. A Steric Antagonism of Actin Polymerization by a Salmonella Virulence Protein. Structure 2006, 14, 1219–1229. [Google Scholar] [CrossRef]
- Vogelsgesang, M.; Pautsch, A.; Aktories, K. C3 exoenzymes, novel insights into structure and action of Rho-ADP-ribosylating toxins. Naunyn. Schmiedebergs. Arch. Pharmacol. 2007, 374, 347–360. [Google Scholar] [CrossRef]
- Heasman, S.J.; Ridley, A.J. Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008, 9, 690–701. [Google Scholar] [CrossRef]
- Nakano, T.; Takahashi-Nakaguchi, A.; Yamamoto, M.; Watanabe, M. Pierisins and CARP-1: ADP-ribosylation of DNA by ARTCs in butterflies and shellfish. In Current Topics in Microbiology and Immunology; Koch-Nolte, F., Ed.; Springer International Publishing: Cham, Switzerland, 2014; Volume 384, pp. 127–149. ISBN 0070-217X. (Print)r0070-217X (Linking). [Google Scholar]
- Takamura-Enya, T.; Watanabe, M.; Totsuka, Y.; Kanazawa, T.; Matsushima-Hibiya, Y.; Koyama, K.; Sugimura, T.; Wakabayashi, K. Mono(ADP-ribosyl)ation of 2’-deoxyguanosine residue in DNA by an apoptosis-inducing protein, pierisin-1, from cabbage butterfly. Proc. Natl. Acad. Sci. USA 2001, 98, 12414–12419. [Google Scholar] [CrossRef]
- Choe, S.; Bennett, M.J.; Fujii, G.; Curmi, P.M.G.; Kantardjieff, K.A.; Collier, R.J.; Eisenberg, D. The crystal structure of diphtheria toxin. Nature 1992, 357, 216–222. [Google Scholar] [CrossRef]
- Weldon, J.E.; Pastan, I. A guide to taming a toxin—Recombinant immunotoxins constructed from Pseudomonas exotoxin A for the treatment of cancer. FEBS J. 2011, 278, 4683–4700. [Google Scholar] [CrossRef]
- Jørgensen, R.; Purdy, A.E.; Fieldhouse, R.J.; Kimber, M.S.; Bartlett, D.H.; Merrill, A.R. Cholix toxin, a novel ADP-ribosylating factor from Vibrio cholerae. J. Biol. Chem. 2008, 283, 10671–10678. [Google Scholar] [CrossRef] [PubMed]
- Schleberger, C.; Hochmann, H.; Barth, H.; Aktories, K.; Schulz, G.E. Structure and Action of the Binary C2 Toxin from Clostridium botulinum. J. Mol. Biol. 2006, 364, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Sixma, T.K.; Kalk, K.H.; van Zanten, B.A.M.; Dauter, Z.; Kingma, J.; Witholt, B.; Hol, W.G.J. Refined structure of Escherichia coli heat-labile enterotoxin, a close relative of cholera toxin. J. Mol. Biol. 1993, 230, 890–918. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.G.; Westbrook, M.L.; Westbrook, E.M.; Scott, D.L.; Otwinowski, Z.; Maulik, P.R.; Reed, R.A.; Shipley, G.G. The 2.4 A crystal structure of cholera toxin B subunit pentamer: Choleragenoid. J. Mol. Biol. 1995, 251, 550–562. [Google Scholar] [CrossRef]
- Rohrbeck, A.; Just, I. Cell Entry of C3 Exoenzyme from Clostridium botulinum. Curr. Top. Microbiol. Immunol. 2017, 406, 97–118. [Google Scholar]
- Fieldhouse, R.J.; Merrill, A.R. Needle in the haystack: Structure-based toxin discovery. Trends Biochem. Sci. 2008, 33, 546–556. [Google Scholar] [CrossRef]
- Domenighini, M.; Rappuoli, R. Three conserved consensus sequences identify the NAD-binding site of ADP-ribosylating enzymes, expressed by eukaryotes, bacteria and T-even bacteriophages. Mol. Microbiol. 1996, 21, 667–674. [Google Scholar] [CrossRef]
- Han, S.; Tainer, J.A. The ARTT motif and a unified structural understanding of substrate recognition in ADP-ribosylating bacterial toxins and eukaryotic ADP-ribosyltransferases. Int. J. Med. Microbiol. 2002, 291, 523–529. [Google Scholar] [CrossRef]
- Fieldhouse, R.J.; Turgeon, Z.; White, D.; Rod Merrill, A. Cholera- and anthrax-like toxins are among several new ADP-Ribosyltransferases. PLoS Comput. Biol. 2010, 6, e1001029. [Google Scholar] [CrossRef]
- Vogelsgesang, M.; Aktories, K. Exchange of glutamine-217 to glutamate of Clostridium limosum exoenzyme C3 turns the asparagine-specific ADP-ribosyltransferase into an arginine-modifying enzyme. Biochemistry 2006, 45, 1017–1025. [Google Scholar] [CrossRef]
- Akiyuki, T.; Toshiharu, T.; Toru, Y.; Yayoi, T.; Tsuge, H. Rho GTPase recognition by C3 exoenzyme based on C3-RhoA complex structure. J. Biol. Chem. 2015, 290, 19423–19432. [Google Scholar] [CrossRef]
- Ménétrey, J.; Flatau, G.; Stura, E.A.; Charbonnier, J.B.; Gas, F.; Teulon, J.M.; Le Du, M.H.; Boquetand, P.; Ménez, A. NAD binding induces conformational changes in Rho ADP-ribosylating Clostridium botulinum C3 exoenzyme. J. Biol. Chem. 2002, 277, 30950–30957. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.; Tremblay, O.; Heney, K.A.; Lugo, M.R.; Ebeling, J.; Genersch, E.; Merrill, A.R. Characterization of C3larvinA, a novel RhoA-targeting ADP-ribosyltransferase toxin produced by the honey bee pathogen, Paenibacillus larvae. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Remmert, M.; Biegert, A.; Hauser, A.; Söding, J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 2011, 9, 173–175. [Google Scholar] [CrossRef]
- Söding, J.; Remmert, M.; Biegert, A.; Lupas, A.N. HHsenser: Exhaustive transitive profile search using HMM-HMM comparison. Nucleic Acids Res. 2006, 34, 374–378. [Google Scholar] [CrossRef]
- Turgeon, Z.; White, D.; Jørgensen, R.; Visschedyk, D.; Fieldhouse, R.J.; Mangroo, D.; Merrill, A.R. Yeast as a tool for characterizing mono-ADP-ribosyltransferase toxins. FEMS Microbiol. Lett. 2009, 300, 97–106. [Google Scholar] [CrossRef]
- Cegelski, L.; Marshall, G.R.; Eldridge, G.R.; Hultgren, S.J. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 2008, 6, 17–27. [Google Scholar] [CrossRef]
- Clatworthy, A.E.; Pierson, E.; Hung, D.T. Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007, 3, 541–548. [Google Scholar] [CrossRef]
- Turgeon, Z.; Jørgensen, R.; Visschedyk, D.; Edwards, P.R.; Legree, S.; McGregor, C.; Fieldhouse, R.J.; Mangroo, D.; Schapira, M.; Merrill, A.R. Newly discovered and characterized antivirulence compounds inhibit bacterial mono-ADP-ribosyltransferase toxins. Antimicrob. Agents Chemother. 2011, 55, 983–991. [Google Scholar] [CrossRef]
- Visschedyk, D.; Rochon, A.; Tempel, W.; Dimov, S.; Park, H.W.; Merrill, A.R. Certhrax toxin, an anthrax-related ADP-ribosyltransferase from Bacillus cereus. J. Biol. Chem. 2012, 287, 41089–41102. [Google Scholar] [CrossRef]
- Juhas, M.; van der Meer, J.R.; Gaillard, M.; Harding, R.M.; Hood, D.W.; Crook, D.W. Genomic islands: Tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009, 33, 376–393. [Google Scholar] [CrossRef] [PubMed]
- Ference, C.M.; Gochez, A.M.; Behlau, F.; Wang, N.; Graham, J.H.; Jones, J.B. Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Mol. Plant Pathol. 2018, 19, 1302–1318. [Google Scholar] [CrossRef] [PubMed]
- Maillard, R.; Petit, E.; Chomel, B.; Lacroux, C.; Schelcher, F.; Vayssier-Taussat, M.; Haddad, N.; Boulouis, H.-J. Endocarditis in cattle caused by Bartonella bovis. Emerg. Infect. Dis. 2007, 13, 1383–1385. [Google Scholar] [CrossRef] [PubMed]
- Maillard, R.; Grimard, B.; Chastant-Maillard, S.; Chomel, B.; Delcroix, T.; Gandoin, C.; Bouillin, C.; Halos, L.; Vayssier-Taussat, M.; Boulouis, H.-J. Effects of cow age and pregnancy on Bartonella infection in a herd of dairy cattle. J. Clin. Microbiol. 2006, 44, 42–46. [Google Scholar] [CrossRef]
- Tay, S.T.; Kho, K.L.; Lye, S.F.; Ngeow, Y.F. Phylogeny and putative virulence gene analysis of Bartonella bovis. J. Vet. Med. Sci. 2018, 80, 653–661. [Google Scholar] [CrossRef]
- Lugo, M.R.; Lyons, B.; Lento, C.; Wilson, D.J.; Merrill, A.R. Dynamics of Scabin toxin. A proposal for the binding mode of the DNA substrate. PLoS ONE 2018, 13, e0194425. [Google Scholar] [CrossRef]
- Pati, N.B.; Doijad, S.P.; Schultze, T.; Mannala, G.K.; Yao, Y.; Jaiswal, S.; Ryan, D.; Suar, M.; Gwozdzinski, K.; Bunk, B.; et al. Enterobacter bugandensis: A novel enterobacterial species associated with severe clinical infection. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Mezzatesta, M.L.; Gona, F.; Stefani, S. Enterobacter cloacae complex: Clinical impact and emerging antibiotic resistance. Future Microbiol. 2012, 7, 887–902. [Google Scholar] [CrossRef]
- Meusch, D.; Gatsogiannis, C.; Efremov, R.G.; Lang, A.E.; Hofnagel, O.; Vetter, I.R.; Aktories, K.; Raunser, S. Mechanism of Tc toxin action revealed in molecular detail. Nature 2014, 508, 61–65. [Google Scholar] [CrossRef]
- Busby, J.N.; Panjikar, S.; Landsberg, M.J.; Hurst, M.R.H.; Lott, J.S. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature 2013, 501, 547–550. [Google Scholar] [CrossRef]
- Lo, W.-S.; Chen, L.-L.; Chung, W.-C.; Gasparich, G.E.; Kuo, C.-H. Comparative genome analysis of Spiroplasma melliferum IPMB4A, a honeybee-associated bacterium. BMC Genom. 2013, 14, 22. [Google Scholar] [CrossRef] [PubMed]
- Alexeev, D.; Kostrjukova, E.; Aliper, A.; Popenko, A.; Bazaleev, N.; Tyakht, A.; Selezneva, O.; Akopian, T.; Prichodko, E.; Kondratov, I.; et al. Application of Spiroplasma melliferum proteogenomic profiling for the discovery of virulence factors and pathogenicity mechanisms in host-associated spiroplasmas. J. Proteome Res. 2012, 11, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Carle, P.; Saillard, C.; Carrère, N.; Carrère, S.; Duret, S.; Eveillard, S.; Gaurivaud, P.; Gourgues, G.; Gouzy, J.; Salar, P.; et al. Partial Chromosome Sequence of Spiroplasma citri Reveals Extensive Viral Invasion and Important Gene Decay. Appl. Environ. Microbiol. 2010, 76, 3420–3426. [Google Scholar] [CrossRef] [PubMed]
- Greub, G.; Raoult, D. Parachlamydiaceae: Potential emerging pathogens. Emerg. Infect. Dis. 2002, 8, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Greub, G.; Mege, J.-L.; Gorvel, J.-P.; Raoult, D.; Meresse, S. Intracellular trafficking of Parachlamydia acanthamoebae. Cell. Microbiol. 2005, 7, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.; Collingro, A.; Walochnik, J.; Wagner, M.; Horn, M. Chlamydia-like bacteria in respiratory samples of community-acquired pneumonia patients. FEMS Microbiol. Lett. 2008, 281, 198–202. [Google Scholar] [CrossRef]
- Meyburgh, C.M.; Bragg, R.R.; Boucher, C.E. Lactococcus garvieae: An emerging bacterial pathogen of fish. Dis. Aquat. Organ. 2017, 123, 67–79. [Google Scholar] [CrossRef]
- Vendrell, D.; Balcazar, J.L.; Ruiz-Zarzuela, I.; de Blas, I.; Girones, O.; Muzquiz, J.L. Lactococcus garvieae in fish: A review. Comp. Immunol. Microbiol. Infect. Dis. 2006, 29, 177–198. [Google Scholar] [CrossRef]
- Eyngor, M.; Zlotkin, A.; Ghittino, C.; Prearo, M.; Douet, D.-G.; Chilmonczyk, S.; Eldar, A. Clonality and diversity of the fish pathogen Lactococcus garvieae in Mediterranean countries. Appl. Environ. Microbiol. 2004, 70, 5132–5137. [Google Scholar] [CrossRef]
- Reguera-Brito, M.; Galan-Sanchez, F.; Blanco, M.M.; Rodriguez-Iglesias, M.; Dominguez, L.; Fernandez-Garayzabal, J.F.; Gibello, A. Genetic analysis of human clinical isolates of Lactococcus garvieae: Relatedness with isolates from foods. Infect. Genet. Evol. 2016, 37, 185–191. [Google Scholar] [CrossRef]
- Aguado-Urda, M.; Gibello, A.; Blanco, M.M.; Lopez-Campos, G.H.; Cutuli, M.T.; Fernandez-Garayzabal, J.F. Characterization of plasmids in a human clinical strain of Lactococcus garvieae. PLoS ONE 2012, 7, e40119. [Google Scholar] [CrossRef] [PubMed]
- Gabrielsen, C.; Brede, D.A.; Hernandez, P.E.; Nes, I.F.; Diep, D.B. Genome sequence of the bacteriocin-producing strain Lactococcus garvieae DCC43. J. Bacteriol. 2012, 194, 6976–6977. [Google Scholar] [CrossRef] [PubMed]
- Malnoy, M.; Martens, S.; Norelli, J.L.; Barny, M.-A.; Sundin, G.W.; Smits, T.H.M.; Duffy, B. Fire Blight: Applied Genomic Insights of the Pathogen and Host. Annu. Rev. Phytopathol. 2011, 50, 475–494. [Google Scholar] [CrossRef] [PubMed]
- Holtappels, M.; Noben, J.-P.; Van Dijck, P.; Valcke, R. Fire blight host-pathogen interaction: Proteome profiles of Erwinia amylovora infecting apple rootstocks. Sci. Rep. 2018, 8, 11689. [Google Scholar] [CrossRef]
- Aćimović, S.G.; Zeng, Q.; McGhee, G.C.; Sundin, G.W.; Wise, J.C. Control of fire blight (Erwinia amylovora) on apple trees with trunk-injected plant resistance inducers and antibiotics and assessment of induction of pathogenesis-related protein genes. Front. Plant Sci. 2015, 6, 16. [Google Scholar] [CrossRef]
- Norelli, J.L.; Jones, A.L.; Aldwinckle, H.S. Fire Blight Management in the Twenty-first Century: Using New Technologies that Enhance Host Resistance in Apple. Plant Dis. 2003, 87, 756–765. [Google Scholar] [CrossRef]
- Mann, R.A.; Smits, T.H.M.; Bühlmann, A.; Blom, J.; Goesmann, A.; Frey, J.E.; Plummer, K.M.; Beer, S.V.; Luck, J.; Duffy, B.; et al. Comparative Genomics of 12 Strains of Erwinia amylovora Identifies a Pan-Genome with a Large Conserved Core. PLoS ONE 2013, 8, e55644. [Google Scholar] [CrossRef]
- Powney, R.; Smits, T.H.M.; Sawbridge, T.; Frey, B.; Blom, J.; Frey, J.E.; Plummer, K.M.; Beer, S.V.; Luck, J.; Duffy, B.; et al. Genome sequence of an Erwinia amylovora strain with pathogenicity restricted to Rubus plants. J. Bacteriol. 2011, 193, 785–786. [Google Scholar] [CrossRef]
- Koskiniemi, S.; Lamoureux, J.G.; Nikolakakis, K.C.; t’Kint de Roodenbeke, C.; Kaplan, M.D.; Low, D.A.; Hayes, C.S. Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. USA 2013, 110, 7032–7037. [Google Scholar] [CrossRef]
- Jackson, A.P.; Thomas, G.H.; Parkhill, J.; Thomson, N.R. Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC Genom. 2009, 10, 584. [Google Scholar] [CrossRef]
- Jamet, A.; Nassif, X. New Players in the Toxin Field: Polymorphic Toxin Systems in Bacteria. MBio 2015, 6, e00285-15. [Google Scholar] [CrossRef] [PubMed]
- Gallique, M.; Bouteiller, M.; Merieau, A. The Type VI Secretion System: A Dynamic System for Bacterial Communication? Front. Microbiol. 2017, 8, 1454. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zou, Y.; She, P.; Wu, Y. Composition, function, and regulation of T6SS in Pseudomonas aeruginosa. Microbiol. Res. 2015, 172, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Mougous, J.D.; Gifford, C.A.; Ramsdell, T.L.; Mekalanos, J.J. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 2007, 9, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-S.; Pissaridou, P.; Wu, H.-H.; Tsai, M.-D.; Filloux, A.; Lai, E.-M. TagF-mediated repression of bacterial type VI secretion systems involves a direct interaction with the cytoplasmic protein Fha. J. Biol. Chem. 2018, 293, 8829–8842. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.B.; Peterson, S.B.; Mougous, J.D. Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 2014, 12, 137–148. [Google Scholar] [CrossRef]
- Yang, X.; Long, M.; Shen, X. Effector–Immunity Pairs Provide the T6SS Nanomachine its Offensive and Defensive Capabilities. Molecules 2018, 23, 1009. [Google Scholar] [CrossRef]
- Alcoforado Diniz, J.; Coulthurst, S.J. Intraspecies Competition in Serratia marcescens Is Mediated by Type VI-Secreted Rhs Effectors and a Conserved Effector-Associated Accessory Protein. J. Bacteriol. 2015, 197, 2350–2360. [Google Scholar] [CrossRef]
- Poole, S.J.; Diner, E.J.; Aoki, S.K.; Braaten, B.A.; t’Kint de Roodenbeke, C.; Low, D.A.; Hayes, C.S. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 2011, 7, e1002217. [Google Scholar] [CrossRef]
- Hachani, A.; Allsopp, L.P.; Oduko, Y.; Filloux, A. The VgrG proteins are “A la carte” delivery systems for bacterial type VI effectors. J. Biol. Chem. 2014, 289, 17872–17884. [Google Scholar] [CrossRef]
- Ma, J.; Sun, M.; Dong, W.; Pan, Z.; Lu, C.; Yao, H. PAAR-Rhs proteins harbor various C-terminal toxins to diversify the antibacterial pathways of type VI secretion systems. Environ. Microbiol. 2017, 19, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, H.; Gao, Z.-Q.; Wang, W.-J.; Liu, G.-F.; Xu, J.-H.; Su, X.-D.; Dong, Y.-H. Structure of the type VI effector-immunity complex (Tae4-Tai4) provides novel insights into the inhibition mechanism of the effector by its immunity protein. J. Biol. Chem. 2013, 288, 5928–5939. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-S.; Hachani, A.; Lin, J.-S.; Filloux, A.; Lai, E.-M. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 2014, 16, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Iyer, L.M.; Aravind, L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011, 39, 4532–4552. [Google Scholar] [CrossRef]
- Lee, K.-Y.; Lee, B.-J. Structure, Biology, and Therapeutic Application of Toxin-Antitoxin Systems in Pathogenic Bacteria. Toxins 2016, 8, 305. [Google Scholar] [CrossRef]
- Kawano, M.; Aravind, L.; Storz, G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007, 64, 738–754. [Google Scholar] [CrossRef]
- Baskaran, S.; Carlson, L.-A.; Stjepanovic, G.; Young, L.N.; Kim, D.J.; Grob, P.; Stanley, R.E.; Nogales, E.; Hurley, J.H. Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Elife 2014, 9, e05115. [Google Scholar] [CrossRef]
- Backer, J.M. The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem. J. 2016, 473, 2251–2271. [Google Scholar] [CrossRef]
- Stjepanovic, G.; Baskaran, S.; Lin, M.G.; Hurley, J.H. Vps34 Kinase Domain Dynamics Regulate the Autophagic PI 3-Kinase Complex. Mol. Cell 2017, 67, 528–534.e3. [Google Scholar] [CrossRef]
- Heenan, E.J.; Vanhooke, J.L.; Temple, B.R.; Betts, L.; Sondek, J.E.; Dohlman, H.G. Structure and function of Vps15 in the endosomal G protein signaling pathway. Biochemistry 2009, 48, 6390–6401. [Google Scholar] [CrossRef]
- Gaur, N.A.; Hasek, J.; Brickner, D.G.; Qiu, H.; Zhang, F.; Wong, C.-M.; Malcova, I.; Vasicova, P.; Brickner, J.H.; Hinnebusch, A.G. Vps factors are required for efficient transcription elongation in budding yeast. Genetics 2013, 193, 829–851. [Google Scholar] [CrossRef] [PubMed]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, P.; Yue, Z. Deregulation of autophagy and vesicle trafficking in Parkinson’s disease. Neurosci. Lett. 2019, 697, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Casanova, J.E. Bacterial Autophagy: Offense and Defense at the Host-Pathogen Interface. Cell. Mol. Gastroenterol. Hepatol. 2017, 4, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Bose, S.; Segovia, J.A.; Somarajan, S.R.; Chang, T.-H.; Kannan, T.R.; Baseman, J.B. ADP-ribosylation of NLRP3 by Mycoplasma pneumoniae CARDS toxin regulates inflammasome activity. MBio 2014, 5, e02186-14. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Kannan, T.R.; Baseman, J.B. Cellular vacuoles induced by Mycoplasma pneumoniae CARDS toxin originate from Rab9-associated compartments. PLoS ONE 2011, 6, e22877. [Google Scholar] [CrossRef]
- Hofius, D.; Munch, D.; Bressendorff, S.; Mundy, J.; Petersen, M. Role of autophagy in disease resistance and hypersensitive response-associated cell death. Cell Death Differ. 2011, 18, 1257–1262. [Google Scholar] [CrossRef]
- Starokadomskyy, P.; Dmytruk, K.V. A bird’s-eye view of autophagy. Autophagy 2013, 9, 1121–1126. [Google Scholar] [CrossRef]
- Hahn, A.; Parey, K.; Bublitz, M.; Mills, D.J.; Zickermann, V.; Vonck, J.; Kühlbrandt, W.; Meier, T. Structure of a Complete ATP Synthase Dimer Reveals the Molecular Basis of Inner Mitochondrial Membrane Morphology. Mol. Cell 2016, 63, 445–456. [Google Scholar] [CrossRef]
- Wagner, K.; Rehling, P.; Sanjuán Szklarz, L.K.; Taylor, R.D.; Pfanner, N.; van der Laan, M. Mitochondrial F1Fo-ATP Synthase: The Small Subunits e and g Associate with Monomeric Complexes to Trigger Dimerization. J. Mol. Biol. 2009, 392, 855–861. [Google Scholar] [CrossRef]
- Reinders, J.; Zahedi, R.P.; Pfanner, N.; Meisinger, C.; Sickmann, A. Toward the complete yeast mitochondrial proteome: Multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 2006, 5, 1543–1554. [Google Scholar] [CrossRef] [PubMed]
- Tarassov, K.; Messier, V.; Landry, C.R.; Radinovic, S.; Serna Molina, M.M.; Shames, I.; Malitskaya, Y.; Vogel, J.; Bussey, H.; Michnick, S.W. An in vivo map of the yeast protein interactome. Science 2008, 320, 1465–1470. [Google Scholar] [CrossRef] [PubMed]
- Segarra, V.A.; Boettner, D.R.; Lemmon, S.K. Atg27 tyrosine sorting motif is important for its trafficking and Atg9 localization. Traffic 2015, 16, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Arlt, H.; Reggiori, F. Chapter 8–The Role of Atg9 in Yeast Autophagy. In Hayat Other Pathologies, Inflammation, Immunity, Infection, and Aging; Hayat, H.A., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 181–192. ISBN 978-0-12-805420-8. [Google Scholar]
- Feyder, S.; De Craene, J.-O.; Bar, S.; Bertazzi, D.L.; Friant, S. Membrane trafficking in the yeast Saccharomyces cerevisiae model. Int. J. Mol. Sci. 2015, 16, 1509–1525. [Google Scholar] [CrossRef] [PubMed]
- Kolaczkowski, M.; Kolaczkowska, A.; Gaigg, B.; Schneiter, R.; Moye-Rowley, W.S. Differential regulation of ceramide synthase components LAC1 and LAG1 in Saccharomyces cerevisiae. Eukaryot. Cell 2004, 3, 880–892. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, S.; Qin, X.; Hou, W.; Dong, H.; Yao, L.; Xiong, L. The pleiotropic roles of sphingolipid signaling in autophagy. Cell Death Amp. Dis. 2014, 5, e1245. [Google Scholar] [CrossRef] [PubMed]
- Pattingre, S.; Bauvy, C.; Levade, T.; Levine, B.; Codogno, P. Ceramide-induced autophagy: To junk or to protect cells? Autophagy 2009, 5, 558–560. [Google Scholar] [CrossRef]
- Siamer, S.; Guillas, I.; Shimobayashi, M.; Kunz, C.; Hall, M.N.; Barny, M.-A. Expression of the Bacterial Type III Effector DspA/E in Saccharomyces cerevisiae Down-regulates the Sphingolipid Biosynthetic Pathway Leading to Growth Arrest. J. Biol. Chem. 2014, 289, 18466–18477. [Google Scholar] [CrossRef]
- Masgrau, A.; Battola, A.; Sanmartin, T.; Pryszcz, L.P.; Gabaldón, T.; Mendoza, M. Distinct roles of the polarity factors Boi1 and Boi2 in the control of exocytosis and abscission in budding yeast. Mol. Biol. Cell 2017, 28, 3082–3094. [Google Scholar] [CrossRef]
- Lecointe, F.; Simos, G.; Sauer, A.; Hurt, E.C.; Motorin, Y.; Grosjean, H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. J. Biol. Chem. 1998, 273, 1316–1323. [Google Scholar] [CrossRef]
- Baladrón, V.; Ufano, S.; Dueñas, E.; Martín-Cuadrado, A.B.; del Rey, F.; Vázquez de Aldana, C.R. Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell 2002, 1, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Krska, D.; Ravulapalli, R.; Fieldhouse, R.J.; Lugo, M.R.; Merrill, A.R. C3larvin Toxin, an ADP-ribosyltransferase from Paenibacillus larvae. J. Biol. Chem. 2015, 290, 1639–1653. [Google Scholar] [CrossRef] [PubMed]
- Dickey, S.W.; Cheung, G.Y.C.; Otto, M. Different drugs for bad bugs: Antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 2017, 16, 457. [Google Scholar] [CrossRef]
- Jankevicius, G.; Ariza, A.; Ahel, M.; Ahel, I. The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA. Mol. Cell 2016, 64, 1109–1116. [Google Scholar] [CrossRef]
- Piscotta, F.J.; Jeffrey, P.D.; Link, A.J. ParST is a widespread toxin–antitoxin module that targets nucleotide metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 826–834. [Google Scholar] [CrossRef] [PubMed]
- De Castro, E.; Sigrist, C.J.A.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H. Predicting Secretory Proteins with SignalP. In Protein Function Prediction: Methods and Protocols; Kihara, D., Ed.; Springer: New York, NY, USA, 2017; pp. 59–73. ISBN 978-1-4939-7015-5. [Google Scholar]
- Bendtsen, J.D.; Kiemer, L.; Fausbøll, A.; Brunak, S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. Advantages of combined transmembrane topology and signal peptide prediction—The Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Kallberg, M.; Margaryan, G.; Wang, S.; Ma, J.; Xu, J. RaptorX server: A resource for template-based protein structure modeling. Methods Mol. Biol. 2014, 1137, 17–27. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007, 35, 3375–3382. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liang, Y.; Zhang, Y. Atomic-Level Protein Structure Refinement Using Fragment-Guided Molecular Dynamics Conformation Sampling. Structure 2011, 19, 1784–1795. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Lindahl, E.; Wallner, B. Improved model quality assessment using ProQ2. BMC Bioinform. 2012, 13, 224. [Google Scholar] [CrossRef] [PubMed]
- Maghrabi, A.H.A.; McGuffin, L.J. ModFOLD6: An accurate web server for the global and local quality estimation of 3D protein models. Nucleic Acids Res. 2017, 45, W416–W421. [Google Scholar] [CrossRef]
- Benatuil, L.; Perez, J.M.; Belk, J.; Hsieh, C.-M. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng. Des. Sel. 2010, 23, 155–159. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
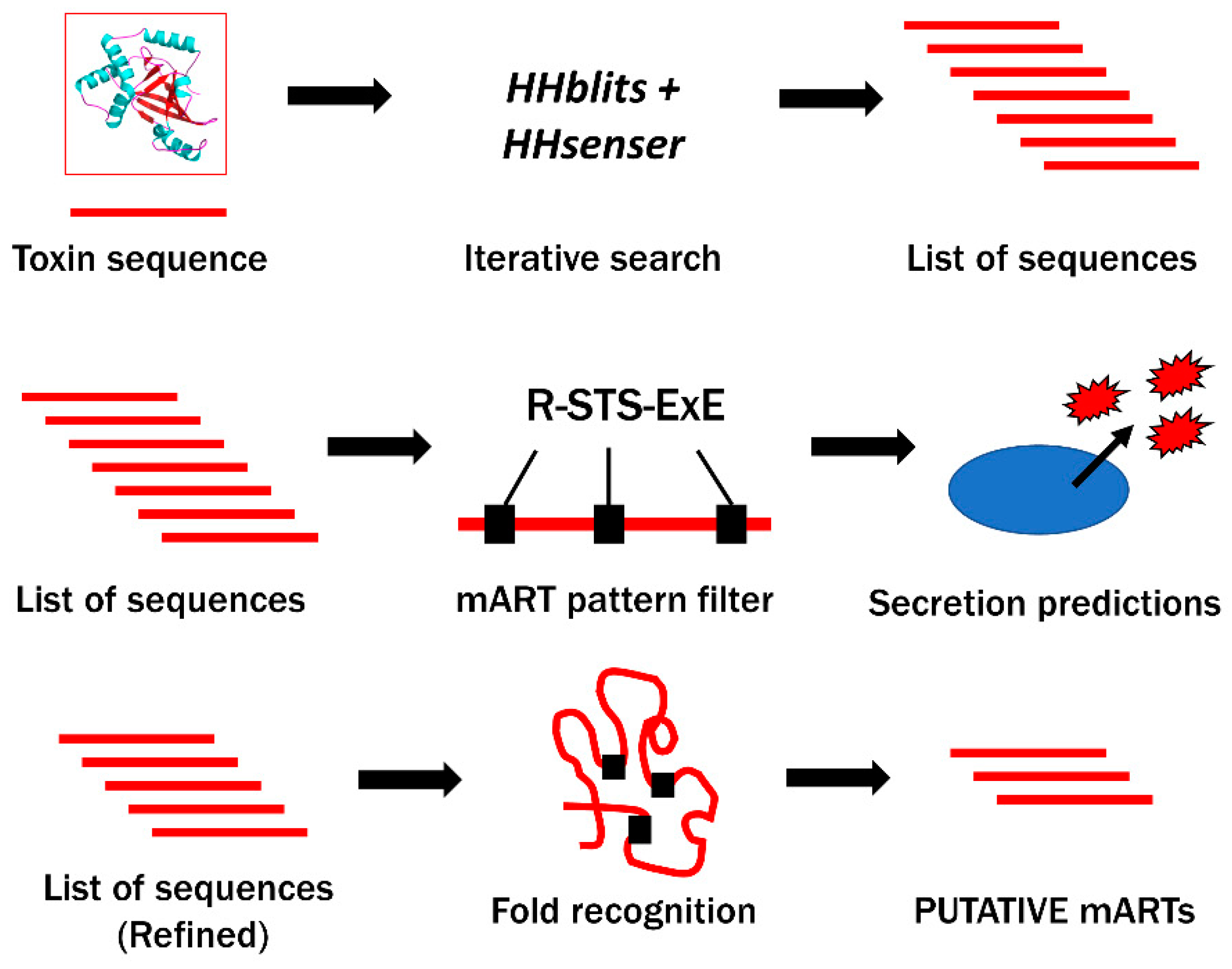
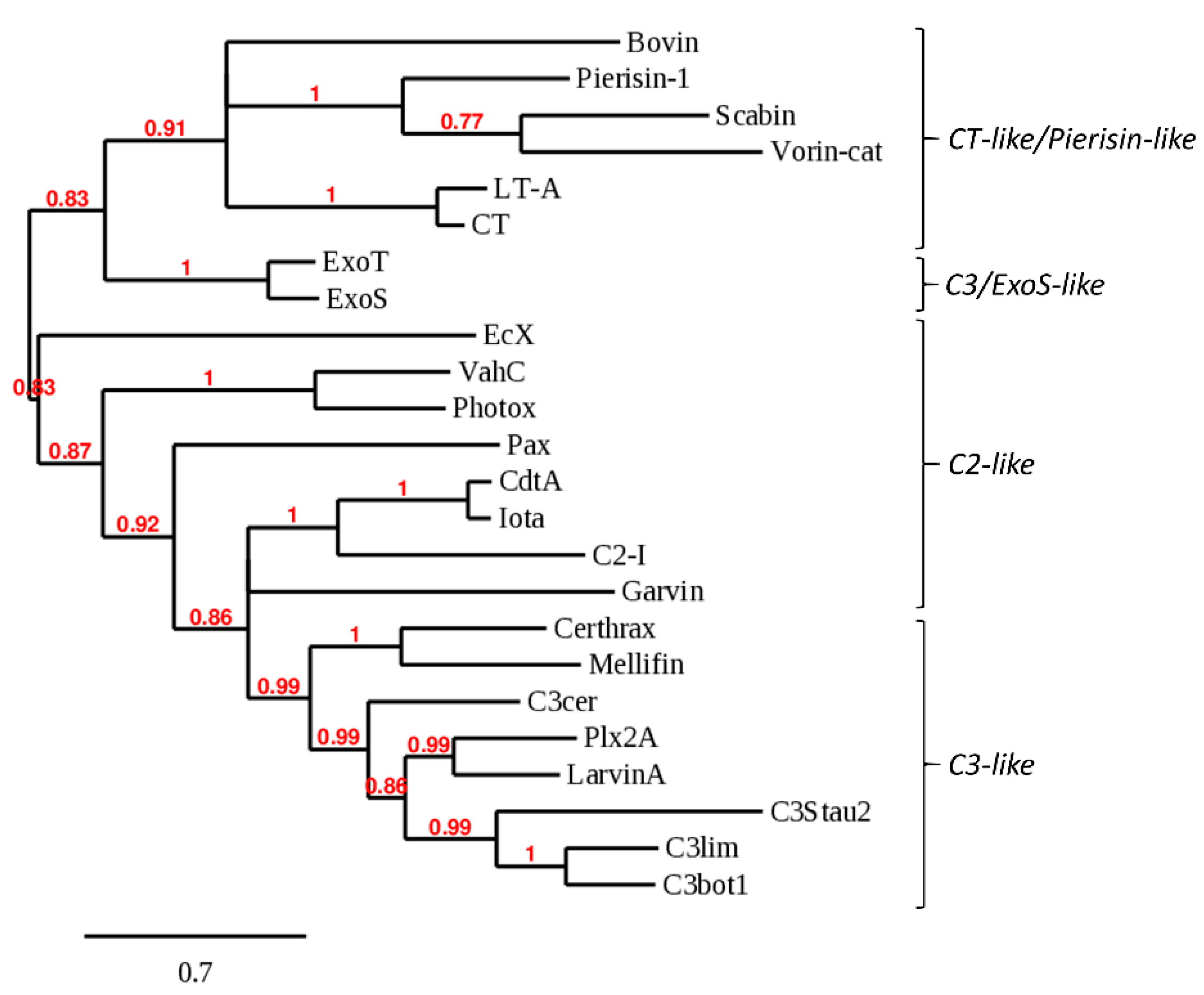
| Organism 1 | UniProt Sequence Identifier | Average Template Coverage | Server Predictions with HIGH Confidence (Out of 10) | Overall Confidence |
|---|---|---|---|---|
| Spiroplasma melliferum | A0A037UQ33 | 0.857 | 10 | High |
| Weissella cibaria | A0A139R9U4 | 0.955 | 10 | High |
| Parachlamydia acanthamoebae | A0A0C1E5A0 | 0.901 | 10 | High |
| Lactobacillus kunkeei | A0A0C3AG37 | 0.854 | 10 | High |
| Escherichia coli | T9AJP6 | 0.914 | 10 | High |
| Fusobacterium necrophorum | A0A064A9T9 | 0.948 | 10 | High |
| Pectobacterium carotovorum | A0A093SYU3 | 0.701 | 6 | Medium |
| Legionella quateirensis | A0A0W0XJR5 | 0.708 | 10 | Medium |
| Enterobacter bugandensis | A0A167JVD8 | 0.857 | 7 | High |
| Photobacterium damselae | D0Z4Y1 | 0.660 | 7 | Medium |
| Providencia alcalifaciens | W3YC38 | 0.864 | 10 | High |
| Erwinia amylovora | E5B8T9 | 0.510 | 10 | Medium |
| Paenibacillus popilliae | M9M5D5 | 0.839 | 10 | High |
| Bartonella bovis | N6VGJ3 | 0.831 | 10 | High |
| Pantoea stuartii | E0LU50 | 0.671 | 3 | Medium |
| Lactococcus garvieae | H2B2R7 | 0.812 | 10 | High |
| Organism | UniProt Sequence Identifier | Residues 1 Modelled (mART Domain) | p-Value | uGDT (GDT) | uSeqID (SeqID) | Templates |
|---|---|---|---|---|---|---|
| Spiroplasma melliferum | A0A037UQ33 | 1–261 | 4.83 × 10−9 | 169(65) | 97(37) | 4fxqA 4fk7A |
| Weissella cibaria | A0A139R9U4 | 1–188 | 6.44 × 10−7 | 131(70) | 38(20) | 1qs2A 1qs1A 5gttA 2j3zA 4fxqA |
| Parachlamydia acanthamoebae | A0A0C1E5A0 | 42–250 | 7.73 × 10−6 | 126(51) | 53(21) | 4xzjA 1qs2A 1qs1A |
| Lactobacillus kunkeei | A0A0C3AG37 | 32–237 | 2.29 × 10−7 | 121(51) | 49(21) | 1qs1A 1qs2A 5gttA 4fxqA 4fk7A |
| Escherichia coli | T9AJP6 | 1–220 | 3.57 × 10−7 | 119(54) | 32(15) | 4xzjA 1qs1A 1qs2A 4elnA 4fxqA |
| Fusobacterium necrophorum | A0A064A9T9 | 306–475 | 1.58 × 10−6 | 119(70) | 36(21) | 4xzjA |
| Legionella quateirensis | A0A0W0XJR5 | 1–324 | 7.80 × 10−7 | 115(36) | 36(11) | 4xzjA 4fxqA 1qs2A 1qs1A 3bw8A |
| Enterobacter bugandensis | A0A167JVD8 | 735–951 | 1.31 × 10−8 | 105(48) | 36(17) | 4xzjA |
| Providencia alcalifaciens | W3YC38 | 102–408 | 1.00 × 10−4 | 102(33) | 41(13) | 4xzjA 1qs1A 1qs2A 5gttA 2wn4A |
| Erwinia amylovora | E5B8T9 | 144–276 | 3.67 × 10−8 | 93(73) | 33(26) | 1tiiA 1xtcA 1lt4A 4z9dA 5ewkA |
| Paenibacillus popilliae | M9M5D5 | 1–222 | 1.22 × 10−6 | 121(54) | 42(19) | 1qs1A 1qs2A 4fk7A 4fxqA 2bovB |
| Bartonella bovis | N6VGJ3 | 1–238 | 3.82 × 10−12 | 141(59) | 68(29) | 1lt4A 1tiiA 1ltiA |
| Lactococcus garvieae | H2B2R7 | 1–239 | 5.56 × 10−7 | 116(48) | 37(15) | 2a78B 5gttA 2bovB 3bw8A 4fxqA |
| Putative mART Toxin | UniProt Accession | mART Homologs (% Sequence ID) | Genomic Neighbours Indicative of Virulence | Research Interest |
|---|---|---|---|---|
| Vorin | E5B8T9 | Pierisin (33%), LT-A (30%) CARDS (29%), Cholera (29%) Scabin (27%) | T6SS subunit homologs, toxin-antitoxin modules | Fire blight in Rubus genus plants |
| Garvin | H2B2R7 | Certhrax (24%) C3bot1 (24%) | Collagen- and mucin-binding proteins | Lactococcosis in salmonid fish; various zoonoses (e.g., UTIs and skin infections) |
| Bovin | N6VGJ3 | LT-IIA/B (40%) Cholera toxin (36%) Pierisin (32%) Scabin (31%) | Possible B-domain subunit (similar to cholera toxin pentameric subunit) | Endocarditis in beef and dairy cattle |
| Mellifin | A0A037UQ33 | Certhrax (42%) C3bot1, C3lim, C3stau1 (29%) | Protective antigen, integrase, PTS glucose transporter | Spiroplasmosis in honey bees? |
| EcX | A0A167JVD8 | Mav (27%) Vis (22%) | Transposases, permease transporter | Antibiotic-resistant nosocomial infections |
| Pax | A0A0C1E5A0 | EFV (29%) Vis (28%) | Mycobacterial two-component regulatory system, chemotaxis protein, LPS synthesis proteins | Community-acquired pneumonia in humans? |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tremblay, O.; Thow, Z.; Geddes-McAlister, J.; Merrill, A.R. Several New Putative Bacterial ADP-Ribosyltransferase Toxins Are Revealed from In Silico Data Mining, Including the Novel Toxin Vorin, Encoded by the Fire Blight Pathogen Erwinia amylovora. Toxins 2020, 12, 792. https://doi.org/10.3390/toxins12120792
Tremblay O, Thow Z, Geddes-McAlister J, Merrill AR. Several New Putative Bacterial ADP-Ribosyltransferase Toxins Are Revealed from In Silico Data Mining, Including the Novel Toxin Vorin, Encoded by the Fire Blight Pathogen Erwinia amylovora. Toxins. 2020; 12(12):792. https://doi.org/10.3390/toxins12120792
Chicago/Turabian StyleTremblay, Olivier, Zachary Thow, Jennifer Geddes-McAlister, and A. Rod Merrill. 2020. "Several New Putative Bacterial ADP-Ribosyltransferase Toxins Are Revealed from In Silico Data Mining, Including the Novel Toxin Vorin, Encoded by the Fire Blight Pathogen Erwinia amylovora" Toxins 12, no. 12: 792. https://doi.org/10.3390/toxins12120792
APA StyleTremblay, O., Thow, Z., Geddes-McAlister, J., & Merrill, A. R. (2020). Several New Putative Bacterial ADP-Ribosyltransferase Toxins Are Revealed from In Silico Data Mining, Including the Novel Toxin Vorin, Encoded by the Fire Blight Pathogen Erwinia amylovora. Toxins, 12(12), 792. https://doi.org/10.3390/toxins12120792





