Insights into the Diversity of Secondary Metabolites of Planktothrix Using a Biphasic Approach Combining Global Genomics and Metabolomics
Abstract
1. Introduction
2. Results and Discussion
2.1. Biosynthetic Gene Cluster Approach Based on Genome Analysis
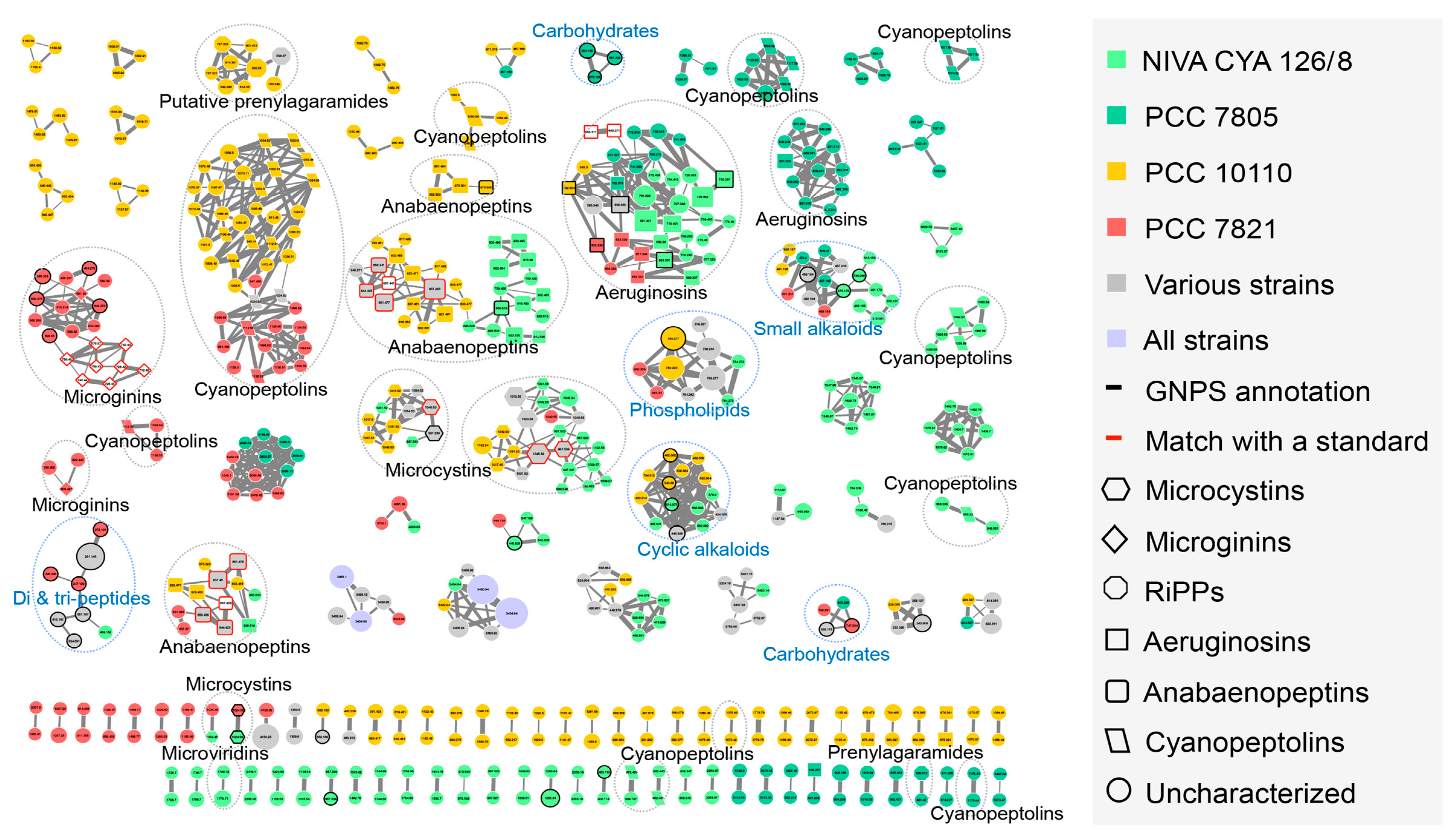
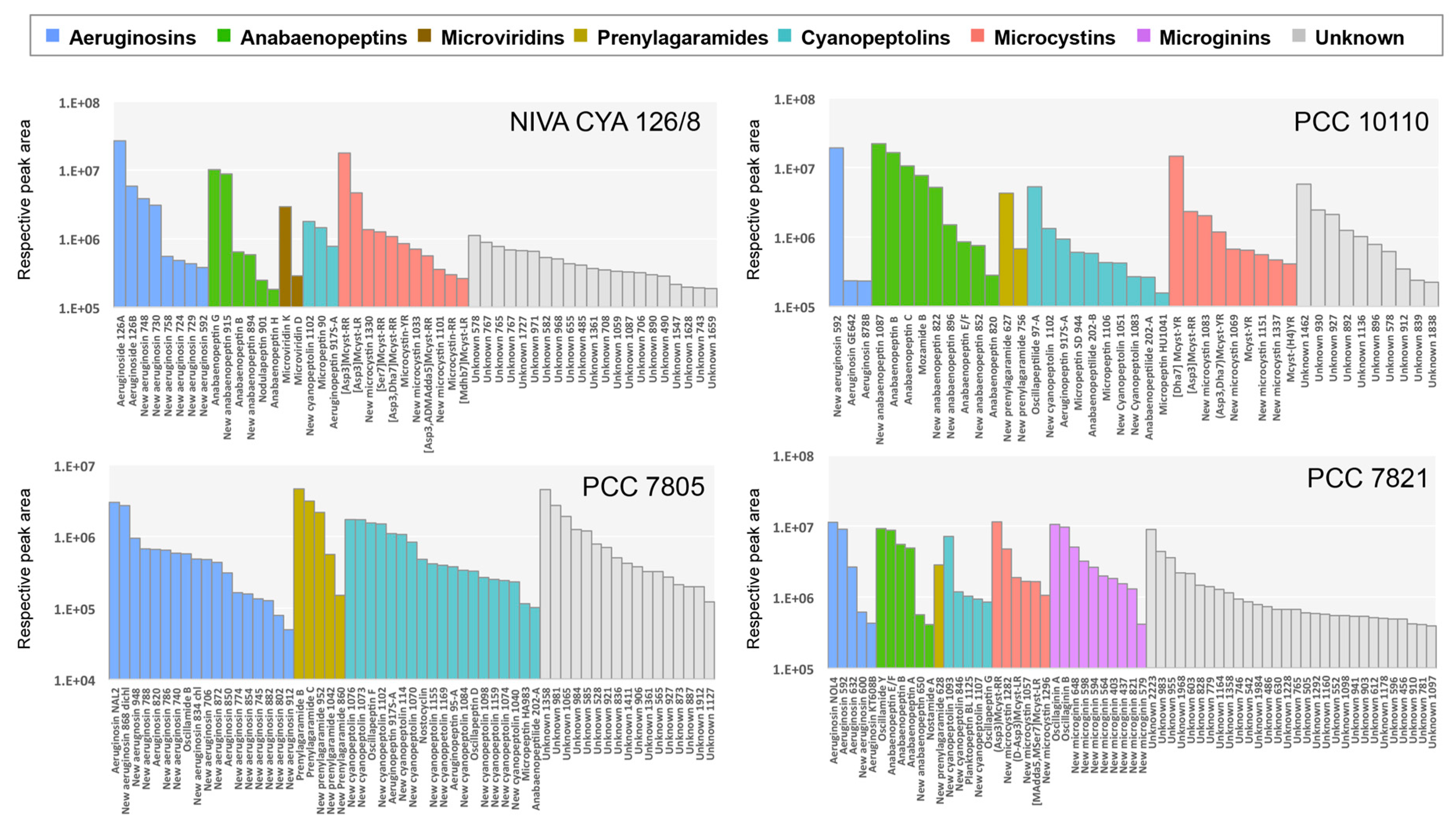
2.2. Insights of the Molecular Networking for the Characterization the Chemical Diversity of the Planktothrix Secondary Metabolite Variants
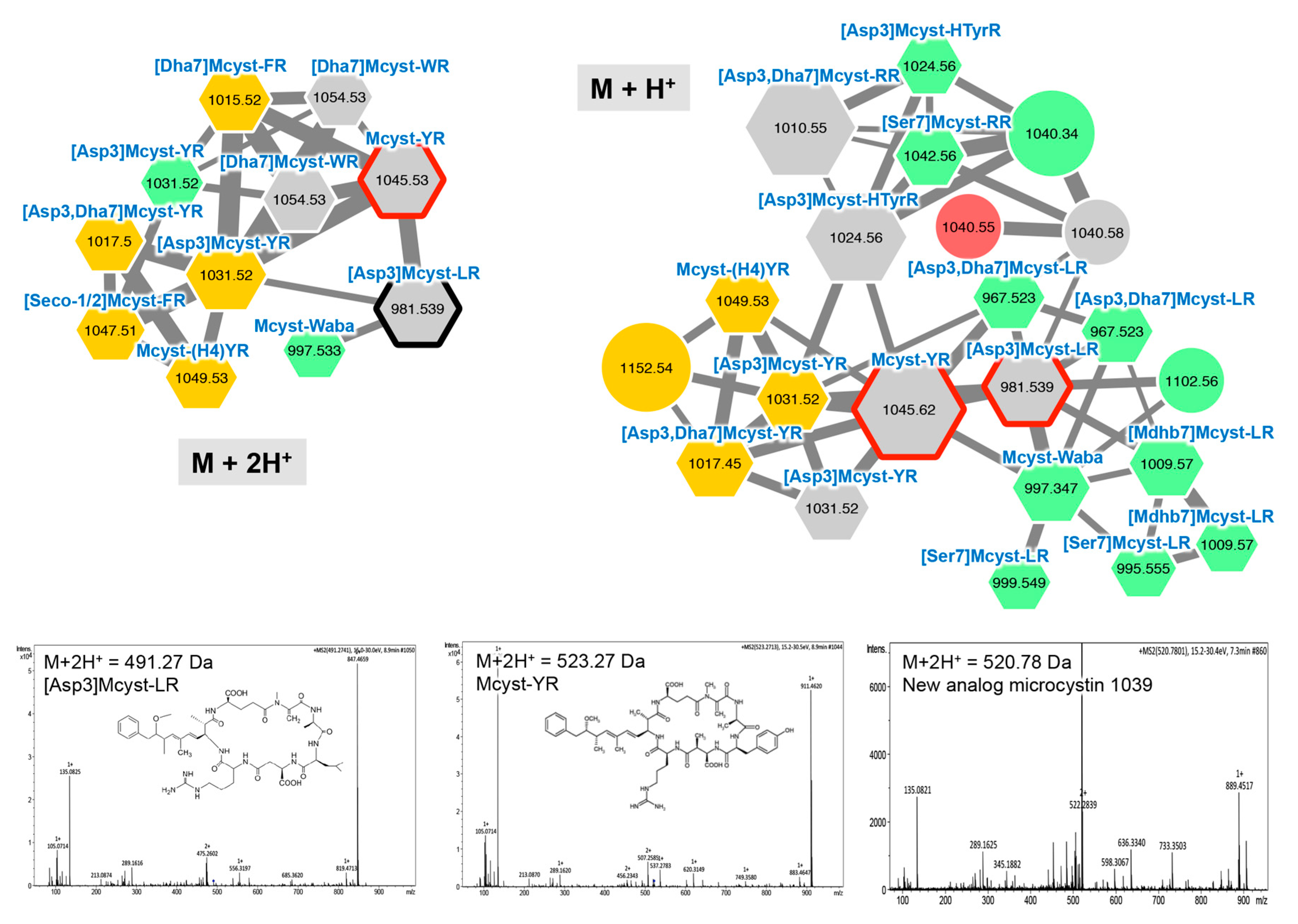
2.2.1. Microcystins
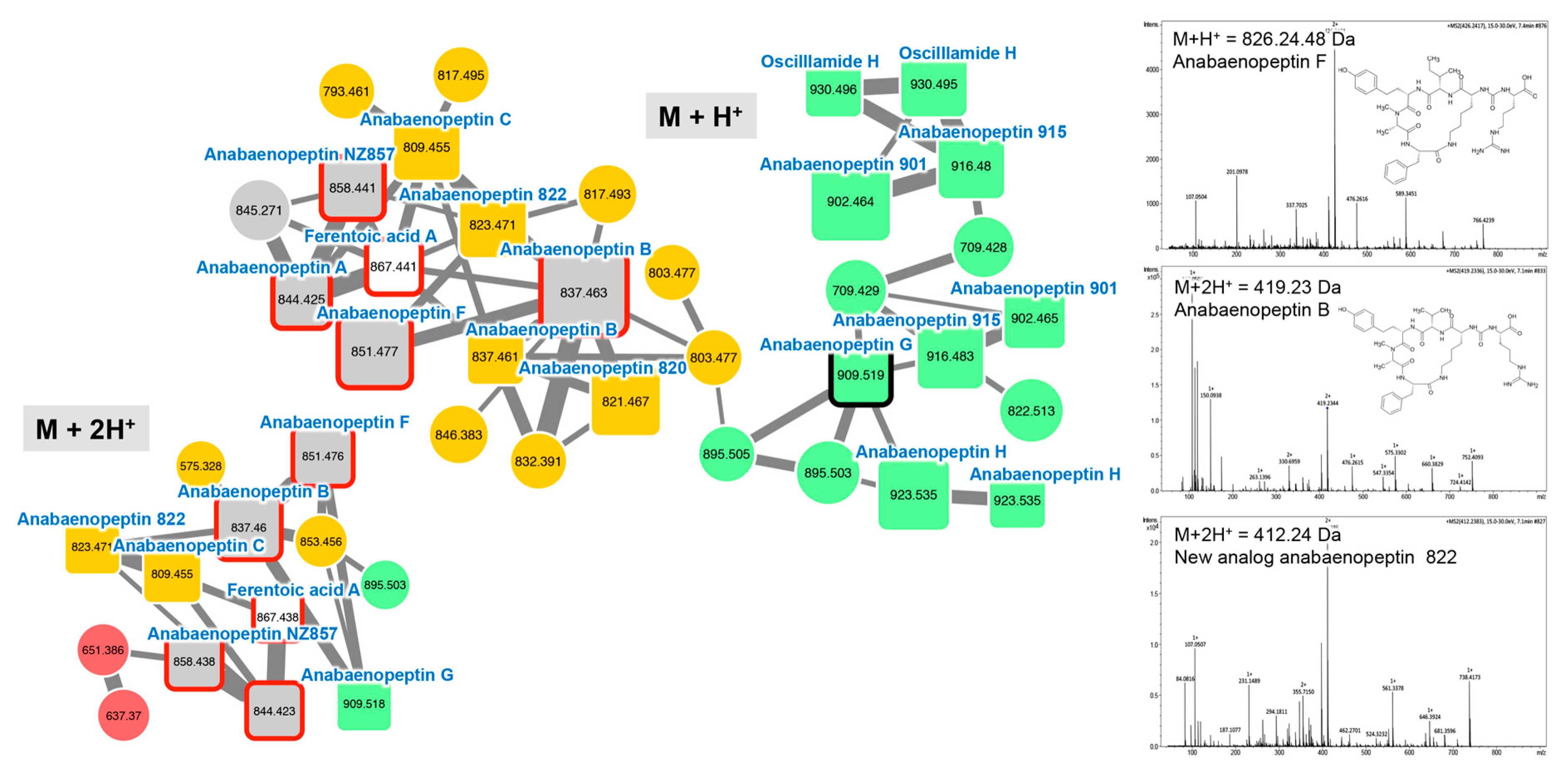
2.2.2. Anabaenopeptins
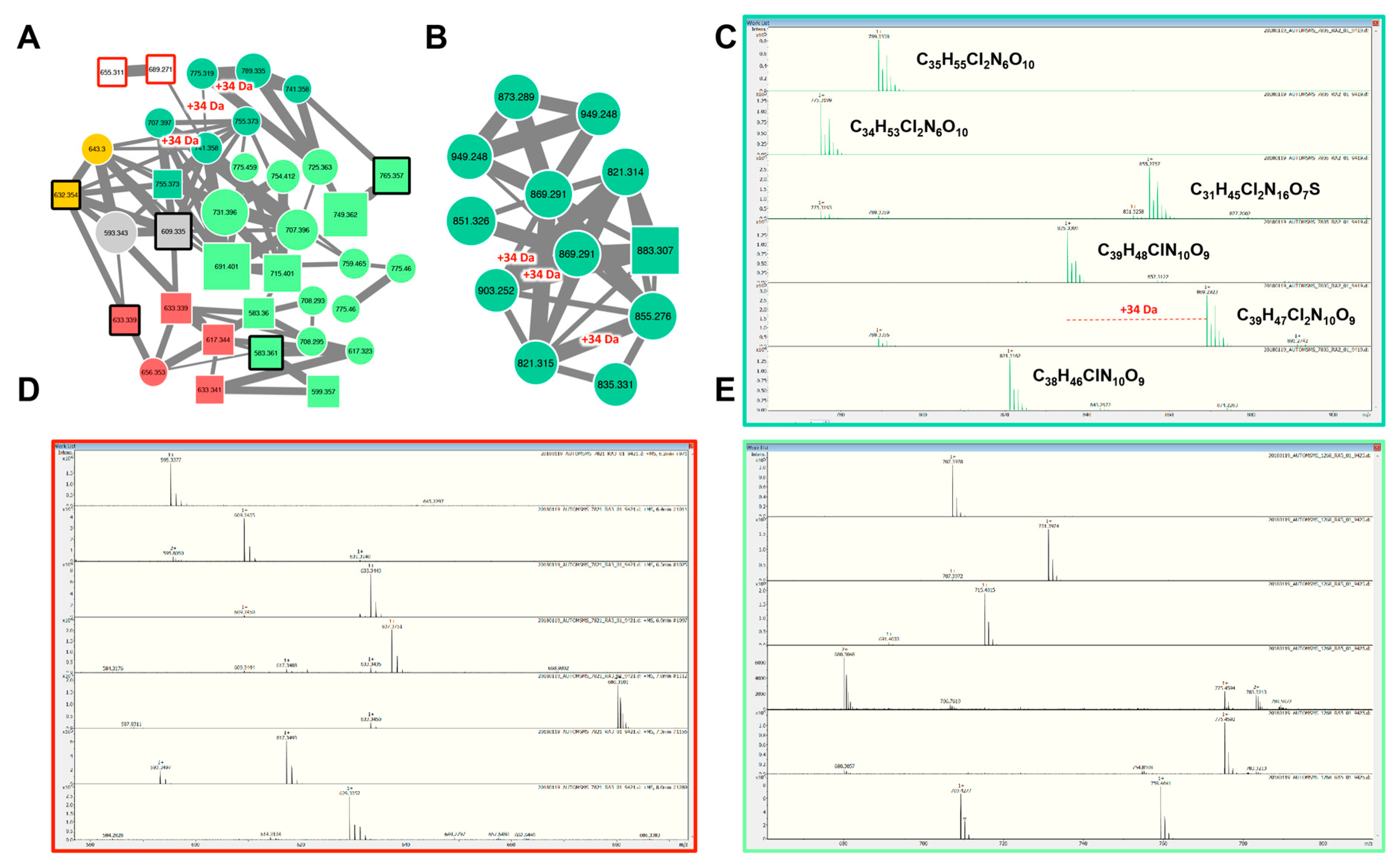
2.2.3. Aeruginosins and Halogenation
2.3. Global Capability to Annotate Cyanobacterial Metabolites
3. Conclusions
4. Materials and Methods
4.1. Planktothrix Strains and Culture
4.2. Description of the DNA Isolation, Sequencing, and Assembling Methods
4.3. In Silico Analyses in MicroScope
4.4. Metabolome Biomass Extraction and Analysis by Mass Spectrometry
4.5. Data Treatment and Molecular Networking
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Whitton, B.A. Ecology of Cyanobacteria II: Their Diversity in Space and Time; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar] [CrossRef]
- Carmichael, W. Cyanobacterial harmful algal blooms: State of the science and research needs. Adv. Exp. Med. Biol. 2008, 619, 831–853. [Google Scholar] [CrossRef]
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Microbiol. 2018, 16, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Pearson, L.; Mihali, T.; Moffitt, M.; Kellmann, R.; Neilan, B. On the chemistry, toxicology and genetics of the cyanobacterial toxins, microcystin, nodularin, saxitoxin and cylindrospermopsin. Mar. Drugs 2010, 8, 1650–1680. [Google Scholar] [CrossRef] [PubMed]
- Leflaive, J.P.; Ten-Hage, L. Algal and cyanobacterial secondary metabolites in freshwaters: A comparison of allelopathic compounds and toxins. Freshw. Biol. 2007, 52, 199–214. [Google Scholar] [CrossRef]
- Zak, A.; Kosakowska, A. Cyanobacterial and microalgal bioactive compounds-the role of secondary metabolites in allelopathic interactions. Oceanol. Hydrobiol. Stud. 2016, 45, 131–143. [Google Scholar] [CrossRef]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural product biosynthetic diversity and comparative genomics of the Cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Burja, A.M.; Banaigs, B.; Abou-Mansour, E.; Burgess, J.G.; Wright, P.C. Marine cyanobacteria—A prolific source of natural products. Tetrahedron 2001, 57, 9347–9377. [Google Scholar] [CrossRef]
- Demay, J.; Bernard, C.; Reinhart, A.; Marie, B. Natural products from Cyanobacteria: Focus on beneficial activities. Mar. Drugs 2019, 17, 320. [Google Scholar] [CrossRef]
- Welker, M.; Dittmann, E.; Von Döhren, H. Cyanobacteria as a source of natural products. Methods Enzymol. 2012, 517, 23–46. [Google Scholar] [CrossRef]
- Jones, A.; Gu, L.; Sorrels, C.; Sherman, D.; Gerwick, W. New tricks from ancient algae: Natural products biosynthesis in marine cyanobacteria. Curr. Opin. Chem. Biol. 2009, 13, 216–223. [Google Scholar] [CrossRef]
- Nikolouli, K.; Mossialos, D. Bioactive compounds synthesized by non-ribosomal peptide synthetases and type-I polyketide synthases discovered through genome-mining and metagenomics. Biotechnol. Lett. 2012, 34, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.M.; Wu, D.; Latifi, A.; Axen, S.D.; Fewer, D.P.; Talla, E.; Calteau, A.; Cai, F.; Rippka, R.; Herdman, M.; et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Rounge, T.B.; Rohrlack, T.; Tooming-Klunderud, A.; Kristensen, T.; Jakobsen, K.S. Comparison of cyanopeptolin genes in Planktothrix, Microcystis, and Anabaena strains: Evidence for independent evolution within each genus. Appl. Environ. Microbiol. 2007, 73, 7322–7330. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.S.; Schmidt, E.W. Linking chemistry and genetics in the growing cyanobactin natural products family. Chem. Biol. 2011, 18, 508–519. [Google Scholar] [CrossRef] [PubMed]
- Starcevic, A.; Zucko, J.; Simunkovic, J.; Long, P.F.; Cullum, J.; Hranueli, D. ClustScan: An integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 2008, 36, 6882–6892. [Google Scholar] [CrossRef] [PubMed]
- Cimermancic, P.; Medema, M.H.; Claesen, J.; Kurita, K.; Brown, L.C.W.; Mavrommatis, K.; Pati, A.; Godfrey, P.A.; Koehrsen, M.; Clardy, J.; et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 2014, 158, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Welker, M.; Erhard, M. Consistency between chemotyping of single filaments of Planktothrix rubescens (cyanobacteria) by MALDI-TOF and the peptide patterns of strains determined by HPLC-MS. J. Mass Spectrom. 2007, 42, 1062–1068. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Boudreau, P.; Monroe, E.; Mehrotra, S.; Desfor, S.; Korabeynikov, A.; Sherman, D.; Murray, T.; Gerwick, L.; Dorrestein, P.; Gerwick, W. Marine cyanobacterium Moorea producens JHB through orthogonal natural products workflows. PLoS ONE 2015, 10, e0133297. [Google Scholar] [CrossRef]
- Mohimani, H.; Pevzner, P.A. Dereplication, sequencing and identification of peptidic natural products: From genome mining to peptidogenomics to spectral networks. Nat. Prod. Rep. 2016, 33, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Kurmayer, R.; Blom, J.F.; Deng, L.; Pernthaler, J. Integrating phylogeny, geographic niche partitioning and secondary metabolite synthesis in bloom-forming Planktothrix. ISME J. 2015, 9, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, G.; Goesmann, A.; Kurmayer, R. Elucidation of insertion elements carried on plasmids and in vitro construction of shuttle vectors from the toxic cyanobacterium Planktothrix. Appl. Environ. Microbiol. 2014, 80, 4887–4897. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pancrace, C.; Barny, M.-A.; Ueoka, R.; Calteau, A.; Scalvenzi, T.; Pédron, J.; Barbe, V.; Piel, J.; Humbert, J.-F.; Gugger, M. Insights into the Planktothrix genus: Genomic and metabolic comparison of benthic and planktic strains. Sci. Rep. 2017, 7, 41181. [Google Scholar] [CrossRef] [PubMed]
- Rounge, T.B.; Rohrlack, T.; Nederbragt, A.J.; Kristensen, T.; Jakobsen, K.S. A genome-wide analysis of nonribosomal peptide synthetase gene clusters and their peptides in a Planktothrix rubescens strain. BMC Genom. 2009, 10, 396. [Google Scholar] [CrossRef]
- Rohrlack, T.; Edvardsen, B.; Skulberg, R.; Halstvedt, C.B.; Utkilen, H.C.; Ptacnik, R.; Skulberg, O.M. Oligopeptide chemotypes of the toxic freshwater cyanobacterium Planktothrix can form sub-populations with dissimilar ecological traits. Limnol. Oceanogr. 2008, 53, 1279–1293. [Google Scholar] [CrossRef]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Dickschat, J.S. AntiSMASH 4.0—Improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, 36–41. [Google Scholar] [CrossRef]
- Christiansen, G.; Fastner, J.; Erhard, M.; Börner, T.; Dittmann, E. Microcystin biosynthesis in Planktothrix: Genes, evolution, and manipulation. J. Bacteriol. 2003, 185, 564–572. [Google Scholar] [CrossRef]
- Ishida, K.; Christiansen, G.; Yoshida, W.Y.; Kurmayer, R.; Welker, M.; Valls, N.; Bonjoch, J.; Hertweck, C.; Börner, T.; Hemscheidt, T.; et al. Biosynthesis and structure of aeruginoside 126A and 126B, cyanobacterial peptide glycosides bearing a 2-carboxy-6-hydroxyoctahydroindole moiety. Chem. Biol. 2007, 14, 565–576. [Google Scholar] [CrossRef]
- Christiansen, G.; Philmus, B.; Hemscheidt, T.; Kurmayer, R. Genetic variation of adenylation domains of the anabaenopeptin synthesis operon and the evolution of substrate promiscuity. J. Bacteriol. 2011, 193, 3822–3831. [Google Scholar] [CrossRef]
- Philmus, B.; Christiansen, G.; Yoshida, W.Y.; Hemscheidt, T.K. Post-translational modification in microviridin biosynthesis. ChemBioChem 2008, 9, 3066–3073. [Google Scholar] [CrossRef]
- Olivon, F.; Allard, P.M.; Koval, A.; Righi, D.; Genta-Jouve, G.; Neyts, J.; Apel, C.; Pannecouque, C.; Nothias, L.F.; Cachet, X.; et al. Bioactive natural products prioritization using massive multi-informational molecular networks. ACS Chem. Biol. 2017, 12, 2644–2651. [Google Scholar] [CrossRef]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; De Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef]
- Shin, H.J.; Matsuda, H.; Murakami, M.; Yamaguchi, K. Agardhipeptins A and B, two new cyclic hepta-and octapeptide, from the cyanobacterium Oscillatoria agardhii (NIES-204). Tetrahedron 1996, 52, 13129–13136. [Google Scholar] [CrossRef]
- Brito, Â.; Gaifem, J.; Ramos, V.; Glukhov, E.; Dorrestein, P.C.; Gerwick, W.H.; Vasconcelos, V.M.; Mendes, M.V.; Tamagnini, P. Bioprospecting Portuguese Atlantic coast cyanobacteria for bioactive secondary metabolites reveals untapped chemodiversity. Algal Res. 2015, 9, 218–226. [Google Scholar] [CrossRef]
- Ding, C.; Pang, L.; Liang, Z.X.; Goh, K.; Glukhov, E.; Gerwick, W.; Tan, L. MS/MS-based molecular networking approach for the detection of aplysiatoxin-related compounds in environmental marine Cyanobacteria. Mar. Drugs 2018, 16, 505. [Google Scholar] [CrossRef]
- Luzzatto-Knaan, T.; Garg, N.; Wang, M.; Glukhov, E.; Peng, Y.; Ackermann, G.; Amir, A.; Duggan, B.M.; Ryazanov, S.; Gerwick, L.; et al. Digitizing mass spectrometry data to explore the chemical diversity and distribution of marine cyanobacteria and algae. Elife 2017, 6, e24214. [Google Scholar] [CrossRef]
- Allard, P.M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.L. Integration of molecular networking and in-silico MS/MS fragmentation for natural products dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef]
- Allard, P.M.; Genta-Jouve, G.; Wolfender, J.L. Deep metabolome annotation in natural products research: Towards a virtuous cycle in metabolite identification. Curr. Opin. Chem. Biol. 2017, 36, 40–49. [Google Scholar] [CrossRef]
- Le Manach, S.; Marie, B.; Duval, C.; Marie, A.; Djediat, C.; Catherine, A.; Edery, M.; Bernard, C.; Marie, B. Global metabolomic characterizations of Microcystis spp. highlights clonal diversity in natural bloom-forming populations and expands metabolite structural diversity. Front. Microbiol. 2019, 10, 791. [Google Scholar] [CrossRef]
- Welker, M.; Christiansen, G.; von Döhren, H. Diversity of coexisting Planktothrix (Cyanobacteria) chemotypes deduced by mass spectral analysis of microystins and other oligopeptides. Arch. Microbiol. 2004, 182, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Welker, M.; Christiansen, G.; Cadel-Six, S.; Bouchier, C.; Dittmann, E.; Hertweck, C.; De Marsac, N.T. Plasticity and evolution of aeruginosin biosynthesis in cyanobacteria. Appl. Environ. Microbiol. 2009, 75, 2017–2026. [Google Scholar] [CrossRef] [PubMed]
- Haruštiaková, D.; Welker, M. Chemotype diversity in Planktothrix rubescens (cyanobacteria) populations is correlated to lake depth. Environ. Microbiol. Rep. 2017, 9, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Briand, J.F.; Jacquet, S.; Flinois, C.; Avois-Jacquet, C.; Maisonnette, C.; Leberre, B.; Humbert, J.F. Variations in the microcystin production of Planktothrix rubescens (Cyanobacteria) assessed from a four-year survey of Lac du Bourget (France) and from laboratory experiments. Microb. Ecol. 2005, 50, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Yéprémian, C.; Gugger, M.F.; Briand, E.; Catherine, A.; Berger, C.; Quiblier, C.; Bernard, C. Microcystin ecotypes in a perennial Planktothrix agardhii bloom. Water Res. 2007, 41, 4446–4456. [Google Scholar] [CrossRef] [PubMed]
- Kurmayer, R.; Deng, L.; Entfellner, E. Role of toxic and bioactive secondary metabolites in colonization and bloom formation by filamentous cyanobacteria Planktothrix. Harmful Algae 2016, 54, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Harke, M.J.; Steffen, M.M.; Gobler, C.J.; Otten, T.G.; Wilhelm, S.W.; Wood, S.A.; Paerl, H.W. A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 2016, 54, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Catherine, A.; Bernard, C.; Spoof, L.; Bruno, M. Microcystins and Nodularins. In Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis; Meriluoto, J., Spoof, L., Codd, G.A., Eds.; Whiley: West Sussex, UK, 2017. [Google Scholar] [CrossRef]
- Miles, C.O. Toxinmasslist COM v15b [Data set]. Unpublished, 2018. [Google Scholar] [CrossRef]
- Entfellner, E.; Frei, M.; Christiansen, G.; Deng, L.; Blom, J.; Kurmayer, R. Evolution of anabaenopeptin peptide structural variability in the cyanobacterium Planktothrix. Front. Microbiol. 2017, 8, 219. [Google Scholar] [CrossRef] [PubMed]
- Welker, M.; Von Döhren, H. Cyanobacterial peptides—Nature’s own combinatorial biosynthesis. FEMS Microbiol. Rev. 2006, 30, 530–563. [Google Scholar] [CrossRef] [PubMed]
- Briand, E.; Reubrecht, S.; Mondeguer, F.; Sibat, M.; Hess, P.; Amzil, Z.; Bormans, M. Chemically mediated interactions between Microcystis and Planktothrix: Impact on their growth, morphology and metabolic profiles. Environ. Microbiol. 2019, 21, 1552–1566. [Google Scholar] [CrossRef] [PubMed]
- Moss, N.A.; Bertin, M.J.; Kleigrewe, K.; Leão, T.F.; Gerwick, L.; Gerwick, W.H. Integrating mass spectrometry and genomics for cyanobacterial metabolite discovery. J. Ind. Microbiol. Biotechnol. 2016, 43, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.R.; Crüsemann, M.; Lechner, A.; Sarkar, A.; Li, J.; Ziemert, N.; Jensen, P.R. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem. Biol. 2015, 22, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Hautbergue, T.; Jamin, E.L.; Debrauwer, L.; Puel, O.; Oswald, I.P. From genomics to metabolomics, moving toward an integrated strategy for the discovery of fungal secondary metabolites. Nat. Prod. Rep. 2018, 35, 147–173. [Google Scholar] [CrossRef] [PubMed]
- Kleigrewe, K.; Almaliti, J.; Tian, I.Y.; Kinnel, R.B.; Korobeynikov, A.; Monroe, E.A.; Duggan, B.M.; Di Marzo, V.; Sherman, D.H.; Dorrestein, P.C.; et al. Combining mass spectrometric metabolic profiling with genomic analysis: A powerful approach for discovering natural products from cyanobacteria. J. Nat. Prod. 2015, 78, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Maansson, M.; Vynne, N.G.; Klitgaard, A.; Nybo, J.L.; Melchiorsen, J.; Nguyen, D.D.; Sanchez, L.M.; Ziemert, N.; Dorrestein, P.C.; Andersen, M.R.; et al. An integrated metabolomic and genomic mining workflow to uncover the biosynthetic potential of bacteria. Msystems 2016, 1, e00028-15. [Google Scholar] [CrossRef] [PubMed]
- Leikoski, N.; Liu, L.; Jokela, J.; Wahlsten, M.; Gugger, M.; Calteau, A.; Permi, P.; Kerfeld, C.A.; Sivonen, K.; Fewer, D.P. Genome mining expands the chemical diversity of the cyanobactin family to include highly modified linear peptides. Chem. Biol. 2013, 20, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Beauxis, Y.; Genta-Jouve, G. Metwork: A web server for natural products anticipation. Bioinformatics 2019, 35, 1795–1796. [Google Scholar] [CrossRef] [PubMed]
- Behsaz, B.; Mohimani, H.; Gurevich, A.; Prjibelski, A.; Fisher, M.; Smarr, L.; Dorrestein, P.C.; Mylne, J.S.; Pevzner, P.A. De novo peptide sequencing reveals a vast cyclopeptidome in human gut and other environments. BioRxiv 2019, 521872, 289–291. [Google Scholar]
- Humbert, J.F.; Barbe, V.; Latifi, A.; Gugger, M.; Calteau, A.; Coursin, T.; Lajus, A.; Castelli, V.; Oztas, S.; Samson, G.; et al. A tribute to disorder in the genome of the bloom-forming freshwater cyanobacterium Microcystis aeruginosa. PLoS ONE 2013, 8, e70747. [Google Scholar] [CrossRef] [PubMed]
- Médigue, C.; Calteau, A.; Cruveiller, S.; Gachet, M.; Gautreau, G.; Josso, A.; Lajus, A.; Langlois, J.; Pereira, H.; Planel, R.; et al. MicroScope—An integrated resource for community expertise of gene functions and comparative analysis of microbial genomic and metabolic data. Brief. Bioinform. 2017, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
| Synthetic Pathways | RiPP | NRPS and/or PKS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Planktothrix Strains/Clusters | Prenylagaramide (pag) | Unknown R1 | Microviridin (mdn) * | Anabaenopeptin (apt) * | Cyanopeptolin (mcn/oci) * | Aeruginosin (aer) | Microcystin (mcy) | Microginin (mic) | Unknown PNL1 | Unknown PNL2 | Unknown PNL3 |
| PCC 7821 | Ref | – | Ref | Ref | Ref | Ref | Ref s | Ref m | – | Ref | – |
| PCC 7805 | 92.1 | – | 92 | Partial | 96.6 | 97.0 h | – | – | Ref m | – | – |
| PCC 10110 | 98.3 | – | 96.4 | 98.2 | 98.7 | 98.3 | 99 | – | – | – | – |
| NIVA CYA 126/8 | 91.7 | Ref | 94.7 | 92.2 | 97.1 | 96.3 | 99.4 | – | 97.3 | – | Ref |
| Synthetic Pathways | RiPP | NRPS and/or PKS | Others | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Planktothrix Strains/Molecular Family | Prenylagaramide | Microviridin | Anabaenopeptin | Cyanopeptolin | Aeruginosin | Microcystin | Microginin | Phospholipid | Small Alkaloid | Carbohydrate |
| PCC 7821 | YES | N.D. | YES | YES | YES | YES | YES | YES | YES | YES |
| PCC 7805 | YES | N.D. | NO | YES | YESh | NO | NO | YES | YES | YES |
| PCC 10110 | YES | N.D. | YES | YES | YES | YES | NO | YES | YES | YES |
| NIVA CYA 126/8 | N.D. | YES | YES | YES | YES | YES | NO | YES | YES | N.D. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim Tiam, S.; Gugger, M.; Demay, J.; Le Manach, S.; Duval, C.; Bernard, C.; Marie, B. Insights into the Diversity of Secondary Metabolites of Planktothrix Using a Biphasic Approach Combining Global Genomics and Metabolomics. Toxins 2019, 11, 498. https://doi.org/10.3390/toxins11090498
Kim Tiam S, Gugger M, Demay J, Le Manach S, Duval C, Bernard C, Marie B. Insights into the Diversity of Secondary Metabolites of Planktothrix Using a Biphasic Approach Combining Global Genomics and Metabolomics. Toxins. 2019; 11(9):498. https://doi.org/10.3390/toxins11090498
Chicago/Turabian StyleKim Tiam, Sandra, Muriel Gugger, Justine Demay, Séverine Le Manach, Charlotte Duval, Cécile Bernard, and Benjamin Marie. 2019. "Insights into the Diversity of Secondary Metabolites of Planktothrix Using a Biphasic Approach Combining Global Genomics and Metabolomics" Toxins 11, no. 9: 498. https://doi.org/10.3390/toxins11090498
APA StyleKim Tiam, S., Gugger, M., Demay, J., Le Manach, S., Duval, C., Bernard, C., & Marie, B. (2019). Insights into the Diversity of Secondary Metabolites of Planktothrix Using a Biphasic Approach Combining Global Genomics and Metabolomics. Toxins, 11(9), 498. https://doi.org/10.3390/toxins11090498





