Distinct Transcriptional Changes in Response to Patulin Underlie Toxin Biosorption Differences in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Results
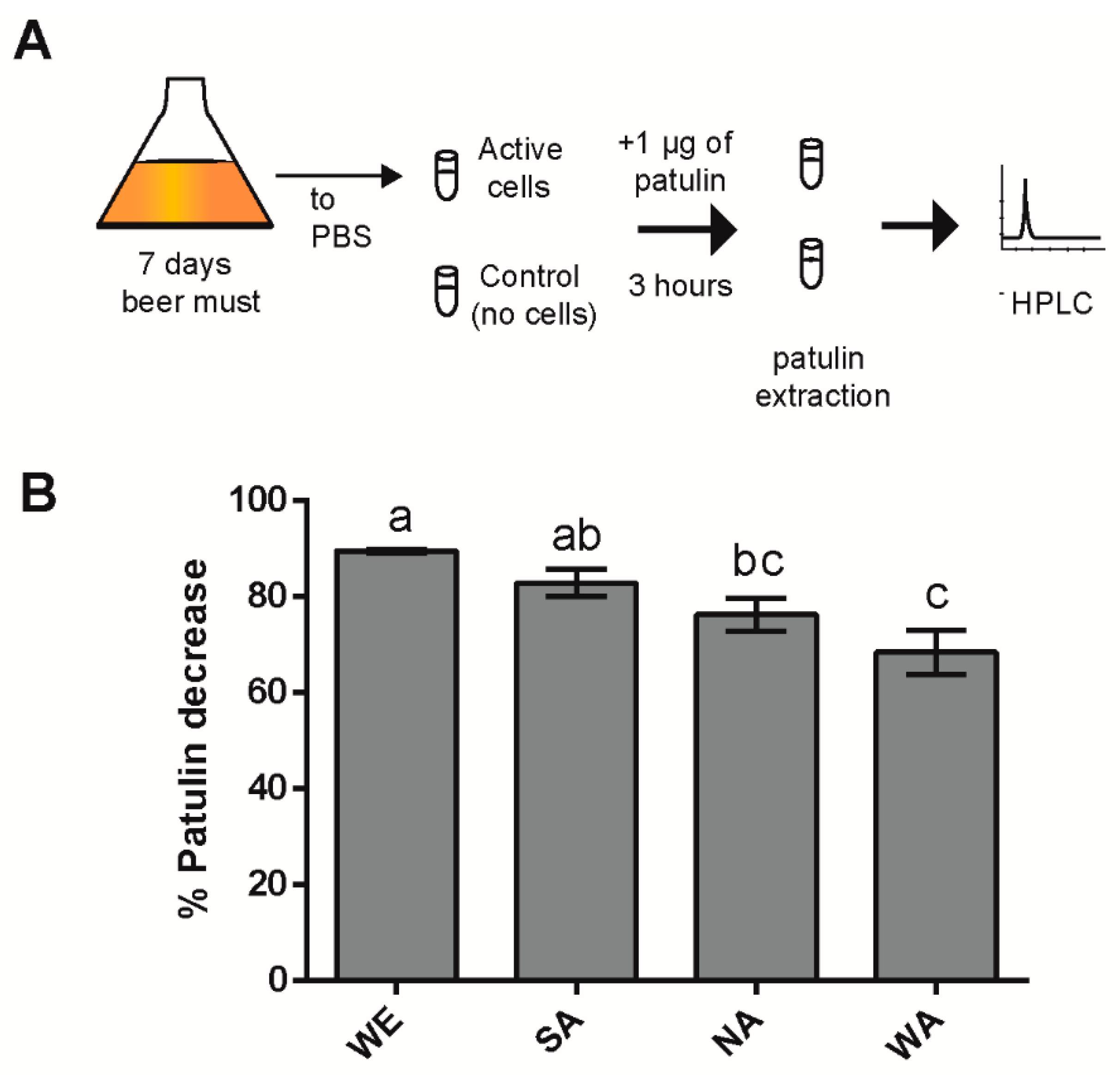
2.1. Natural Variation in PAT Biocontrol in Saccharomyces Cerevisiae
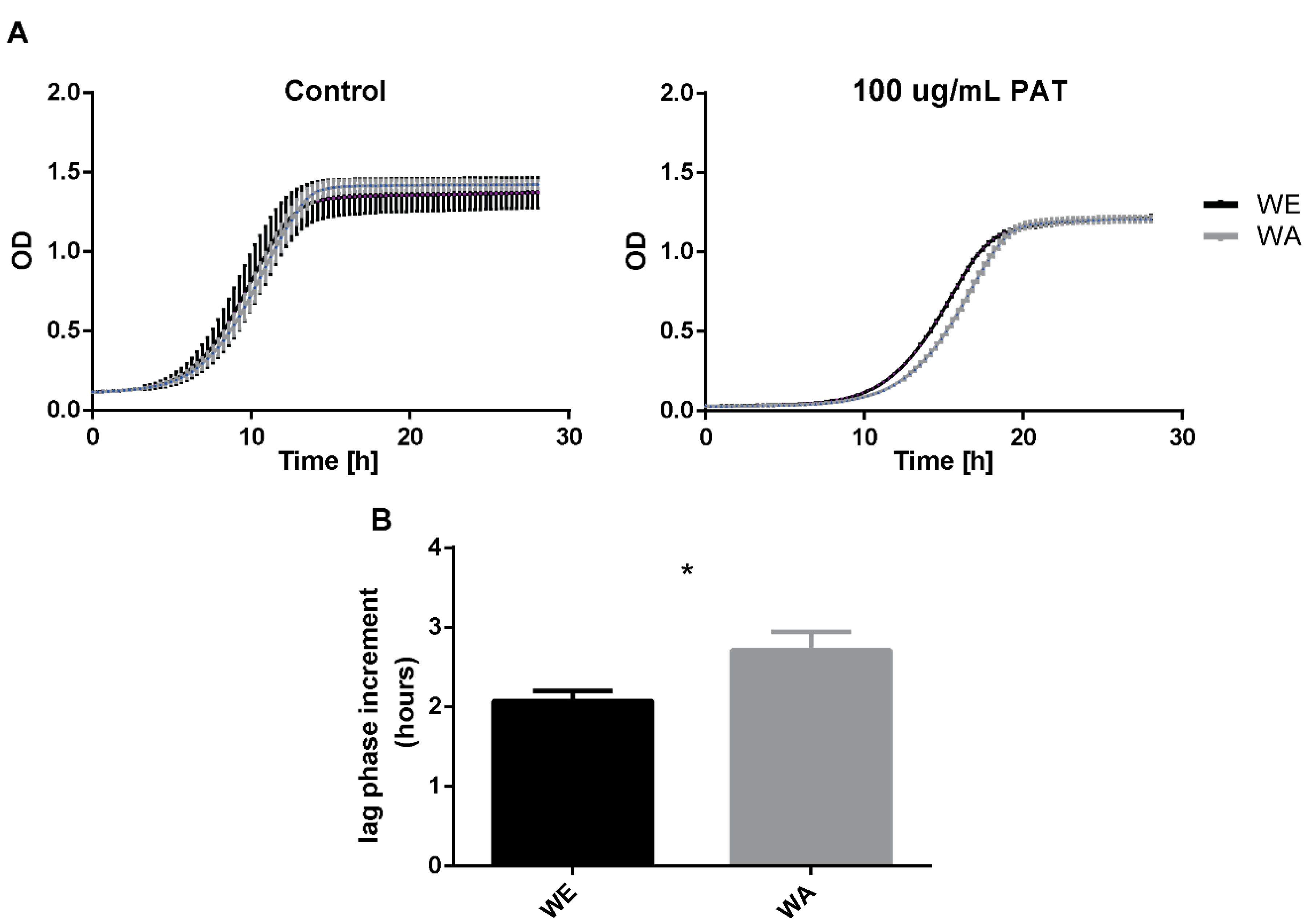
2.2. Effect of PAT on Growth of WE and WA Strains
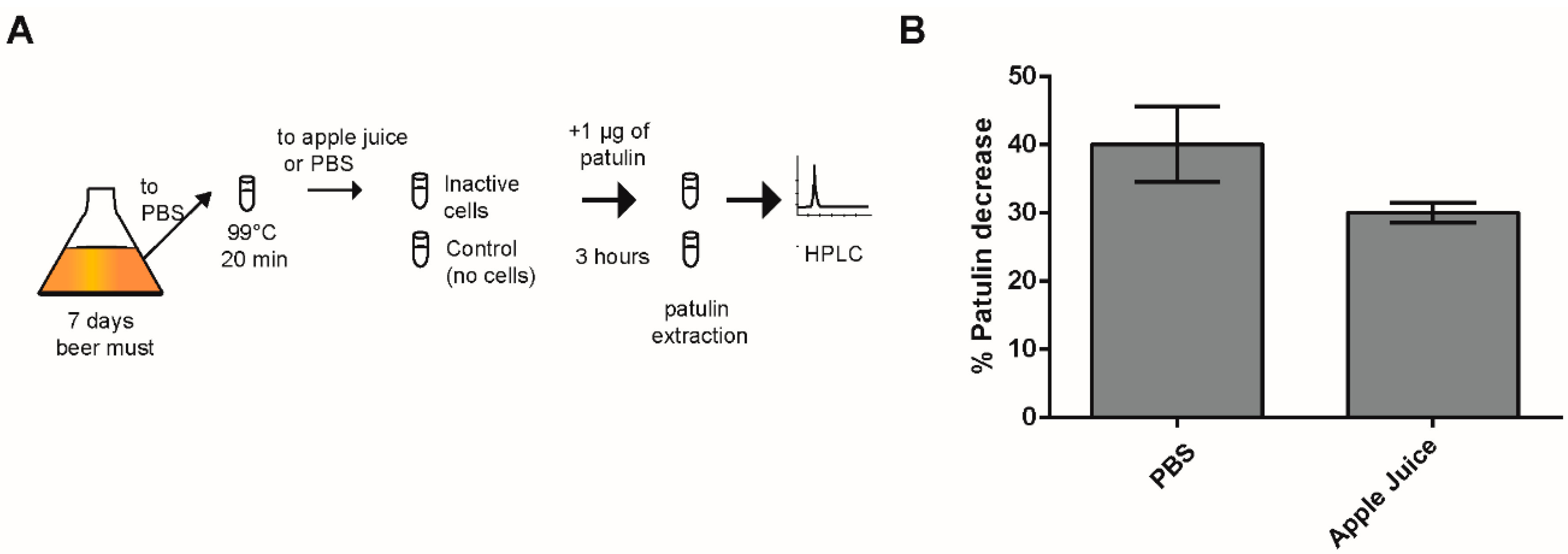
2.3. Effect of Heat-Killed Cells on PAT Biocontrol in Apple Juice
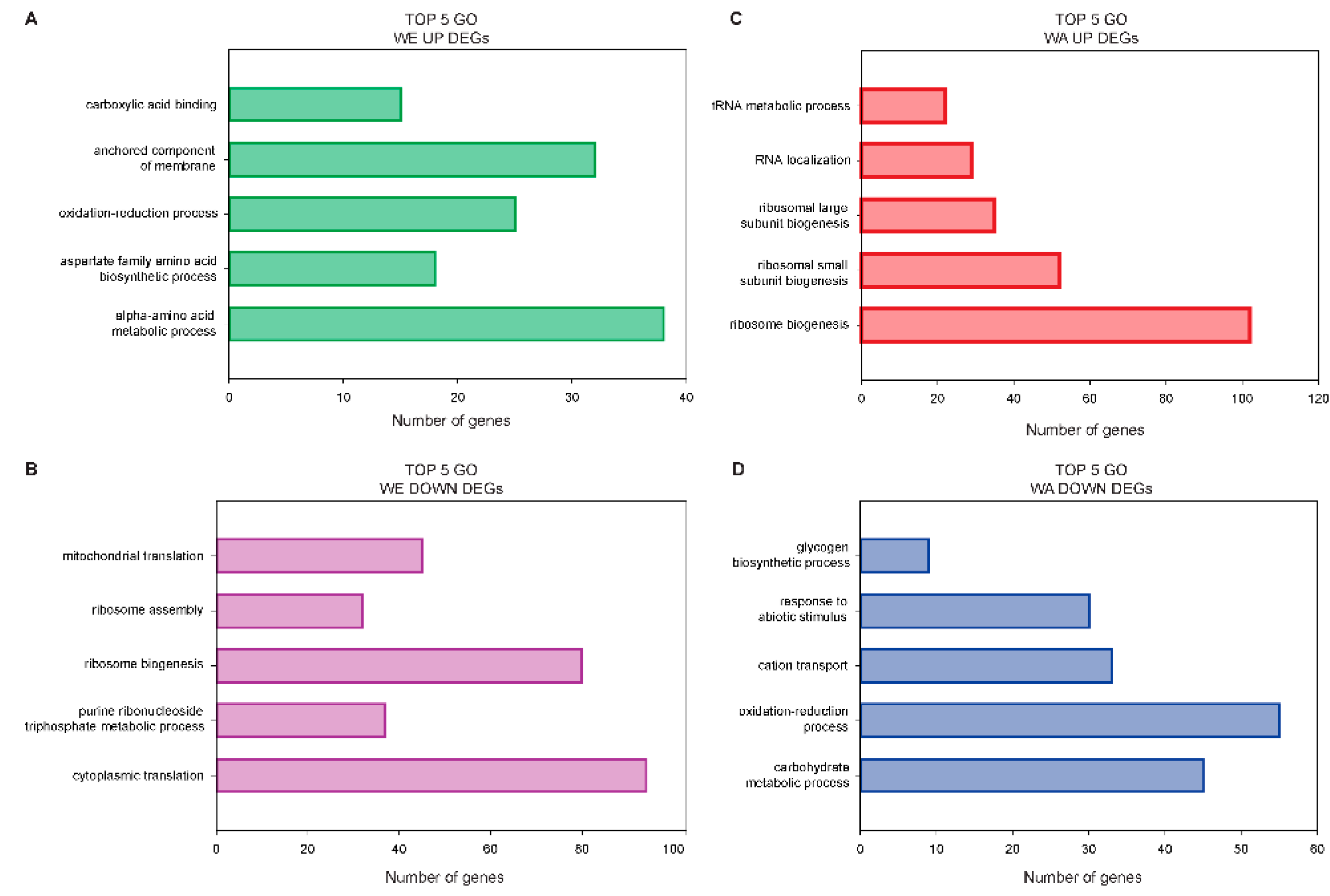
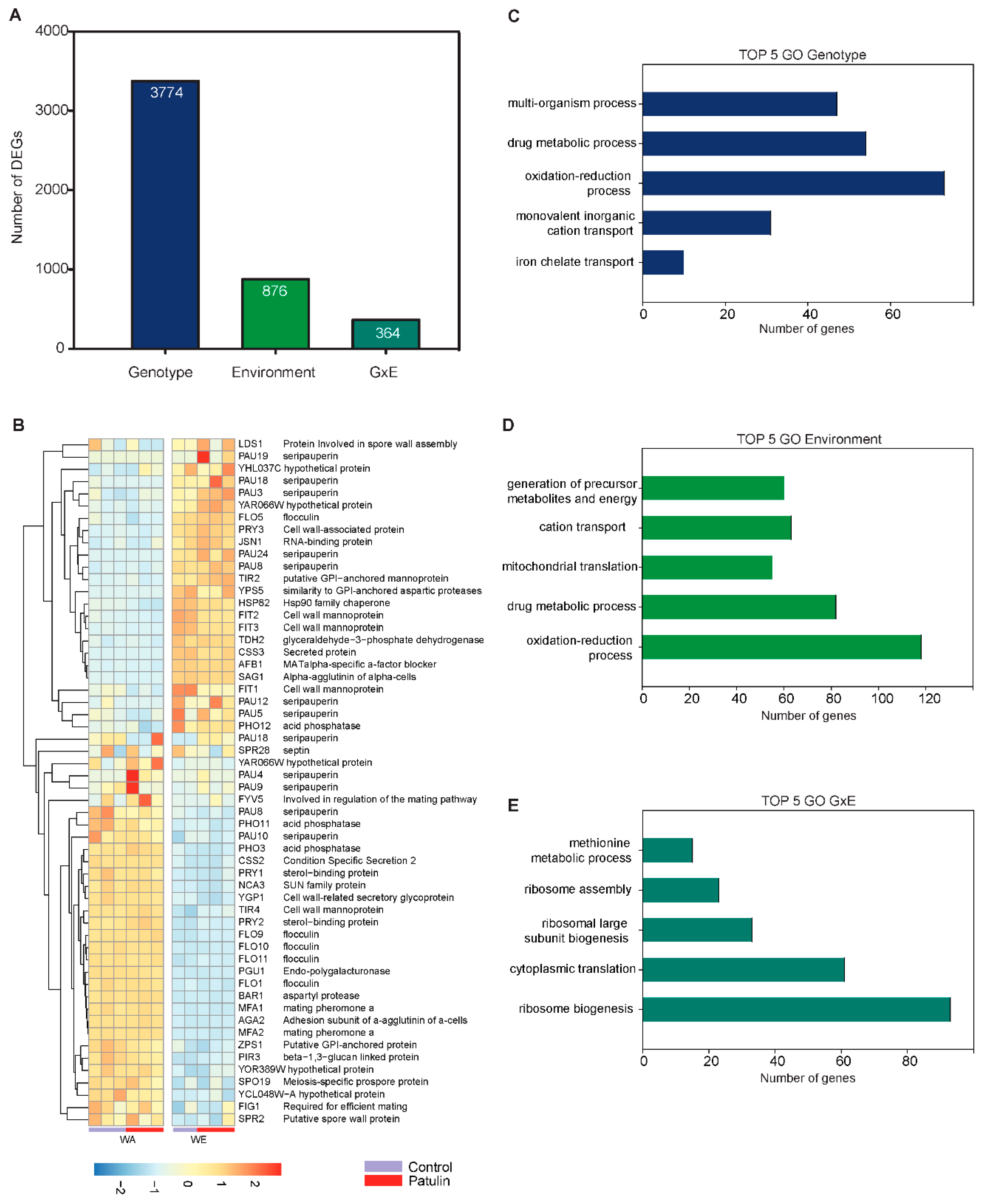
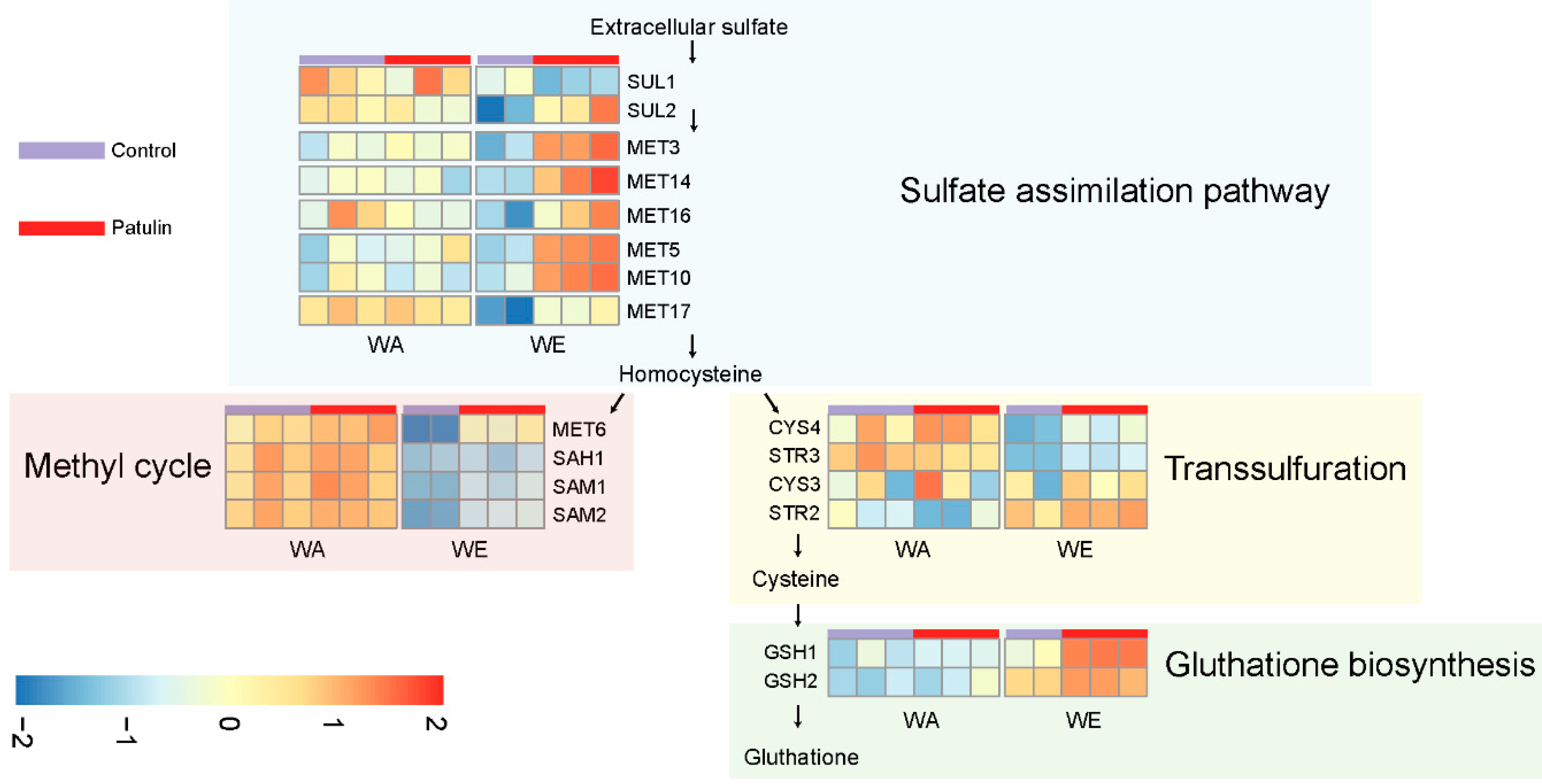
2.4. Contrasting Transcriptomic Changes after PAT Exposure in WA and WE Strains
3. Discussion
3.1. Contrasting Transcriptome Responses after PAT Exposure of Two Yeast Strains
3.2. Basal and Induced Expression of Plasma Membrane Protein Components Could Determine Differences in PAT Biosorption
3.3. PAT Biocontrol Using Inactive WE Cells
4. Materials and Methods
4.1. Yeast Strains and PAT Stock Solutions
4.2. Growth Assays
4.3. PAT Biocontrol Assay
4.4. Apple Juice Biocontrol Assay
4.5. RNA-Seq
4.6. HPLC PAT Quantification
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Capcarova, M.; Zbynovska, K.; Kalafova, A.; Bulla, J.; Bielik, P. Environment contamination by mycotoxins and their occurrence in food and feed: Physiological aspects and economical approach. J. Environ. Sci. Health B 2016, 51, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Dalie, D.K.; Deschamps, A.M.; Atanasova-Penichon, V.; Richard-Forget, F. Potential of Pediococcus pentosaceus (L006) isolated from maize leaf to suppress fumonisin-producing fungal growth. J. Food Prot. 2010, 73, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Groopman, J.D.; Pestka, J.J. Public health impacts of foodborne mycotoxins. Annu. Rev. Food Sci. Technol. 2014, 5, 351–372. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Cruz, M.L.; Mansilla, M.L.; Tadeo, J.L. Mycotoxins in fruits and their processed products: Analysis, occurrence and health implications. J. Adv. Res. 2010, 1, 113–122. [Google Scholar] [CrossRef]
- Moss, M.O. Fungi, quality and safety issues in fresh fruits and vegetables. J. Appl. Microbiol. 2008, 104, 1239–1243. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, G.; McCay, J.A.; Brown, R.D.; Musgrove, D.L.; Butterworth, L.F.; Munson, A.E.; White, K.L., Jr. Immunological evaluation of the mycotoxin patulin in female B6C3F1 mice. Food Chem. Toxicol. 1998, 36, 1107–1115. [Google Scholar] [CrossRef]
- Mahfoud, R.; Maresca, M.; Garmy, N.; Fantini, J. The mycotoxin patulin alters the barrier function of the intestinal epithelium: Mechanism of action of the toxin and protective effects of glutathione. Toxicol. Appl. Pharmacol. 2002, 181, 209–218. [Google Scholar] [CrossRef]
- Puel, O.; Galtier, P.; Oswald, I.P. Biosynthesis and Toxicological Effects of Patulin. Toxins 2010, 2, 613–631. [Google Scholar] [CrossRef]
- European Commission. Commission regulation (EC) No 1881/2006 of 19 december 2006 setting maximum levels for certain contaminants in foodstuffs. Off. J. Eur. Union 2006, 364, 5–24. [Google Scholar]
- Food and Drug Administration (FDA). Compliance policy guidance for FDA staff. Sec. 510.150 Apple juice, apple juice concentrates, and apple juice products—Adulteration with patulin. In Food and Drug Administration (FDA) Compliance Policy Guide; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2004. [Google Scholar]
- Zhong, L.; Carere, J.; Lu, Z.; Lu, F.; Zhou, T. Patulin in Apples and Apple-Based Food Products: The Burdens and the Mitigation Strategies. Toxins 2018, 10, 475. [Google Scholar] [CrossRef]
- Bandoh, S.; Takeuchi, M.; Ohsawa, K.; Higashihara, K.; Kawamoto, Y.; Goto, T. Patulin distribution in decayed apple and its reduction. Int. Biodeter. Biodegr. 2009, 63, 379–382. [Google Scholar] [CrossRef]
- Ioi, J.D.; Zhou, T.; Tsao, R.; Marcone, M.F. Mitigation of Patulin in Fresh and Processed Foods and Beverages. Toxins 2017, 9, 157. [Google Scholar] [CrossRef] [PubMed]
- Errampalli, D. Effect of fludioxonil on germination and growth of Penicillium expansum and decay in apple cvs. Empire and Gala. Crop Prot. 2004, 23, 811–817. [Google Scholar] [CrossRef]
- Sharma, R.R.; Singh, D.; Singh, R. Biological control of postharvest diseases of fruits and vegetables by microbial antagonists: A review. Biol. Control. 2009, 50, 205–221. [Google Scholar] [CrossRef]
- Funes, G.J.; Gomez, P.L.; Resnik, S.L.; Alzamora, S.M. Application of pulsed light to patulin reduction in McIlvaine buffer and apple products. Food Control 2013, 30, 405–410. [Google Scholar] [CrossRef]
- Dong, Q.; Manns, D.C.; Feng, G.; Yue, T.; Churey, J.J.; Worobo, R.W. Reduction of patulin in apple cider by UV radiation. J. Food Prot. 2010, 73, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Sands, D.C.; McIntyre, J.L.; Walton, G.S. Use of activated charcoal for the removal of patulin from cider. Appl. Environ. Microbiol. 1976, 32, 388–391. [Google Scholar]
- Topcu, A.; Bulat, T.; Wishah, R.; Boyaci, I.H. Detoxification of aflatoxin B1 and patulin by Enterococcus faecium strains. Int. J. Food Microbiol. 2010, 139, 202–205. [Google Scholar] [CrossRef]
- Yue, T.; Dong, Q.; Guo, C.; Worobo, R.W. Reducing patulin contamination in apple juice by using inactive yeast. J. Food Prot. 2011, 74, 149–153. [Google Scholar] [CrossRef]
- Coelho, A.R.; Celli, M.G.; Ono, E.Y.S.; Hoffmann, F.L.; Pagnocca, F.C.; Garcia, S.; Sabino, M.; Harada, K.I.; Wosiacki, G.; Hirooka, E.Y. Patulin biodegradation using Pichia ohmeri and Saccharomyces cerevisiae. World Mycotoxin J. 2008, 1, 325–331. [Google Scholar] [CrossRef]
- Stinson, E.E.; Osman, S.F.; Huhtanen, C.N.; Bills, D.D. Disappearance of patulin during alcoholic fermentation of apple juice. Appl. Environ. Microbiol. 1978, 36, 620–622. [Google Scholar] [PubMed]
- Moss, M.O.; Long, M.T. Fate of patulin in the presence of the yeast Saccharomyces cerevisiae. Food Addit. Contam. 2002, 19, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Tannous, J.; Snini, S.P.; El Khoury, R.; Canlet, C.; Pinton, P.; Lippi, Y.; Alassane-Kpembi, I.; Gauthier, T.; El Khoury, A.; Atoui, A.; et al. Patulin transformation products and last intermediates in its biosynthetic pathway, E- and Z-ascladiol, are not toxic to human cells. Arch. Toxicol. 2017, 91, 2455–2467. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.; Bahuguna, A.; Kim, M. The effects of mycotoxin patulin on cells and cellular components. Trends Food Sci. Technol. 2019, 83, 99–113. [Google Scholar] [CrossRef]
- Pfenning, C.; Esch, H.L.; Fliege, R.; Lehmann, L. The mycotoxin patulin reacts with DNA bases with and without previous conjugation to GSH: Implication for related α, β-unsaturated carbonyl compounds? Arch. Toxicol. 2016, 90, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Ianiri, G.; Idnurm, A.; Castoria, R. Transcriptomic responses of the basidiomycete yeast Sporobolomyces sp. to the mycotoxin patulin. BMC Genom. 2016, 17, 210. [Google Scholar] [CrossRef] [PubMed]
- Iwahashi, Y.; Hosoda, H.; Park, J.-H.; Lee, J.-H.; Suzuki, Y.; Kitagawa, E.; Murata, S.M.; Jwa, N.-S.; Gu, M.-B.; Iwahashi, H. Mechanisms of Patulin Toxicity under Conditions That Inhibit Yeast Growth. J. Agric. Food Chem. 2006, 54, 1936–1942. [Google Scholar] [CrossRef]
- Yiannikouris, A.; François, J.; Poughon, L.; Dussap, C.-G.; Bertin, G.; Jeminet, G.; Jouany, J.-P. Alkali Extraction of β-d-Glucans from Saccharomyces cerevisiae Cell Wall and Study of Their Adsorptive Properties toward Zearalenone. J. Agric. Food Chem. 2004, 52, 3666–3673. [Google Scholar] [CrossRef]
- Yiannikouris, A.; André, G.; Poughon, L.; François, J.; Dussap, C.-G.; Jeminet, G.; Bertin, G.; Jouany, J.-P. Chemical and Conformational Study of the Interactions Involved in Mycotoxin Complexation with β-d-Glucans. Biomacromolecules 2006, 7, 1147–1155. [Google Scholar] [CrossRef]
- Guo, C.; Yuan, Y.; Yue, T.; Hatab, S.; Wang, Z. Binding mechanism of patulin to heat-treated yeast cell. Lett. Appl. Microbiol. 2012, 55, 453–459. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, J.; Liu, B.; Wang, Z.; Yuan, Y.; Yue, T. Effect of Yeast Cell Morphology, Cell Wall Physical Structure and Chemical Composition on Patulin Adsorption. PLoS ONE 2015, 10, e0136045. [Google Scholar] [CrossRef] [PubMed]
- Castoria, R.; Mannina, L.; Durán-Patrón, R.; Maffei, F.; Sobolev, A.P.; De Felice, D.V.; Pinedo-Rivilla, C.; Ritieni, A.; Ferracane, R.; Wright, S.A.I. Conversion of the Mycotoxin Patulin to the Less Toxic Desoxypatulinic Acid by the Biocontrol Yeast Rhodosporidium kratochvilovae Strain LS11. J. Agric. Food Chem. 2011, 59, 11571–11578. [Google Scholar] [CrossRef] [PubMed]
- Cubillos, F.A. Exploiting budding yeast natural variation for industrial processes. Curr. Genet. 2016, 62, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Liti, G.; Carter, D.M.; Moses, A.M.; Warringer, J.; Parts, L.; James, S.A.; Davey, R.P.; Roberts, I.N.; Burt, A.; Koufopanou, V.; et al. Population genomics of domestic and wild yeasts. Nature 2009, 458, 337–341. [Google Scholar] [CrossRef]
- Wang, L.; Yue, T.L.; Yuan, Y.H.; Wang, Z.L.; Ye, M.Q.; Cal, R. A new insight into the adsorption mechanism of patulin by the heat-inactive lactic acid bacteria cells. Food Control 2015, 50, 104–110. [Google Scholar] [CrossRef]
- Bergstrom, A.; Simpson, J.T.; Salinas, F.; Barre, B.; Parts, L.; Zia, A.; Nguyen Ba, A.N.; Moses, A.M.; Louis, E.J.; Mustonen, V.; et al. A high-definition view of functional genetic variation from natural yeast genomes. Mol. Biol. Evol. 2014, 31, 872–888. [Google Scholar] [CrossRef] [PubMed]
- Jara, M.; Cubillos, F.A.; Garcia, V.; Salinas, F.; Aguilera, O.; Liti, G.; Martinez, C. Mapping genetic variants underlying differences in the central nitrogen metabolism in fermenter yeasts. PLoS ONE 2014, 9, e86533. [Google Scholar] [CrossRef] [PubMed]
- Kessi-Perez, E.I.; Araos, S.; Garcia, V.; Salinas, F.; Abarca, V.; Larrondo, L.F.; Martinez, C.; Cubillos, F.A. RIM15 antagonistic pleiotropy is responsible for differences in fermentation and stress response kinetics in budding yeast. FEMS Yeast Res. 2016, 16, fow021. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zimmer, A.; Durand, C.; Loira, N.; Durrens, P.; Sherman, D.J.; Marullo, P. QTL dissection of Lag phase in wine fermentation reveals a new translocation responsible for Saccharomyces cerevisiae adaptation to sulfite. PLoS ONE 2014, 9, e86298. [Google Scholar] [CrossRef]
- Quispe, X.; Tapia, S.M.; Villarroel, C.; Oporto, C.; Abarca, V.; Garcia, V.; Martinez, C.; Cubillos, F.A. Genetic basis of mycotoxin susceptibility differences between budding yeast isolates. Sci. Rep. 2017, 7, 9173. [Google Scholar] [CrossRef] [PubMed]
- Peter, J.; De Chiara, M.; Friedrich, A.; Yue, J.X.; Pflieger, D.; Bergstrom, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; et al. Genome evolution across 1011 Saccharomyces cerevisiae isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Dickens, F.; Jones, H.E. Carcinogenic activity of a series of reactive lactones and related substances. Br. J. Cancer 1961, 15, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Moreno, A.; Hanna, J. Induction of proteotoxic stress by the mycotoxin patulin. Toxicol. Lett. 2017, 276, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.; Francke Johannesen, P. Cysteine is essential for transcriptional regulation of the sulfur assimilation genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000, 263, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Thorsen, M.; Lagniel, G.; Kristiansson, E.; Junot, C.; Nerman, O.; Labarre, J.; Tamás, M.J. Quantitative transcriptome, proteome, and sulfur metabolite profiling of the Saccharomyces cerevisiae response to arsenite. Physiol. Genom. 2007, 30, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Momose, Y.; Iwahashi, H. Bioassay of cadmium using a DNA microarray: Genome-wide expression patterns of Saccharomyces cerevisiae response to cadmium. Environ. Toxicol. Chem. 2001, 20, 2353–2360. [Google Scholar] [CrossRef] [PubMed]
- Piper, P.; Mahé, Y.; Thompson, S.; Pandjaitan, R.; Holyoak, C.; Egner, R.; Mühlbauer, M.; Coote, P.; Kuchler, K. The pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 1998, 17, 4257–4265. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, A.J.; Brauer, M.J.; Botstein, D. Nutritional homeostasis in batch and steady-state culture of yeast. Mol. Biol. Cell 2004, 15, 4089–4104. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Sirisattha, S.; Mori, K.; Iwahashi, Y. Mycotoxin Toxicity in Saccharomyces cerevisiae Differs Depending on Gene Mutations. Food Sci. Technol. Res. 2009, 15, 453–458. [Google Scholar] [CrossRef]
- Abramova, N.; Sertil, O.; Mehta, S.; Lowry, C.V. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol. 2001, 183, 2881–2887. [Google Scholar] [CrossRef]
- Abe, F. Induction of DAN/TIR yeast cell wall mannoprotein genes in response to high hydrostatic pressure and low temperature. FEBS Lett. 2007, 581, 4993–4998. [Google Scholar] [CrossRef] [PubMed]
- Protchenko, O.; Ferea, T.; Rashford, J.; Tiedeman, J.; Brown, P.O.; Botstein, D.; Philpott, C.C. Three Cell Wall Mannoproteins Facilitate the Uptake of Iron in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 49244–49250. [Google Scholar] [CrossRef] [PubMed]
- Cubillos, F.A.; Billi, E.; Zorgo, E.; Parts, L.; Fargier, P.; Omholt, S.; Blomberg, A.; Warringer, J.; Louis, E.J.; Liti, G. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol. Ecol. 2011, 20, 1401–1413. [Google Scholar] [CrossRef] [PubMed]
- Cubillos, F.A.; Louis, E.J.; Liti, G. Generation of a large set of genetically tractable haploid and diploid Saccharomyces strains. FEMS Yeast Res. 2009, 9, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G.; Acar, H.; Nandipati, A.; Barlow, M. Growth Rates Made Easy. Mol. Biol. Evol. 2013, 31, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Bovo, F.; Corassin, C.H.; Rosim, R.E.; de Oliveira, C.A.F. Efficiency of Lactic Acid Bacteria Strains for Decontamination of Aflatoxin M1 in Phosphate Buffer Saline Solution and in Skimmed Milk. Food Bioprocess Technol. 2012, 6, 2230–2234. [Google Scholar] [CrossRef]
- Oliveira, P.M.; Zannini, E.; Arendt, E.K. Cereal fungal infection, mycotoxins, and lactic acid bacteria mediated bioprotection: From crop farming to cereal products. Food Microbiol. 2014, 37, 78–95. [Google Scholar] [CrossRef]
- Shedlovskiy, D.; Shcherbik, N.; Pestov, D.G. One-step hot formamide extraction of RNA from Saccharomyces cerevisiae. RNA Biol. 2017, 14, 1722–1726. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oporto, C.I.; Villarroel, C.A.; Tapia, S.M.; García, V.; Cubillos, F.A. Distinct Transcriptional Changes in Response to Patulin Underlie Toxin Biosorption Differences in Saccharomyces cerevisiae. Toxins 2019, 11, 400. https://doi.org/10.3390/toxins11070400
Oporto CI, Villarroel CA, Tapia SM, García V, Cubillos FA. Distinct Transcriptional Changes in Response to Patulin Underlie Toxin Biosorption Differences in Saccharomyces cerevisiae. Toxins. 2019; 11(7):400. https://doi.org/10.3390/toxins11070400
Chicago/Turabian StyleOporto, Christian I., Carlos A. Villarroel, Sebastián M. Tapia, Verónica García, and Francisco A. Cubillos. 2019. "Distinct Transcriptional Changes in Response to Patulin Underlie Toxin Biosorption Differences in Saccharomyces cerevisiae" Toxins 11, no. 7: 400. https://doi.org/10.3390/toxins11070400
APA StyleOporto, C. I., Villarroel, C. A., Tapia, S. M., García, V., & Cubillos, F. A. (2019). Distinct Transcriptional Changes in Response to Patulin Underlie Toxin Biosorption Differences in Saccharomyces cerevisiae. Toxins, 11(7), 400. https://doi.org/10.3390/toxins11070400





