A Short Peptide Derived from the ZorO Toxin Functions as an Effective Antimicrobial
Abstract
1. Introduction
2. Results
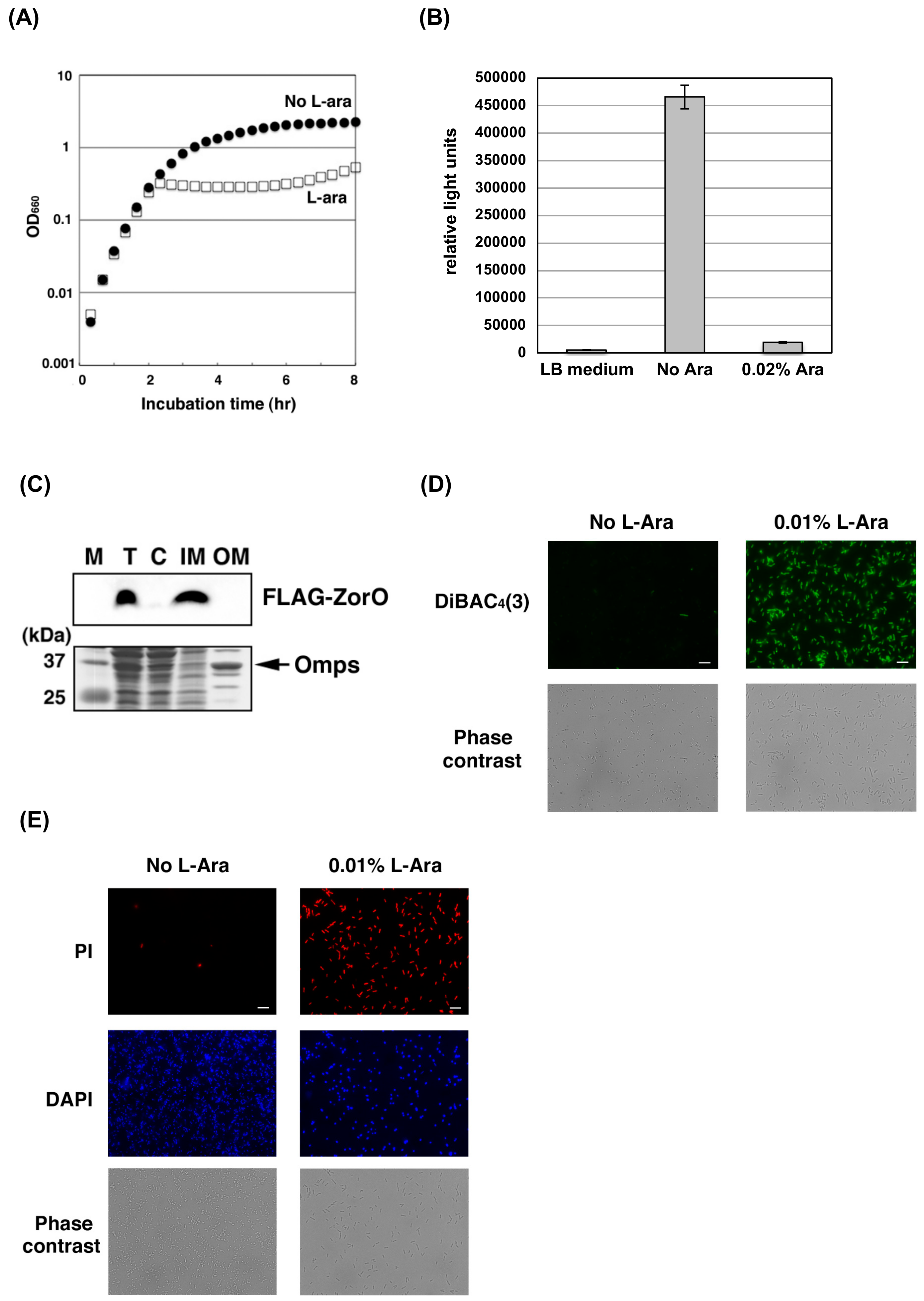
2.1. ZorO Affects the Plasma Membrane Potential and Integrity
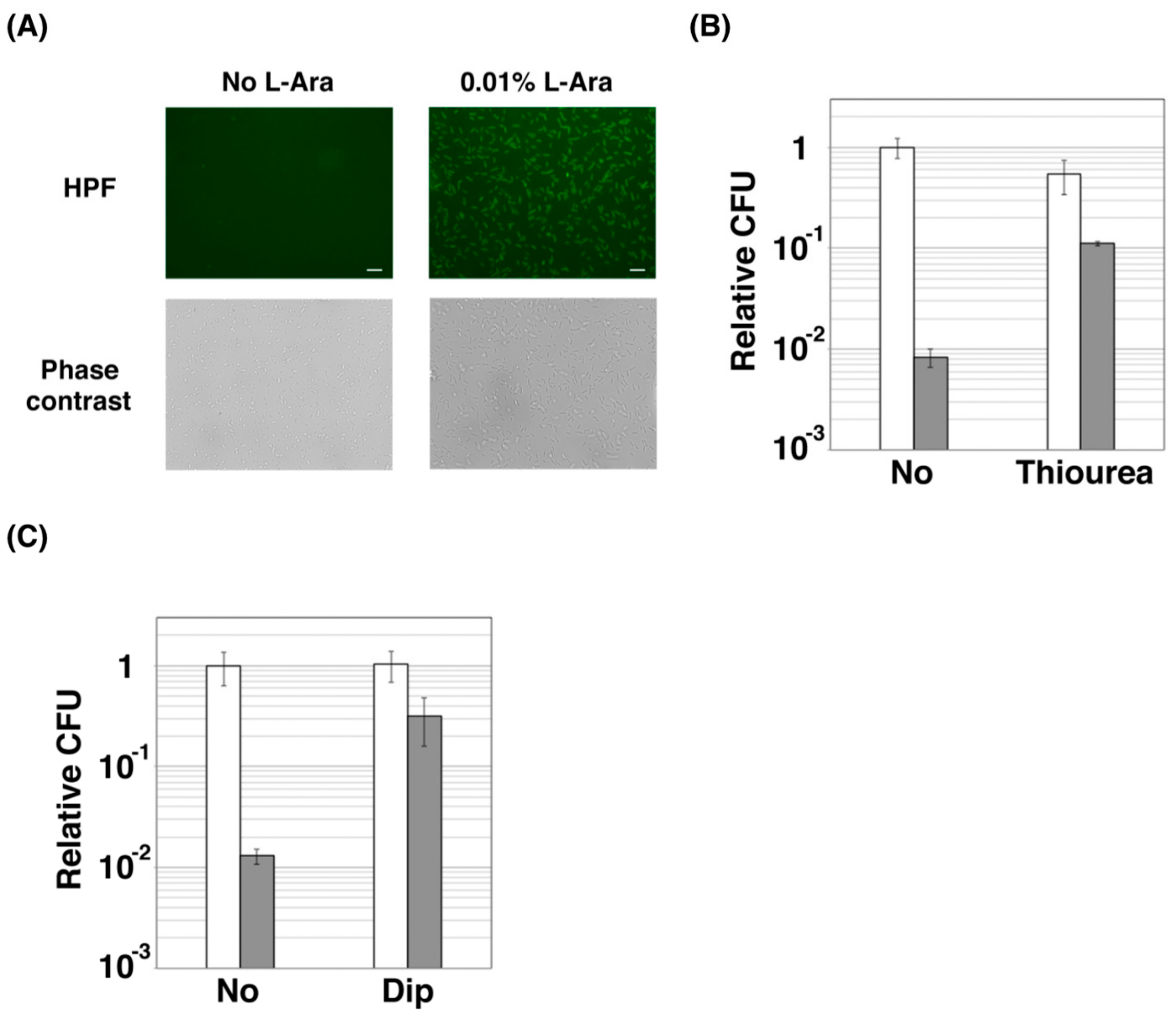
2.2. ZorO Affects Cell Viability Though Production of Hydroxyl Radicals
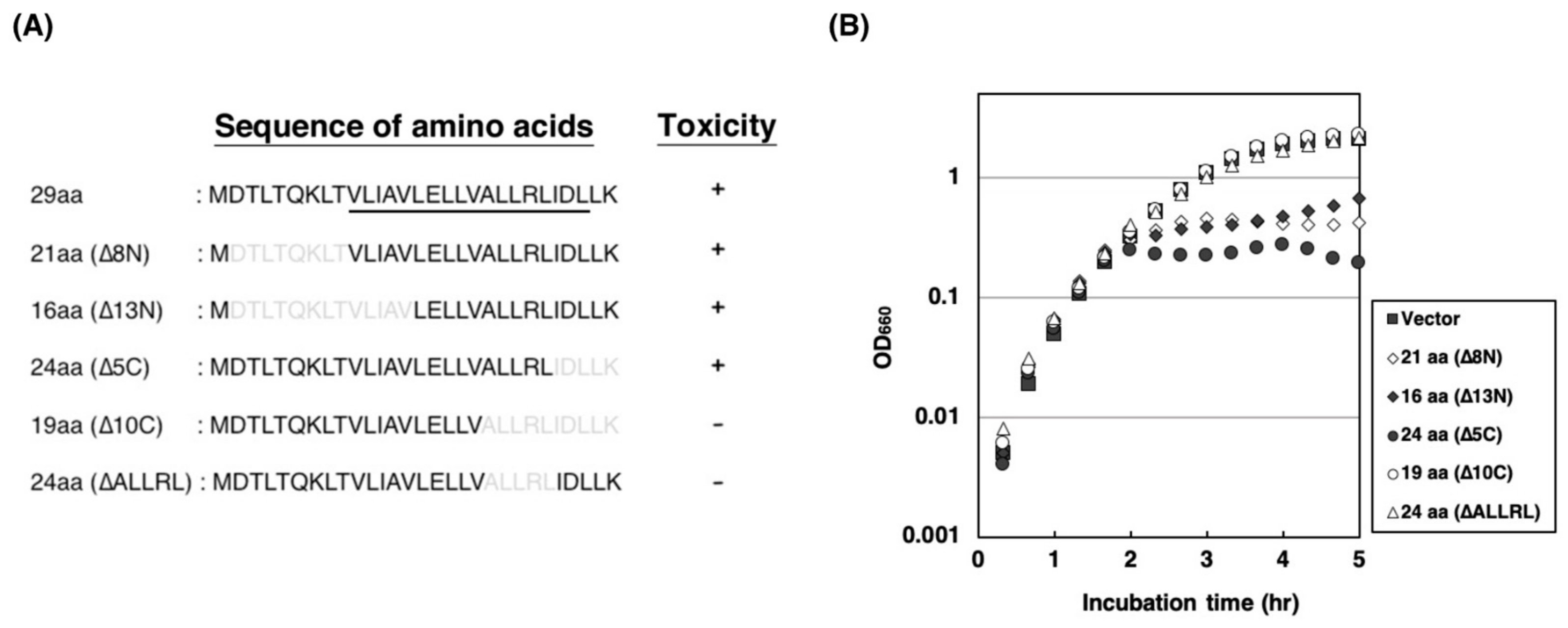
2.3. Amino Acid Residues of ALLRL Are Required for the Toxicity of ZorO
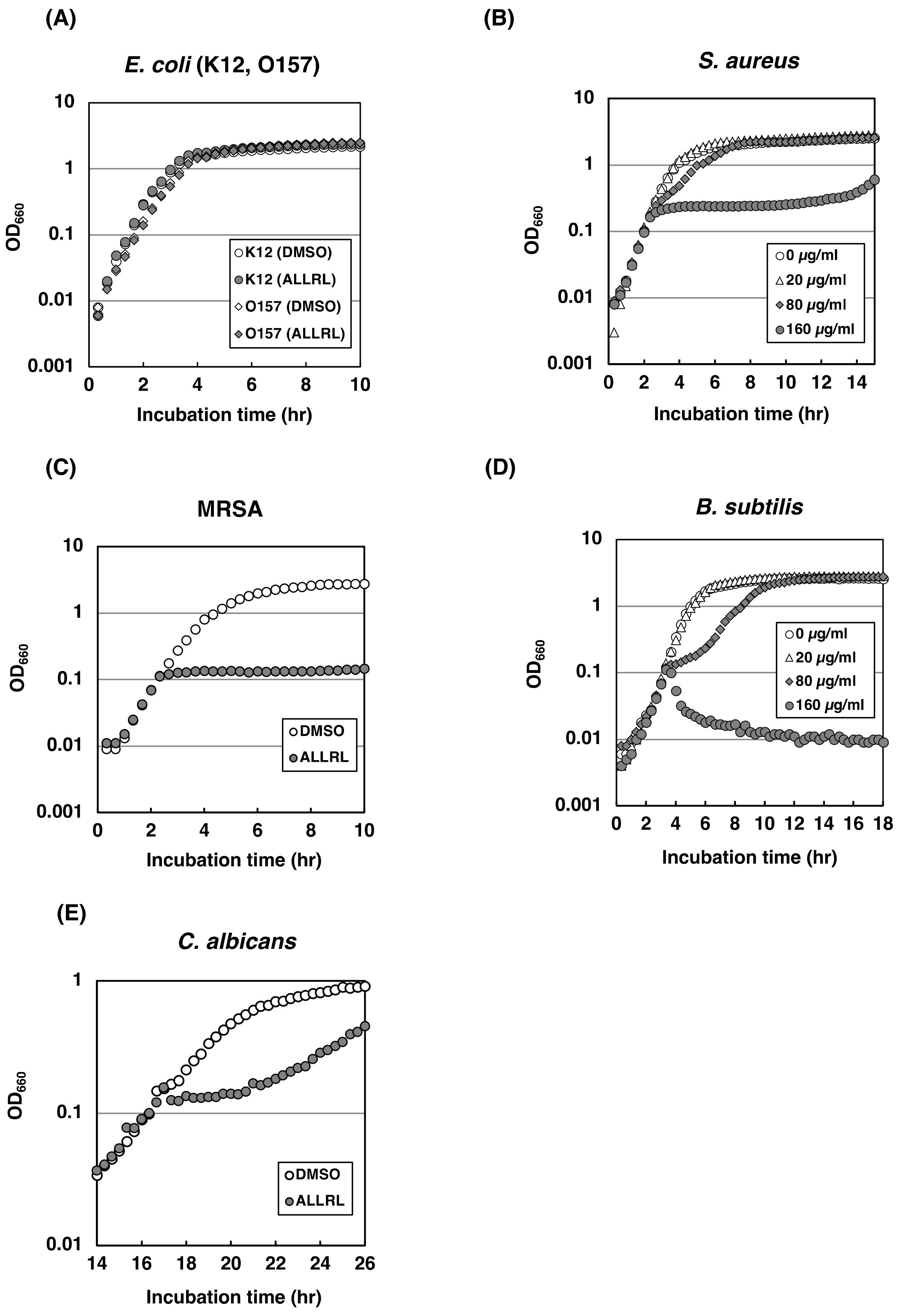
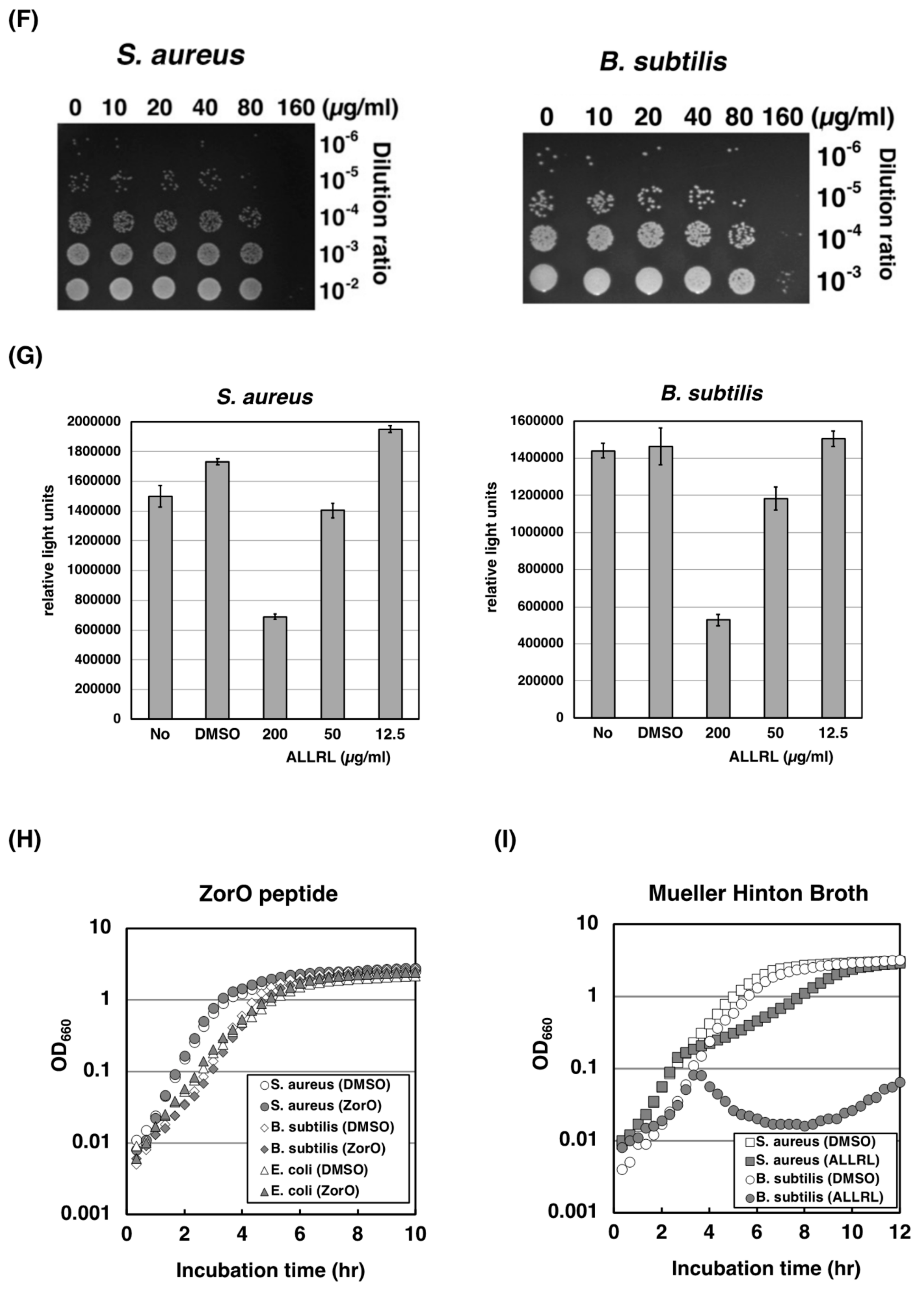
2.4. The ALLRL Peptide Exhibits Antimicrobial Activity against Gram-Positive Bacteria and Candida
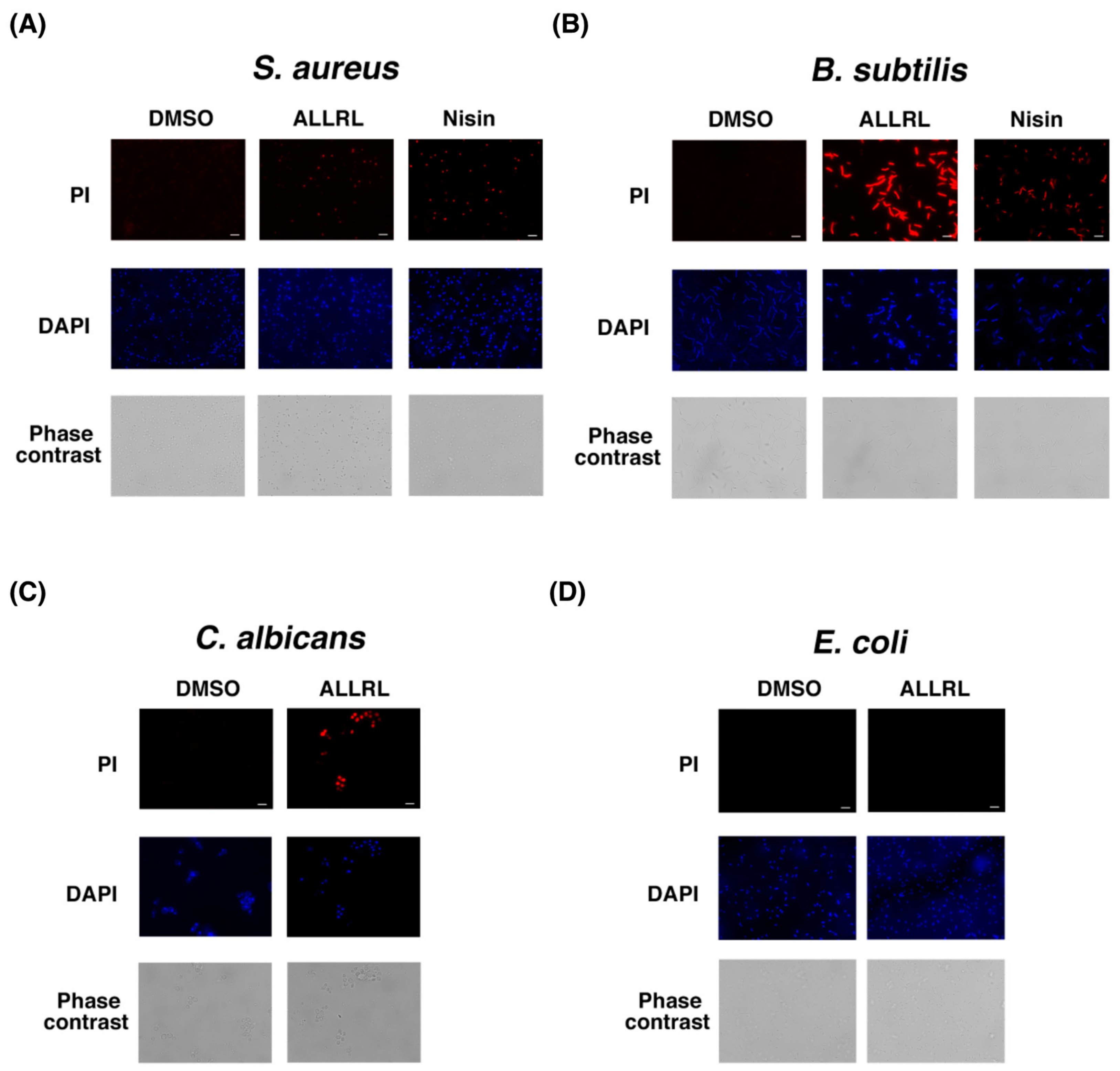
2.5. The ALLRL Peptide Induces the Plasma Membrane Damage
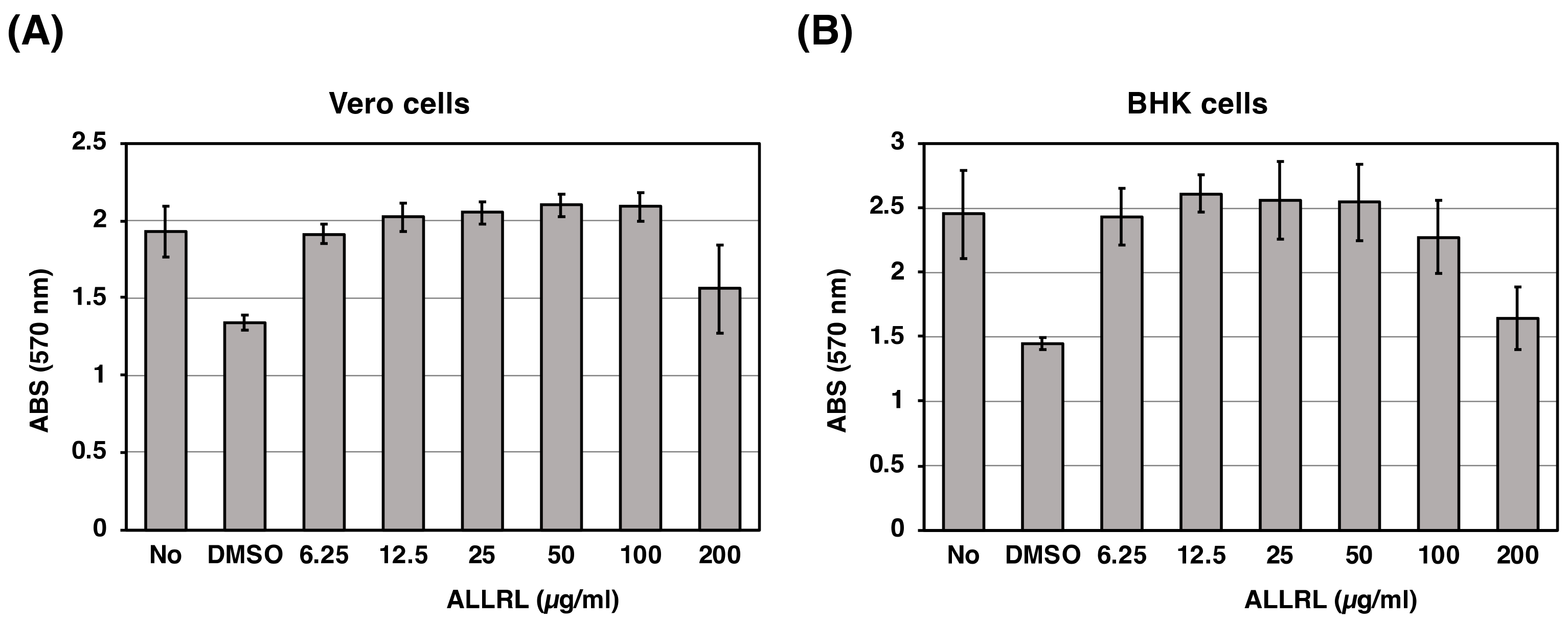
2.6. The ALLRL Peptide Has no Cytotoxicity Against Mammalian Cells
3. Discussion
4. Materials and Methods
4.1. Bacterial and Fungus Strains and Mammalian Cultured Cells
4.2. Construction of Plasmids
4.3. Peptides
4.4. Growth of Bacteria and Fungus
4.5. Microbial Cell Viability Assay
4.6. Subcellular Fractionation and Western Blotting Analysis
4.7. CFU Assay
4.8. Microscopy
4.9. Cytotoxicity Assay (MTT Assay)
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Kjos, M.; Nes, I.F.; Diep, D.B.; Lotfipour, F. Natural antimicrobial peptides from bacteria: Characteristics and potential applications to fight against antibiotic resistance. J. Appl. Microbiol. 2012, 113, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.N.; Ferre, R.; Castanho, M.A.R.B. Antimicrobial peptides: Linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009, 7, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Gomes, B.; Augusto, M.T.; Felício, M.R.; Hollmann, A.; Franco, O.L.; Gonçalves, S.; Santos, N.C. Designing improved active peptides for therapeutic approaches against infectious diseases. Biotechnol. Adv. 2018, 36, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.; Cho, Y.; Dinh, N.N.; Waring, A.J.; Lehrer, R.I. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob. Agents Chemother. 1998, 42, 2206–2214. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.L.; Yu, H.Y.; Yip, B.S.; Chih, Y.H.; Liang, C.W.; Cheng, H.T.; Cheng, J.W. Boosting salt resistance of short antimicrobial peptides. Antimicrob. Agents Chemother. 2013, 57, 4050–4052. [Google Scholar] [CrossRef]
- Goldman, M.J.; Anderson, G.M.; Stolzenberg, E.D.; Kari, U.P.; Zasloff, M.; Wilson, J.M. Human β-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 1997, 88, 553–560. [Google Scholar] [CrossRef]
- Gerdes, K.; Christensen, S.K.; Løbner-Olesen, A. Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 2005, 3, 371–382. [Google Scholar] [CrossRef]
- Yarmolinsky, M.B. Programmed cell death in bacterial populations. Science 1995, 267, 836–837. [Google Scholar] [CrossRef]
- Ogura, T.; Hiraga, S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 1983, 80, 4784–4788. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kim, Y.; Hong, S.H.; Ma, Q.; Brown, B.L.; Pu, M.; Tarone, A.M.; Benedik, M.J.; Peti, W.; Page, R.; et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 2011, 7, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Benedik, M.J.; Wood, T.K. Antitoxin DinJ influences the general stress response through transcript stabilizer CspE. Environ. Microbiol. 2012, 14, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Pecota, D.C.; Wood, T.K. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 1996, 178, 2044–2050. [Google Scholar] [CrossRef] [PubMed]
- Koga, M.; Otsuka, Y.; Lemire, S.; Yonesaki, T. Escherichia coli rnlA and rnlB compose a novel toxin–antitoxin systems. Genetics 2011, 187, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Fineran, P.C.; Blower, T.R.; Foulds, I.J.; Humphreys, D.P.; Lilley, K.S.; Salmond, G.P.C. The phage abortive infection system, ToxIN, functions as a protein–RNA toxin–antitoxin pair. Proc. Natl. Acad. Sci. USA 2009, 106, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Tan, Q.; Awano, N.; Wu, K.P.; Inouye, M. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 2012, 84, 979–989. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lord, D.M.; Cheng, H.Y.; Osbourne, D.O.; Hong, S.H.; Sanchez-Torres, V.; Quiroga, C.; Zheng, K.; Herrmann, T.; Peti, W.; et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 2012, 8, 855–861. [Google Scholar] [CrossRef]
- Aakre, C.D.; Phung, T.N.; Huang, D.; Laub, M.T. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the β sliding clamp. Mol. Cell 2013, 52, 617–628. [Google Scholar] [CrossRef]
- Fozo, E.M.; Hemm, M.R.; Storz, G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008, 72, 579–589. [Google Scholar] [CrossRef]
- Fozo, E.M.; Makarova, K.S.; Shabalina, S.A.; Yutin, N.; Koonin, E.V.; Stroz, G. Abundance of type I toxin–antitoxin systems in bacteria: Searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010, 38, 3743–3759. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Won, D.; Fozo, E.M. The ZorO-OrzO type I toxin–antitoxin locus: Repression by the OrzO antitoxin. Nucleic Acids Res. 2014, 42, 1930–1946. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, F.; Fuchs, R.T.; Storz, G. Membrane localization of small proteins in Escherichia coli. J. Biol. Chem. 2011, 286, 32464–32474. [Google Scholar] [CrossRef] [PubMed]
- Sayed, N.; Nonin-Lecomte, S.; Réty, S.; Felden, B. Functional and structural insights of a Staphylococcus aureus apoptotic-like membrane peptide from a toxin–antitoxin module. J. Biol. Chem. 2012, 287, 43454–43463. [Google Scholar] [CrossRef]
- Jepras, R.I.; Paul, F.E.; Pearson, S.C.; Wilkinson, M.J. Rapid assessment of antibiotic effects on Escherichia coli by bis-(1,3-dibutylbarbituric acid) trimethine oxonol and flow cytometry. Antimicrob. Agents Chemother. 1997, 41, 2001–2005. [Google Scholar] [CrossRef]
- Davies, B.W.; Kohanski, M.A.; Simmons, L.A.; Winkler, J.A.; Collins, J.J.; Walker, G.C. Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Mol. Cell 2009, 36, 845–860. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef]
- Breukink, E.; Wiedemann, I.; van Kraaij, C.; Kuipers, O.P.; Sahl, H.G.; de Kruijff, B. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 1999, 286, 2361–2364. [Google Scholar] [CrossRef]
- Gurnev, P.A.; Ortenberg, R.; Dörr, T.; Lewis, K.; Bezrukov, S.M. Persister-promoting bacterial toxin TisB produces anion-selective pores in planar lipid bilayers. FEBS Lett. 2012, 586, 2529–2534. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Soo, V.W.; Islam, S.; McAnulty, M.J.; Benedik, M.J.; Wood, T.K. Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ. Microbiol. 2014, 16, 1741–1754. [Google Scholar] [CrossRef]
- Wilmaerts, D.; Bayoumi, M.; Dewachter, L.; Knapen, W.; Mika, J.T.; Hofkens, J.; Dedecker, P.; Maglia, G.; Verstraeten, N.; Michiels, J. The Persistence-inducing toxin HokB forms dynamic pores that cause ATP leakage. MBio 2018, 9, e00744-18. [Google Scholar] [CrossRef] [PubMed]
- Le Lay, C.; Akerey, B.; Fliss, I.; Subirade, M.; Rouabhia, M. Nisin Z inhibits the growth of Candida albicans and its transition from blastospore to hyphal form. J. Appl. Microbiol. 2008, 105, 1630–1639. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, J.; Montville, T.J.; Nes, I.F.; Chikindas, M.L. Bacteriocins: Safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 2001, 71, 1–20. [Google Scholar] [CrossRef]
- Wiedemann, I.; Breukink, E.; van Kraaij, C.; Kuipers, O.P.; Bierbaum, G.; de Kruijff, B.; Sahl, H.G. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 2001, 276, 1772–1779. [Google Scholar] [CrossRef] [PubMed]
- Hasper, H.E.; Kramer, N.E.; Smith, J.L.; Hillman, J.D.; Zachariah, C.; Kuipers, O.P.; de Kruijff, B.; Breukink, E. An alternative bactericidal mechanism of action for lantibaiotic peptides that target lipid II. Science 2006, 313, 1636–1637. [Google Scholar] [CrossRef] [PubMed]
- Pecota, D.C.; Osapay, G.; Selsted, M.E.; Wood, T.K. Antimicrobial properties of the Escherichia coli R1 plasmid host killing peptide. J. Biotechnol. 2003, 100, 1–12. [Google Scholar] [CrossRef]
- Stevens, K.A.; Sheldon, B.W.; Klapes, N.A.; Klaenhammer, T.R. Nisin treatment for inactivation of Salmonella species and other gram-negative bacteria. Appl. Environ. Microbiol. 1991, 57, 3613–3615. [Google Scholar]
- Grenier, F.; Matteau, D.; Baby, V.; Rodrigue, S. Complete genome sequence of Escherichia coli BW25113. Genome Announc. 2014, 2, e01038-14. [Google Scholar] [CrossRef]
- Morita, M.; Tanji, Y.; Mizoguchi, K.; Akitsu, T.; Kijima, N.; Unno, H. Characterization of a virulent bacteriophage specific for Escherichia coli O157:H7 and analysis of its cellular receptor and two tail fiber genes. FEMS Microbiol. Lett. 2002, 211, 77–83. [Google Scholar] [CrossRef]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
| Primer Name | Sequence (5′–3′) | |
|---|---|---|
| Construction of plasmids | ||
| YO-57 | GCCTGCAGCTACTTCAACAAATCAATCAACCG | |
| YO-69 | AGGAATTCACCATGGATTACAAGGATGACGACGATAAGATGGACTCGCTGACACAAAAG | |
| YO-76 | GCAAGCTTCTACTTCAACAAATCAATCAACC | |
| YO-79 | GCCTGCAGCTACAACCGTAACAGAGC | |
| YO-81 | AGGAATTCGTTGGGACGTTGCGCCGGATCGA | |
| YO-82 | AGATTAGCGGATCCTACCTG | |
| YO-284 | CACTAATAACTCCAGTACGGCAATGAGCAC | |
| YO-285 | ATTGATTTGTTGAAGTAGCTGCAGGCATGC | |
| YO-370 | AGGAATTCACCATGGTGCTCATTGCCGTACTG | |
| YO-371 | AGGAATTCACCATGGACACGCTGACAC | |
| YO-372 | AGGAATTCACCATGCTGGAGTTATTAG | |
| YO-373 | GCCTGCAGCTACACTAATAACTCCAG | |
| Sequencing for bacterial 16S rRNA gene or C. albicans 18S rRNA gene | ||
| 9F | GAGTTTGATCCTGGCTCAG | (for bacteria) |
| 785F | GGATTAGATACCCTGGTAGTC | (for bacteria) |
| 1510R | GGCTACCTTGTTACGA | (for bacteria) |
| YO-301 | TATCTGGTTGATCCTGCCAGTAGTC | (for C. albicans) |
| YO-303 | TTGATGCGTACTGGACCCAGCCG | (for C. albicans) |
| YO-304 | GCGATAACGAACGAGACCTTAACC | (for C. albicans) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otsuka, Y.; Ishikawa, T.; Takahashi, C.; Masuda, M. A Short Peptide Derived from the ZorO Toxin Functions as an Effective Antimicrobial. Toxins 2019, 11, 392. https://doi.org/10.3390/toxins11070392
Otsuka Y, Ishikawa T, Takahashi C, Masuda M. A Short Peptide Derived from the ZorO Toxin Functions as an Effective Antimicrobial. Toxins. 2019; 11(7):392. https://doi.org/10.3390/toxins11070392
Chicago/Turabian StyleOtsuka, Yuichi, Tomohiro Ishikawa, Chisato Takahashi, and Michiaki Masuda. 2019. "A Short Peptide Derived from the ZorO Toxin Functions as an Effective Antimicrobial" Toxins 11, no. 7: 392. https://doi.org/10.3390/toxins11070392
APA StyleOtsuka, Y., Ishikawa, T., Takahashi, C., & Masuda, M. (2019). A Short Peptide Derived from the ZorO Toxin Functions as an Effective Antimicrobial. Toxins, 11(7), 392. https://doi.org/10.3390/toxins11070392




