Proteomic Analysis of Novel Components of Nemopilema nomurai Jellyfish Venom: Deciphering the Mode of Action
Abstract
1. Introduction
2. Results
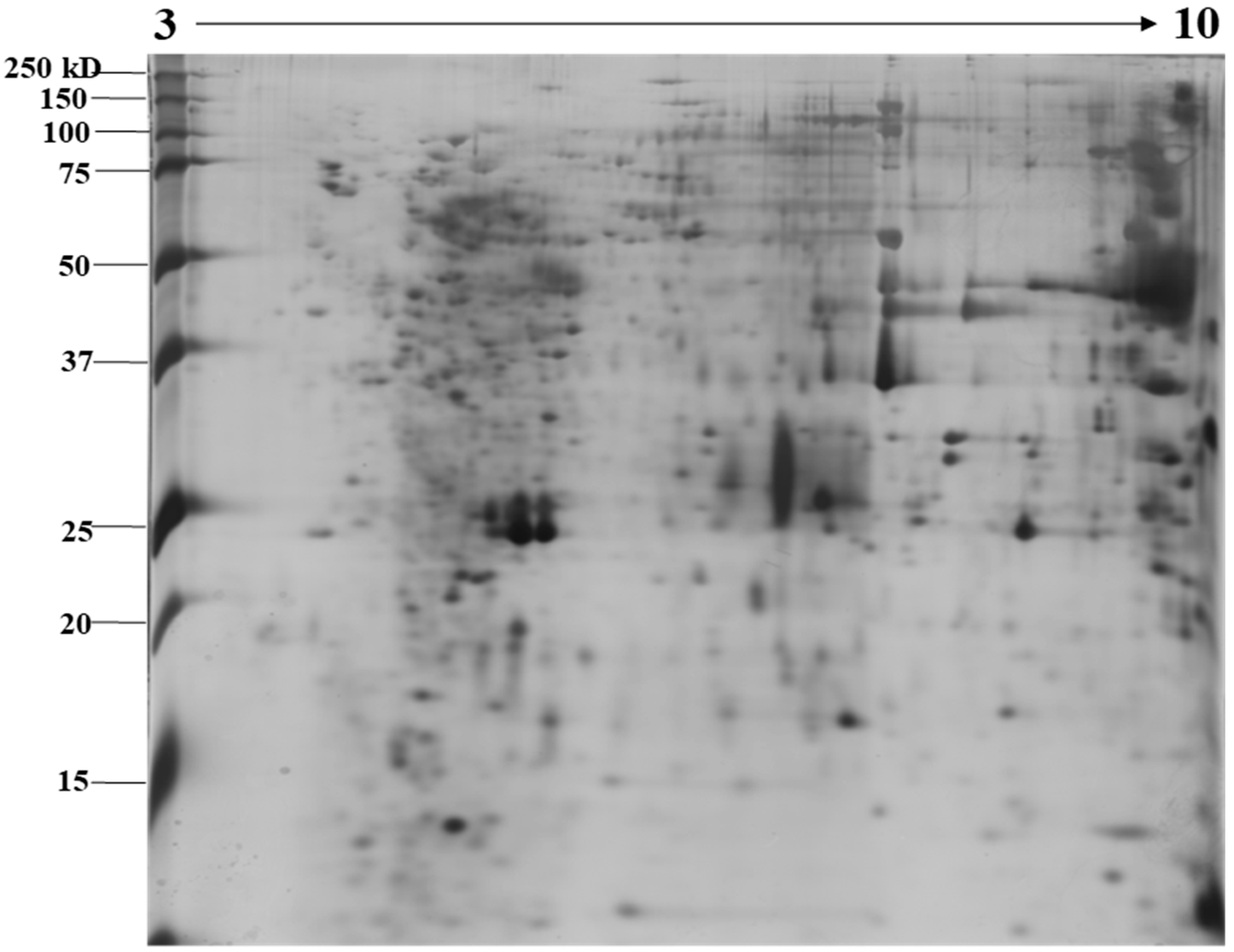
2.1. Identification of N. nomurai Nematocyst Proteins by Proteomic Characterization
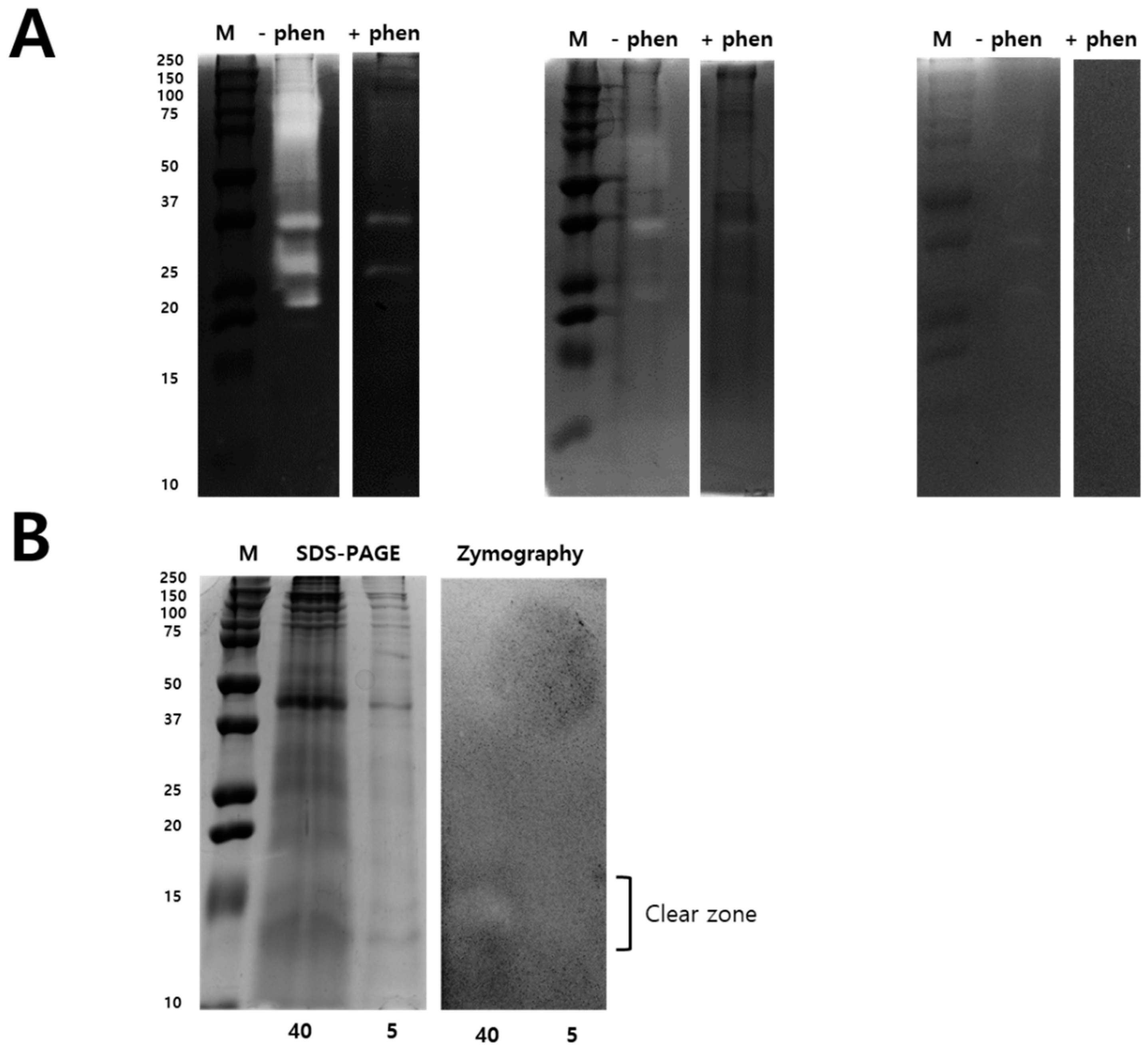
2.2. Modified Zymography Identify Metalloproteinase and PLA2 in NnV
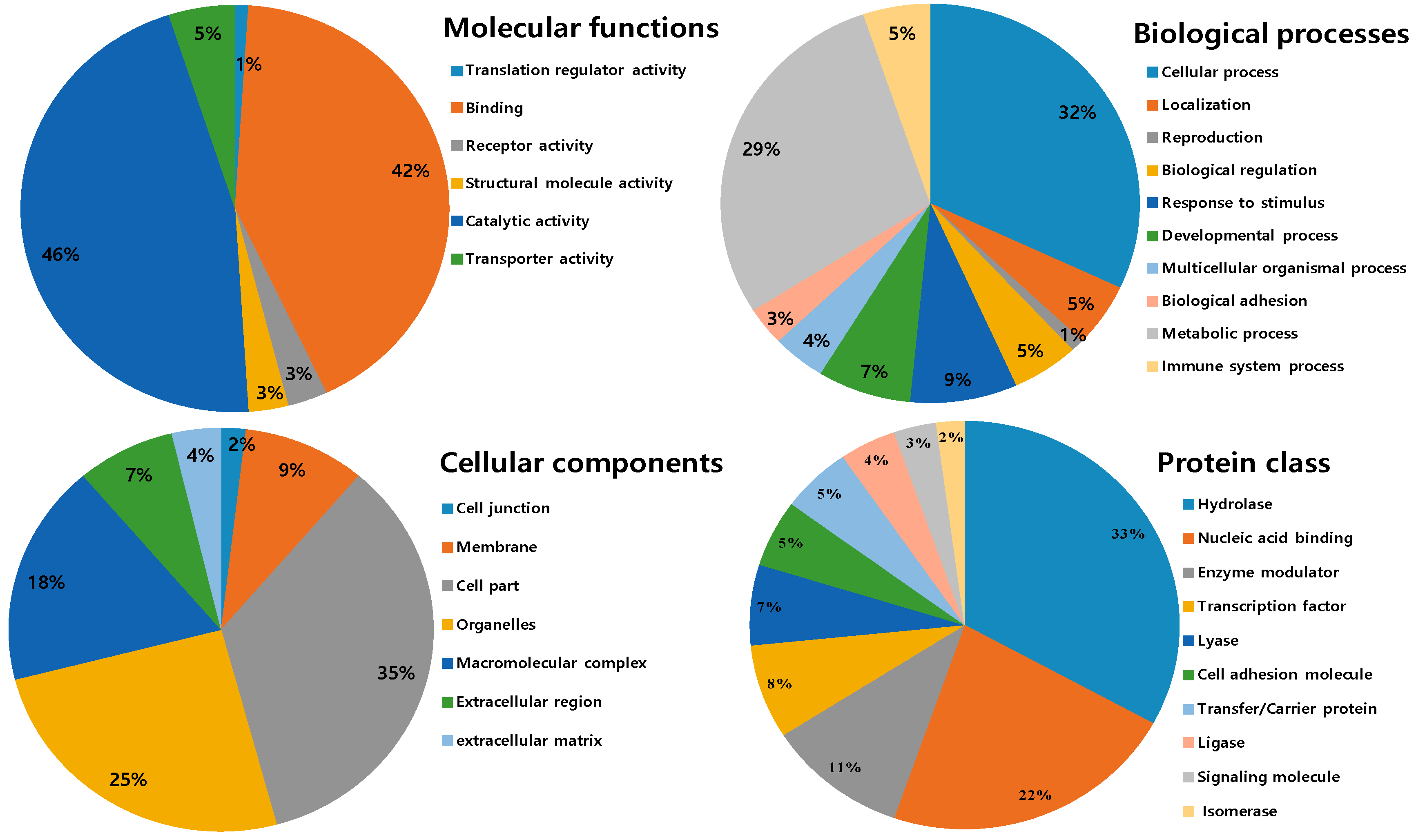
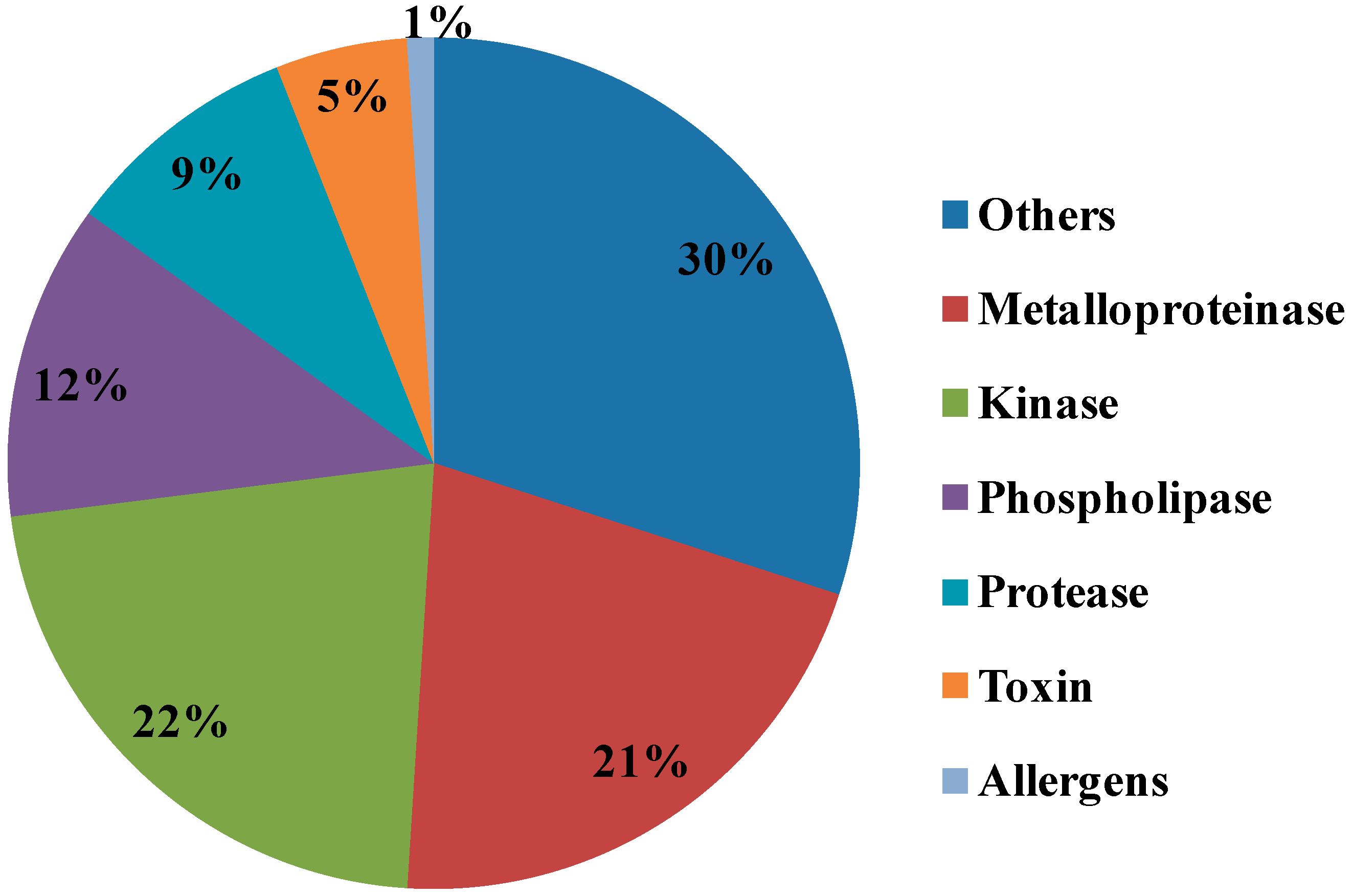
2.3. Ontological Classification of Differentially Expressed Proteins
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Reagents
5.2. Sample Collection and Preparation
5.3. Venom Extraction
5.4. Two-Dimensional Gel Electrophoresis under Reducing Conditions and Image Analysis
5.5. In-gel Digestion
5.6. MALDI/TOF/MS Analysis and Database Searching
5.7. Proteolytic Activity Assay
5.8. Modified Zymography Assays to Identify PLA2 Activity in NnV
5.9. Two-Dimensional Gel Electrophoresis under Non-Reducing Conditions and 2-DE Gelatin Zymography
5.10. Gene Ontology Analysis of the Identified Venom Proteins
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Uye, S.-I. Blooms of the giant jellyfish Nemopilema nomurai: A threat to the fisheries sustainability of the East Asian Marginal Seas. Plankton Benthos Res. 2008, 3, 125–131. [Google Scholar] [CrossRef]
- Dong, Z.; Liu, D.; Keesing, J.K. Jellyfish blooms in China: Dominant species, causes and consequences. Mar. Pollut. Bull. 2010, 60, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Tibballs, J. Australian venomous jellyfish, envenomation syndromes, toxins and therapy. Toxicon 2006, 48, 830–859. [Google Scholar] [CrossRef] [PubMed]
- Burnett, J.W.; Fenner, P.J.; Rifkin, J.F. Venomous and Poisonous Marine Animals: A Medical and Biological Handbook; UNSW Press: Sydney, Australia, 1996. [Google Scholar]
- Kim, E.; Lee, S.; Kim, J.-S.; Yoon, W.D.; Lim, D.; Hart, A.J.; Hodgson, W.C. Cardiovascular effects of Nemopilema nomurai (Scyphozoa: Rhizostomeae) jellyfish venom in rats. Toxicol. Lett. 2006, 167, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, I.; Lee, H.; Pyo, M.-J.; Heo, Y.; Bae, S.K.; Kwon, Y.C.; Yoon, W.D.; Kang, C.; Kim, E. Proteomics approach to examine the cardiotoxic effects of Nemopilema nomurai jellyfish venom. J. Proteom. 2015, 128, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Munawir, A.; Cha, M.; Sohn, E.-T.; Lee, H.; Kim, J.-S.; Yoon, W.D.; Lim, D.; Kim, E. Cytotoxicity and hemolytic activity of jellyfish Nemopilema nomurai (Scyphozoa: Rhizostomeae) venom. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2009, 150, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, I.; Lee, H.; Pyo, M.-J.; Heo, Y.; Bae, S.K.; Kwon, Y.C.; Yoon, W.D.; Kang, C.; Kim, E. Nemopilema nomurai jellyfish venom treatment leads to alterations in rat cardiomyocytes proteome. Data Brief 2015, 5, 884–887. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Kim, Y.K.; Lee, H.; Cha, M.; Sohn, E.-T.; Jung, E.-S.; Song, C.; Kim, M.; Lee, H.C.; Kim, J.-S. Target organ identification of jellyfish envenomation using systemic and integrative analyses in anesthetized dogs. J. Pharmacol. Toxicol. Methods 2011, 64, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, M.; Uye, S.; Burnett, J.; Mianzan, H. Stings of edible jellyfish (Rhopilema hispidum, Rhopilema esculentum and Nemopilema nomurai) in Japanese waters. Toxicon 2006, 48, 713–716. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, S.A.M.; Magalhães, M.R.; de Oliveira, L.P.; da Cunha, L.C. Identification of antinociceptive fraction of snake venom from Crotalus durissus collilineatus crotamine-negative and its acute toxicity evaluation. Toxicon 2016, 122, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.C.; Dekan, Z.; Rosengren, K.J.; Erickson, A.; Vetter, I.; Deuis, J.; Herzig, V.; Alewood, P.; King, G.F.; Lewis, R.J. Identification and characterization of ProTx-III [μ-TRTX-Tp1a], a new voltage-gated sodium channel inhibitor from venom of the tarantula Thrixopelma pruriens. Mol. Pharmacol. 2015, 88, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Lima, P.C.; Bordon, K.C.; Pucca, M.B.; Cerni, F.A.; Zoccal, K.F.; Faccioli, L.H.; Arantes, E.C. Partial purification and functional characterization of Ts19 Frag-I, a novel toxin from Tityus serrulatus scorpion venom. J. Venom. Anim. Toxins Incl. Trop. Dis. 2015, 21, 49. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.; Le Caer, J.-P.; Aráoz, R.; Thai, R.; Lamthanh, H.; Benoit, E.; Molgó, J. Isolation, purification and functional characterization of α-BnIA from Conus bandanus venom. Toxicon 2014, 91, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, H.; Yue, Y.; Liu, S.; Xing, R.; Chen, X.; Li, P. Combined proteomics and transcriptomics identifies sting-related toxins of jellyfish Cyanea nozakii. J. Proteom. 2016, 148, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, D.L.; Jia, X.; Potriquet, J.; Kumar, D.; Dash, D.; Kvaskoff, D.; Mulvenna, J. Transcriptome and venom proteome of the box jellyfish Chironex fleckeri. BMC Genom. 2015, 16, 407. [Google Scholar] [CrossRef] [PubMed]
- Weston, A.J.; Chung, R.; Dunlap, W.C.; Morandini, A.C.; Marques, A.C.; Moura-da-Silva, A.M.; Ward, M.; Padilla, G.; da Silva, L.F.; Andreakis, N. Proteomic characterisation of toxins isolated from nematocysts of the South Atlantic jellyfish Olindias sambaquiensis. Toxicon 2013, 71, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Bloom, D.A.; Burnett, J.W.; Alderslade, P. Partial purification of box jellyfish (Chironex fleckeri) nematocyst venom isolated at the beachside. Toxicon 1998, 36, 1075–1085. [Google Scholar] [CrossRef]
- Nomura, J.T.; Sato, R.L.; Ahern, R.M.; Snow, J.L.; Kuwaye, T.T.; Yamamoto, L.G. A randomized paired comparison trial of cutaneous treatments for acute jellyfish (Carybdea alata) stings. Am. J. Emerg. Med. 2002, 20, 624–626. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.; Koh, C. Metalloproteases affecting blood coagulation, fibrinolysis and platelet aggregation from snake venoms: Definition and nomenclature of interaction sites. Toxins 2016, 8, 284. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Jung, E.-S.; Kang, C.; Yoon, W.D.; Kim, J.-S.; Kim, E. Scyphozoan jellyfish venom metalloproteinases and their role in the cytotoxicity. Toxicon 2011, 58, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Jin, Y.B.; Kwak, J.; Jung, H.; Yoon, W.D.; Yoon, T.-J.; Kim, J.-S.; Kim, E. protective effect of tetracycline against dermal toxicity induced by Jellyfish venom. PLoS ONE 2013, 8, e57658. [Google Scholar] [CrossRef] [PubMed]
- da Silveira, R.B.; Pigozzo, R.B.; Chaim, O.M.; Appel, M.H.; Silva, D.T.; Dreyfuss, J.L.; Toma, L.; Dietrich, C.P.; Nader, H.B.; Veiga, S.S. Two novel dermonecrotic toxins LiRecDT4 and LiRecDT5 from brown spider (Loxosceles intermedia) venom: From cloning to functional characterization. Biochimie 2007, 89, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Moreira, D.; Souza, F.N.; Fogaça, R.T.; Mangili, O.C.; Gremski, W.; Senff-Ribeiro, A.; Chaim, O.M.; Veiga, S.S. The relationship between calcium and the metabolism of plasma membrane phospholipids in hemolysis induced by brown spider venom phospholipase-D toxin. J. Cell. Biochem. 2011, 112, 2529–2540. [Google Scholar] [CrossRef] [PubMed]
- Ghorbanpur, M.; Zare Mirakabadi, A.; Zokaee, F.; Zolfagarrian, H.; Rabiei, H. Purification and partial characterization of a coagulant serine protease from the venom of the Iranian snake Agkistrodon halys. J. Venom. Anim. Toxins Incl. Trop. Dis. 2009, 15, 411–423. [Google Scholar] [CrossRef]
- Möller, C.; Vanderweit, N.; Bubis, J.; Marí, F. Comparative analysis of proteases in the injected and dissected venom of cone snail species. Toxicon 2013, 65, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Veiga, S.S.; da Silveira, R.B.; Dreyfuss, J.L.; Haoach, J.; Pereira, A.M.; Mangili, O.C.; Gremski, W. Identification of high molecular weight serine-proteases in Loxosceles intermedia (brown spider) venom. Toxicon 2000, 38, 825–839. [Google Scholar] [CrossRef]
- Ma, Y.; Zhao, R.; He, Y.; Li, S.; Liu, J.; Wu, Y.; Cao, Z.; Li, W. Transcriptome analysis of the venom gland of the scorpion Scorpiops jendeki: Implication for the evolution of the scorpion venom arsenal. BMC Genom. 2009, 10, 290. [Google Scholar] [CrossRef] [PubMed]
- Rojas, A.; Doolittle, R.F. The occurrence of type S1A serine proteases in sponge and jellyfish. J. Mol. Evol. 2002, 55, 790–794. [Google Scholar] [CrossRef]
- He, J.; Chen, S.; Gu, J. Identification and characterization of Harobin, a novel fibrino (geno) lytic serine protease from a sea snake (Lapemis hardwickii). FEBS Lett. 2007, 581, 2965–2973. [Google Scholar] [CrossRef] [PubMed]
- Zaqueo, K.D.; Kayano, A.M.; Simões-Silva, R.; Moreira-Dill, L.S.; Fernandes, C.F.; Fuly, A.L.; Maltarollo, V.G.; Honório, K.M.; da Silva, S.L.; Acosta, G. Isolation and biochemical characterization of a new thrombin-like serine protease from Bothrops pirajai snake venom. BioMed Res. Int. 2014, 2014, 595186. [Google Scholar] [CrossRef] [PubMed]
- Markland, F.S. Snake venoms and the hemostatic system. Toxicon 1998, 36, 1749–1800. [Google Scholar] [CrossRef]
- Tsai, I.-H.; Wang, Y.-M.; Cheng, A.C.; Starkov, V.; Osipov, A.; Nikitin, I.; Makarova, Y.; Ziganshin, R.; Utkin, Y. cDNA cloning, structural, and functional analyses of venom phospholipases A2 and a Kunitz-type protease inhibitor from steppe viper Vipera ursinii renardi. Toxicon 2011, 57, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Fahrenkrog, B. Nma111p, the Pro-Apoptotic HtrA-Like Nuclear Serine Protease in Saccharomyces Cerevisiae: A Short Survey; In Portland Press Limited: London, UK, 2011. [Google Scholar]
- Lu, J.; Yang, H.; Yu, H.; Gao, W.; Lai, R.; Liu, J.; Liang, X. A novel serine protease inhibitor from Bungarus fasciatus venom. Peptides 2008, 29, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.-H.; He, Q.-Y.; Peng, K.; Diao, J.-B.; Jiang, L.-P.; Tang, X.; Liang, S.-P. Discovery of a distinct superfamily of Kunitz-type toxin (KTT) from tarantulas. PLoS ONE 2008, 3, e3414. [Google Scholar] [CrossRef]
- Zhao, R.; Dai, H.; Qiu, S.; Li, T.; He, Y.; Ma, Y.; Chen, Z.; Wu, Y.; Li, W.; Cao, Z. SdPI, the first functionally characterized Kunitz-type trypsin inhibitor from scorpion venom. PLoS ONE 2011, 6, e27548. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, Z.; Xiao, Y.; Li, Y.; Rong, M.; Liang, S.; Zhang, Z.; Yu, H.; King, G.F.; Lai, R. Chemical punch packed in venoms makes centipedes excellent predators. Mol. Cell. Proteom. 2012, 11, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Mourão, C.; Schwartz, E. Protease inhibitors from marine venomous animals and their counterparts in terrestrial venomous animals. Mar. Drugs 2013, 11, 2069–2112. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Lee, K.S.; Kim, B.Y.; Zou, F.M.; Yoon, H.J.; Je, Y.H.; Li, J.; Jin, B.R. A spider-derived Kunitz-type serine protease inhibitor that acts as a plasmin inhibitor and an elastase inhibitor. PLoS ONE 2013, 8, e53343. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M.; Doley, R. Structure, function and evolution of three-finger toxins: Mini proteins with multiple targets. Toxicon 2010, 56, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Corrêa-Netto, C.; de LM Junqueira-de-Azevedo, I.; Silva, D.A.; Ho, P.L.; Leitão-de-Araújo, M.; Alves, M.L.M.; Sanz, L.; Foguel, D.; Zingali, R.B.; Calvete, J.J. Snake venomics and venom gland transcriptomic analysis of Brazilian coral snakes, Micrurus altirostris and M. corallinus. J. Proteom. 2011, 74, 1795–1809. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, H.; Xue, W.; Yue, Y.; Liu, S.; Xing, R.; Li, P. Jellyfish venomics and venom gland transcriptomics analysis of Stomolophus meleagris to reveal the toxins associated with sting. J. Proteom. 2014, 106, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, D.; Burnell, J. Identification, cloning and sequencing of two major venom proteins from the box jellyfish, Chironex fleckeri. Toxicon 2007, 50, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, D.; Burnell, J. Partial purification of cytolytic venom proteins from the box jellyfish, Chironex fleckeri. Toxicon 2008, 51, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Lassen, S.; Helmholz, H.; Ruhnau, C.; Prange, A. A novel proteinaceous cytotoxin from the northern Scyphozoa Cyanea capillata (L.) with structural homology to cubozoan haemolysins. Toxicon 2011, 57, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Takuwa, K.; Nakao, M.; Sakamoto, B.; Crow, G.L.; Nakajima, T. Isolation and characterization of a novel protein toxin from the Hawaiian box jellyfish (sea wasp) Carybdea alata. Biochem. Biophys. Res. Commun. 2000, 275, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Azuma, H.; Sekizaki, S.; Satoh, A.; Nakajima, T. Platelet aggregation caused by carybdea rastonii toxins (CrTX-I, II, and III) obtained from a jellyfish, carybdea rastonii. Proc. Soc. Exp. Biol. Med. 1986, 182, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Takuwa, K.; Nakao, M.; Ito, E.; Miyake, M.; Noda, M.; Nakajima, T. Novel proteinaceous toxins from the box jellyfish (sea wasp) Carybdea rastoni. Biochem. Biophys. Res. Commun. 2000, 275, 582–588. [Google Scholar] [CrossRef] [PubMed]
- König, W.; Faltin, Y.; Scheffer, J.; Schöffler, H.; Braun, V. Role of cell-bound hemolysin as a pathogenicity factor for Serratia infections. Infect. Immunity 1987, 55, 2554–2561. [Google Scholar]
- Poole, K.; Schiebel, E.; Braun, V. Molecular characterization of the hemolysin determinant of Serratia marcescens. J. Bacteriol. 1988, 170, 3177–3188. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Ma, D.; Shi, J.; Chengappa, M. Molecular characterization of a leukotoxin gene from a Pasteurella haemolytica-like organism, encoding a new member of the RTX toxin family. Infect. Immunity 1993, 61, 2089–2095. [Google Scholar]
- Singh, M.B.; Hough, T.; Theerakulpisut, P.; Avjioglu, A.; Davies, S.; Smith, P.M.; Taylor, P.; Simpson, R.J.; Ward, L.D.; McCluskey, J. Isolation of cDNA encoding a newly identified major allergenic protein of rye-grass pollen: Intracellular targeting to the amyloplast. Proc. Natl. Acad. Sci. USA 1991, 88, 1384–1388. [Google Scholar] [CrossRef] [PubMed]
- Bungy, G.A.; Rodda, S.; Roitt, I.; Brostoff, J. Mapping of T cell epitopes of the major fraction of rye grass using peripheral blood mononuclear cells from atopics and non-atopics. II. Isoallergen clone 5A of Loliuum perenne group I (Lol p I). Eur. J. Immunol. 1994, 24, 2098–2103. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, A.; Ishimaru, N.; Seto, A.; Yamada, H.; Aki, T.; Shigeta, S.; Wada, T.; Jyo, T.; Murooka, Y.; Oka, S. Cloning and characterization of a new allergen, Mag 3, from the house dust mite, Dermatophagoides farinae: Cross-reactivity with high-molecular-weight allergen. Mol. Immunol. 1996, 33, 311–319. [Google Scholar] [CrossRef]
- Arlian, L.G. House-dust-mite allergens: A review. Exp. Appl. Acarol. 1991, 10, 167–186. [Google Scholar] [CrossRef] [PubMed]
- Aki, T.; Ono, K.; Paik, S.-Y.; Wada, T.; Jyo, T.; Shigeta, S.; Murooka, Y.; Oka, S. Cloning and characterization of cDNA coding for a new allergen from the house dust mite, Dermatophagoides farinae. Int. Arch. Allergy Immunol. 1994, 103, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.S.; Chung, M.C.; Jeyaseelan, K.; Kini, R.M. Amino Acid Sequence of Trocarin, a Prothrombin Activator From Tropidechis carinatus Venom: Its Structural Similarity to Coagulation Factor Xa. Blood 1999, 94, 621–631. [Google Scholar] [PubMed]
- Jansen, R.; Briaire, J.; Kamp, E.; Gielkens, A.; Smits, M. Structural analysis of the Actinobacillus pleuropneumoniae-RTX-toxin I (ApxI) operon. Infect. Immunity 1993, 61, 3688–3695. [Google Scholar]
- Kamp, E.M.; Popma, J.K.; Anakotta, J.; Smits, M.A. Identification of hemolytic and cytotoxic proteins of Actinobacillus pleuropneumoniae by use of monoclonal antibodies. Infect. Immunity 1991, 59, 3079–3085. [Google Scholar]
- De Grandis, S.; Ginsberg, J.; Toone, M.; Climie, S.; Friesen, J.; Brunton, J. Nucleotide sequence and promoter mapping of the Escherichia coli Shiga-like toxin operon of bacteriophage H-19B. J. Bacteriol. 1987, 169, 4313–4319. [Google Scholar] [CrossRef] [PubMed]
- Karmali, M.A.; Petric, M.; Lim, C.; Fleming, P.C.; Arbus, G.S.; Lior, H. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 1985, 151, 775–782. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, A.; Lively, T.; Chen, M.; Rothman, S.; Formal, S. Escherichia coli 0157: H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae 1 (Shiga) like cytotoxin. Lancet 1983, 321, 702. [Google Scholar] [CrossRef]
- Kanki, K.; Akechi, Y.; Ueda, C.; Tsuchiya, H.; Shimizu, H.; Ishijima, N.; Toriguchi, K.; Hatano, E.; Endo, K.; Hirooka, Y. Biological and clinical implications of retinoic acid-responsive genes in human hepatocellular carcinoma cells. J. Hepatol. 2013, 59, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- García-Santisteban, I.; Bañuelos, S.; Rodríguez, J.A. A global survey of CRM1-dependent nuclear export sequences in the human deubiquitinase family. Biochem. J. 2012, 441, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Hartwieg, E.; Horvitz, H.R. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 2001, 104, 43–56. [Google Scholar] [CrossRef]
- Ellis, R.E.; Jacobson, D.M.; Horvitz, H.R. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 1991, 129, 79–94. [Google Scholar] [PubMed]
- Yu, X.; Odera, S.; Chuang, C.-H.; Lu, N.; Zhou, Z. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev. Cell 2006, 10, 743–757. [Google Scholar] [CrossRef] [PubMed]
- Carrette, T.; Seymour, J. A rapid and repeatable method for venom extraction from Cubozoan nematocysts. Toxicon 2004, 44, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Mortz, E.; Krogh, T.N.; Vorum, H.; Görg, A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 2001, 1, 1359–1363. [Google Scholar] [CrossRef]
- Shevchenko, A.; Jensen, O.N.; Podtelejnikov, A.V.; Sagliocco, F.; Wilm, M.; Vorm, O.; Mortensen, P.; Shevchenko, A.; Boucherie, H.; Mann, M. Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 1996, 93, 14440–14445. [Google Scholar] [CrossRef] [PubMed]
- Choi, N.-S.; Kim, S.-H. Application of fibrin zymography for determining the optimum culture time for protease activity. Biotechnol. Tech. 1999, 13, 899–901. [Google Scholar] [CrossRef]
- d’Avila-Levy, C.M.; Santos, A.L.; Cuervo, P.; de Jesus, J.B.; Branquinha, M.H. Applications of zymography (substrate-SDS-PAGE) for peptidase screening in a post-genomic era. In Gel Electrophoresis-Advanced Techniques; InTech: Rijeka, Croatia, 2012. [Google Scholar]
- Díaz-García, A.; Ruiz-Fuentes, J.L.; Yglesias-Rivera, A.; Rodríguez-Sánchez, H.; Garlobo, Y.R.; Martinez, O.F.; Castro, J.A.F. Enzymatic analysis of venom from Cuban scorpion Rhopalurus junceus. J. Venom Res. 2015, 6, 11. [Google Scholar] [PubMed]
- Muller, V.D.; Soares, R.O.; dos Santos-Junior, N.N.; Trabuco, A.C.; Cintra, A.C.; Figueiredo, L.T.; Caliri, A.; Sampaio, S.V.; Aquino, V.H. Phospholipase A2 isolated from the venom of Crotalus durissus terrificus inactivates dengue virus and other enveloped viruses by disrupting the viral envelope. PLoS ONE 2014, 9, e112351. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551. [Google Scholar] [CrossRef] [PubMed]
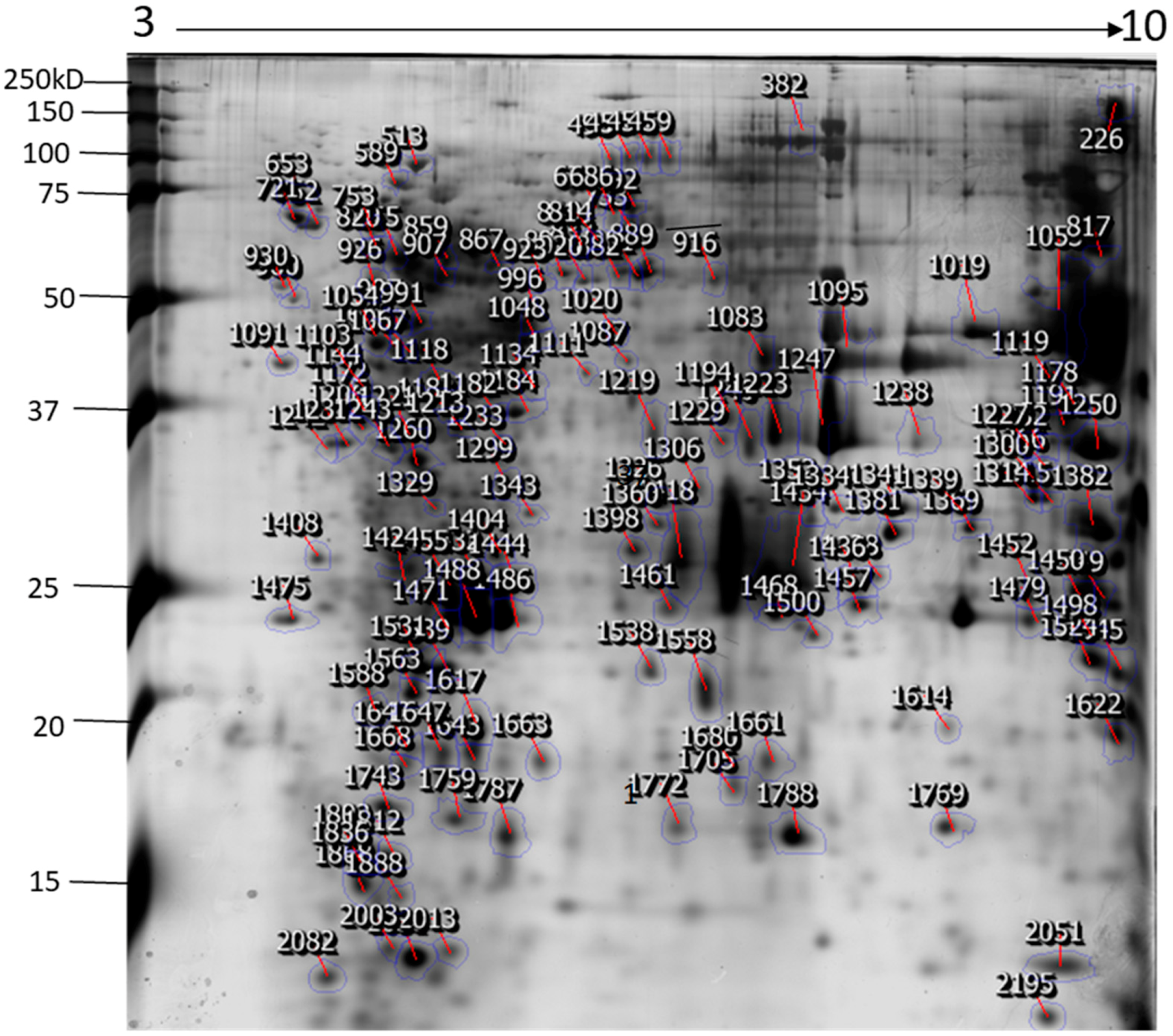
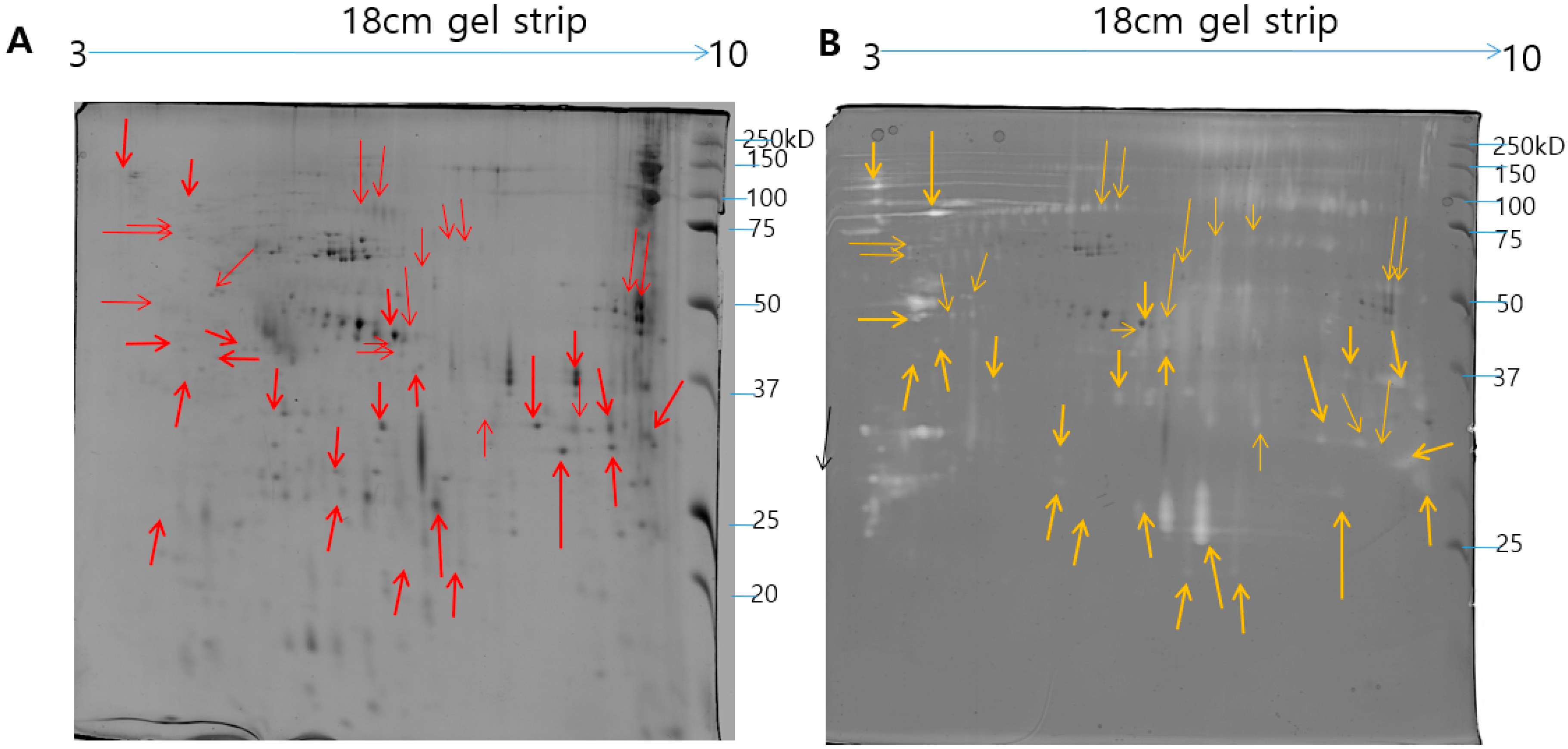
| Spot No | Accession Number 1 | Protein Name | Uniprot ID | Therotical MW/Pi 2 | Organism | Matched Peptide 3 | MOWSE Score | Biological Process |
|---|---|---|---|---|---|---|---|---|
| 1450 | P23897 | Heat-stable enterotoxin recepter | GUC2C_RAT | 123,468/6.4 | Rattus norvegicus | 8% | 4851 | Intracellular signal, transduction, regulation of cell proliferation |
| 1053 | P55128 | RTX-I toxin determinant A from serotypes 1/9 | RTXII_ACTPL | 110,194/5.5 | Actinobacillus pleuropneumoniae | 9.20% | 4636 | Hemolysis in other organism, pathogenesis |
| 1622 | P15321 | Hemolysin transporter, protein ShlB | HLYB_SERMA | 61,917/9.2 | Serratia marcescens | 15.30% | 3414 | Hemolysis in other organism, pathogenesis protein transmembrane transport |
| 1260 | Q1W694 | Phospholipase D LiSicTox-betaIDI | B1Q_LOXIN | 34,831/7.6 | Loxosceles intermedia | 22.00% | 154 | Pathogenesis, Hemolysis in other organism, phospholipid catabolic process |
| 1338 | P39673 | Allergen Mag (fragment) | MAG_DERFA | 39,668/6.9 | Dermatophagoides farinae | 11.70% | 320 | |
| 721 | P55123 | Leukotoxin | LKTA_PASSP | 101,560/5.6 | Pasteurella haemo-lytica-like sp. (strain 5943b) | 8.40% | 1331 | Hemolysis in other organism, pathogenesis |
| 1213 | B2BS84 | Putative Kunitz-type serine protease inhibitor | VKT_AUSLA | 27,571/7.9 | Austrelaps labialis | 25.00% | 7.14 × 102 | |
| 1486 | P0C845 | Turripeptide Gsp9.1 | C91_GEMSP | 9290/9.1 | Gemmula speciosa | 26.80% | 1802 | |
| 837 | F5CPD3 | Three-finger toxin MALT0044C | 3SX4_MICAT | 9398/8.4 | Micrurus altirostris | 54.10% | 8534 | Pathogenesis |
| 1643 | P81428 | Venom prothrombin activator trocarin-D | FAXD_TROCA | 51,407/8.1 | Tropidechis carinatus | 9.90% | 294 | Blood coagulation, envenomation resulting in positive regulation of blood coagulation in other organism |
| 889 | P55130 | RTX-III toxin determinant A from serotype 2 | RTX31_ACTPL | 112,492/5.8 | Actinobacillus pleuropneumoniae | 13.00% | 6.89 × 106 | Cytolysis, pathogenesis |
| 930 | O59824 | ATP-dependent zinc metalloprotease YME1 homolog | YME1_SCHPO | 78,219/8.5 | Schizosaccharomyces pombe (strain 972/ATCC 24843) | 6.20% | 1335 | proteolysis |
| 1614 | Q7S9D2 | Pro-apoptotic serine protease nma111 | NM111_NEUCR | 113,312/5.7 | Neurospora crassa | 10.80% | 30913 | Apoptotic process |
| 1182 | P08026 | Shiga-like toxin 1 subunit A | STXA_BPH19 | 34,800/9.6 | Enterobacteria phage H19B | 21% | 2329 | negative regulation of translation, pathogenesis |
| 1450 | Q2FZP2 | Serine protease HtrA-like | HTRAL_STAA8 | 86,460/6.5 | Staphylococcus aureus (strain NCTC 8325) | 20.20% | 876,777 | |
| 1647 | E5AJX2 | Snake venom serine protease nikobin | VSP_VIPNI | 28,216/8.0 | Vipera nikolskii | 18.20% | 425 | |
| 801 | Q2QA02 | Zinc metalloproteinase-disintegrin-like alternative name Snake venom metalloproteinase | VM3_CRODD | 68,292/5.1 | Crotalus durissus durissus | 19.50% | 57,907 | |
| 1233 | Q40240 | Major pollen allergen Lol p 5a | MPA5A_LOLPR | 30,888/5.4 | Lolium perenne | 16.90% | 707 | type I hypersensitivity |
| 1339 | P54319 | Phospholipase A-2-activating protein | PLAP_RAT | 87,085/5.7 | Rattus norvegicus | 11.30% | 1961 | Inflammatory response |
| 1343 | P23636 | Major serine/threonine-protein phosphatase PP2A_2 catalytic subunit | PP2A2_SCHPO | 36,489/4.7 | Schizosaccharomyces pombe (strain 972/ATCC 24843) | 19.60% | 2620 | Cell division, signal transduction, mitotic nuclear division |
| 1206 | Q9TT93 | A disintegrin and metallo-proteinase with thrombo-spondin motifs 4 | ATS4_BOVIN | 90,281/8.6 | Bos taurus | 11.90% | 3.76 × 104 | Proteolysis, Angiogenesis |
| 1500 | P97570 | 85/88 kDa calcium-independent phospholipase A2 | PLPL_RAT | 89,556/6.7 | Rattus norvegicus | 12% | 19,976 | ATP dependent protein binding, chemotaxis, positive regulation of vasodilation |
| 1315 | P58459 | A disintegrin and metalloproteinase with thrombospondin motifs 10 | ATS10_MOUSE | 121,087/8.4 | Mus musculus | 11.30% | 40,176 | Microfibrils assembly |
| 1382 | Q9R1V7 | A disintegrin and metalloproteinase with thrombospondin motifs 23 | ADA_23 MOUSE | 91,548/7.9 | Mus musculus | 14% | 2773 | Cell adhesion |
| 1461 | Q9XWD6 | Cell death abnormality protein 1 | CED1_CAEEL | 118,805/5.5 | Caenorhabditis elegans | 10.10% | 6333 | Programmed cell death, apoptotic cell death, receptor-mediated endocytosis, |
| 1272 | Q9T051 | Phospholipase D gamma 2 | PLDG2_ARATH | 96,024/8.3 | Arabidopsis thaliana | 12.60% | 11,703 | Response to stress, membrane lipid metabolic process, phosphotidylcholine metabolic process |
| 1334 | Q10743 | A disintegrin and metalloproteinase with thrombospondin motifs 10 fragment | ADA10_RAT | 60,445/8.4 | Rattus norvegicus | 15.10% | 5386 | Negative regulation of cell adhesion, Notch signaling pathway, protein phosphorylation |
| 1498 | Q83XX3 | ATP-dependent zinc metalloprotease FtsH | FTSH_OENOE | 78,070/9.3 | Oenococcus oeni | 10.30% | 447 | Protein catabolic process |
| 867 | P78536 | Disintegrin and metalloproteinase domain-containing protein 17 | ADA17_HUMAN | 93,022/5.5 | Homo sapiens | 10.70% | 2397 | Positive regulation of cell growth and cell migration, negative regulation of transforming growth factor beta receptor signaling |
| 1434 | A8XEZ1 | Cell death abnormality protein 12 | CED12_CAEBR | 83,692/5.2 | Saccharomyces cerevisiae (strain ATCC 204508/S288c) | 9.00% | 3371 | Engulfment of apoptotic cell, cell migration, apoptotic process, phagocytosis, positive regulation of GTPase activity |
| 1558 | Q9S5Z2 | ATP-dependent Clp protease ATP-binding subunit ClpE | PLPL8_MOUSE | 87,382/9.3 | Mus musculus | 9.10% | 2564 | Cell death, arachidonic acid, secretion, phosphatidylcholine catabolic process |
| 1418 | Q8K1N1 | Calcium-independent phospholipase A2-gamma | HTRAL_STAA8 | 86,460/6.5 | Staphylococcus aureus (strain NCTC 8325) | 20.20% | 876,777 | |
| 1183 | O93654 | Tricorn protease-interacting factor F2 | NAS8_CAEEL | 46,096/5.8 | Caenorhabditis elegans | 13.40% | 104 | |
| 1087 | Q18439 | Zinc metalloproteinase nas-8 | CYM1_YEAST | 112,181/6.0 | Saccharomyces cerevisiae (strain ATCC 204508/S288c) | 18.20% | 383,182 | Protein processing, proteolysis, 54 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choudhary, I.; Hwang, D.H.; Lee, H.; Yoon, W.D.; Chae, J.; Han, C.H.; Yum, S.; Kang, C.; Kim, E. Proteomic Analysis of Novel Components of Nemopilema nomurai Jellyfish Venom: Deciphering the Mode of Action. Toxins 2019, 11, 153. https://doi.org/10.3390/toxins11030153
Choudhary I, Hwang DH, Lee H, Yoon WD, Chae J, Han CH, Yum S, Kang C, Kim E. Proteomic Analysis of Novel Components of Nemopilema nomurai Jellyfish Venom: Deciphering the Mode of Action. Toxins. 2019; 11(3):153. https://doi.org/10.3390/toxins11030153
Chicago/Turabian StyleChoudhary, Indu, Du Hyeon Hwang, Hyunkyoung Lee, Won Duk Yoon, Jinho Chae, Chang Hoon Han, Seungshic Yum, Changkeun Kang, and Euikyung Kim. 2019. "Proteomic Analysis of Novel Components of Nemopilema nomurai Jellyfish Venom: Deciphering the Mode of Action" Toxins 11, no. 3: 153. https://doi.org/10.3390/toxins11030153
APA StyleChoudhary, I., Hwang, D. H., Lee, H., Yoon, W. D., Chae, J., Han, C. H., Yum, S., Kang, C., & Kim, E. (2019). Proteomic Analysis of Novel Components of Nemopilema nomurai Jellyfish Venom: Deciphering the Mode of Action. Toxins, 11(3), 153. https://doi.org/10.3390/toxins11030153