Exploring the Diversity and Novelty of Toxin Genes in Naja sumatrana, the Equatorial Spitting Cobra from Malaysia through De Novo Venom-Gland Transcriptomics
Abstract
1. Introduction
2. Results and Discussion
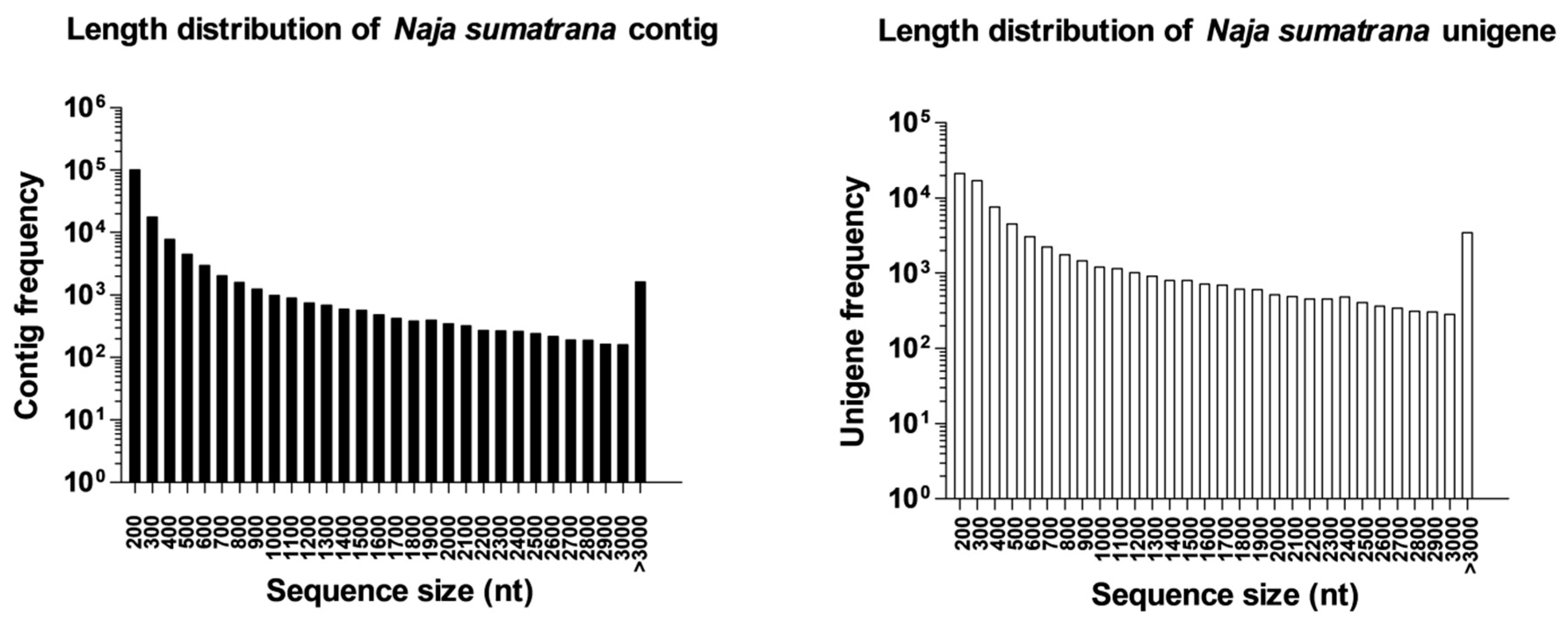
2.1. Sequencing Output Statistics and De Novo Transcriptome Assembly
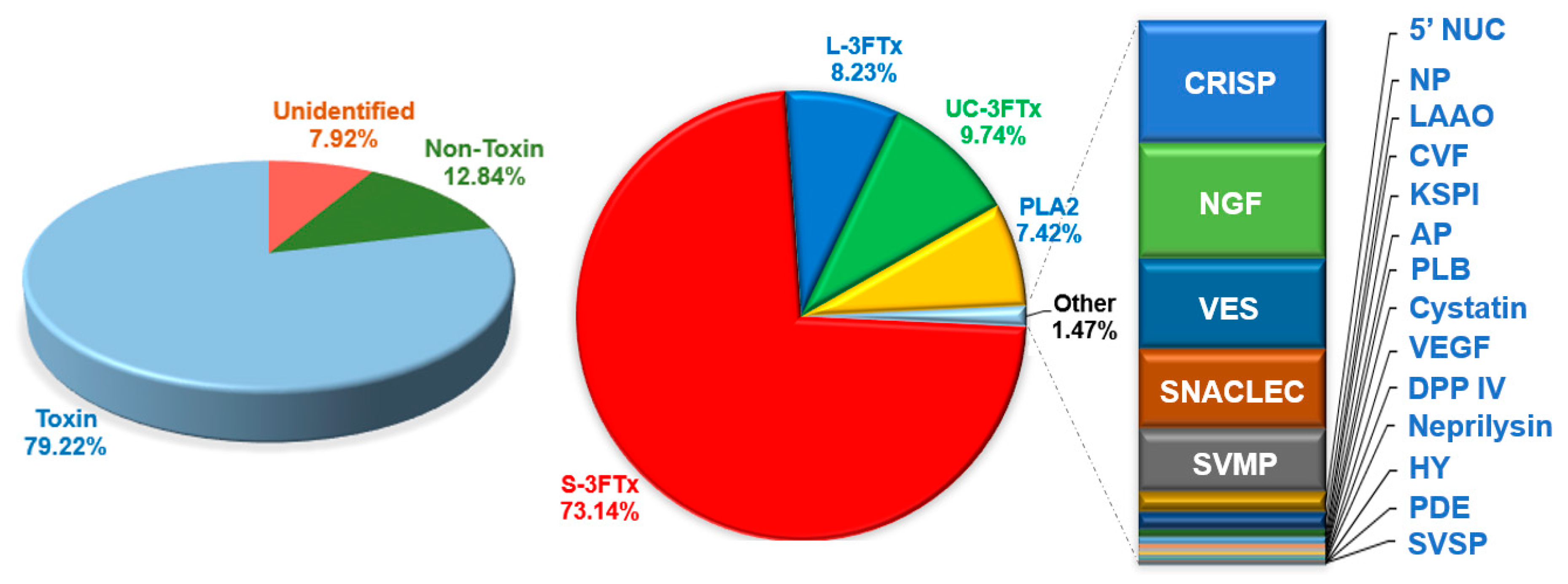
2.2. Categorization of Transcripts and Gene Expression
2.3. Complexity of N. sumatrana Venom-Gland Transcriptome
2.4. Diversity of Toxin Transcripts and Major Venom Constituents
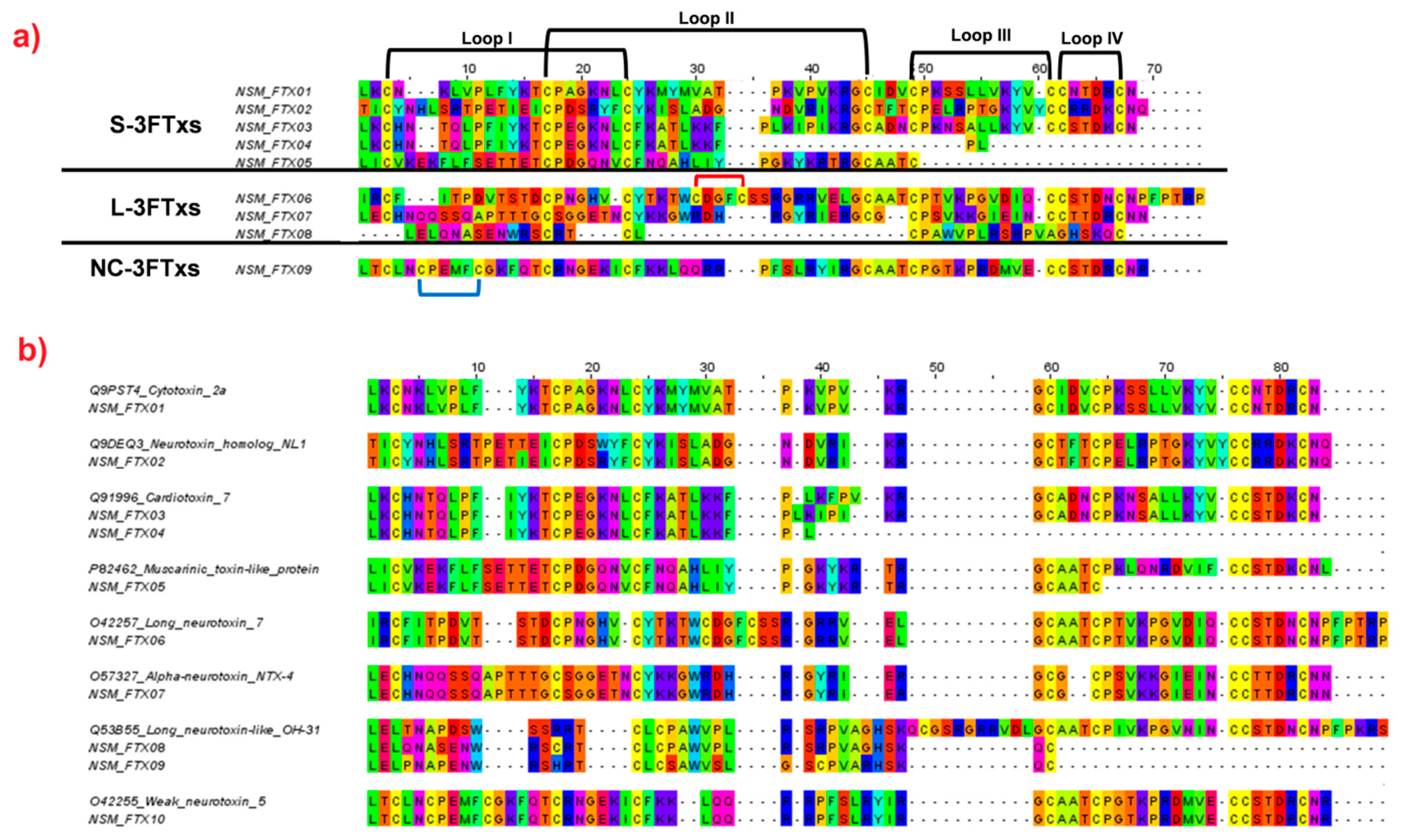
2.4.1. Three-finger Toxins (3FTxs)
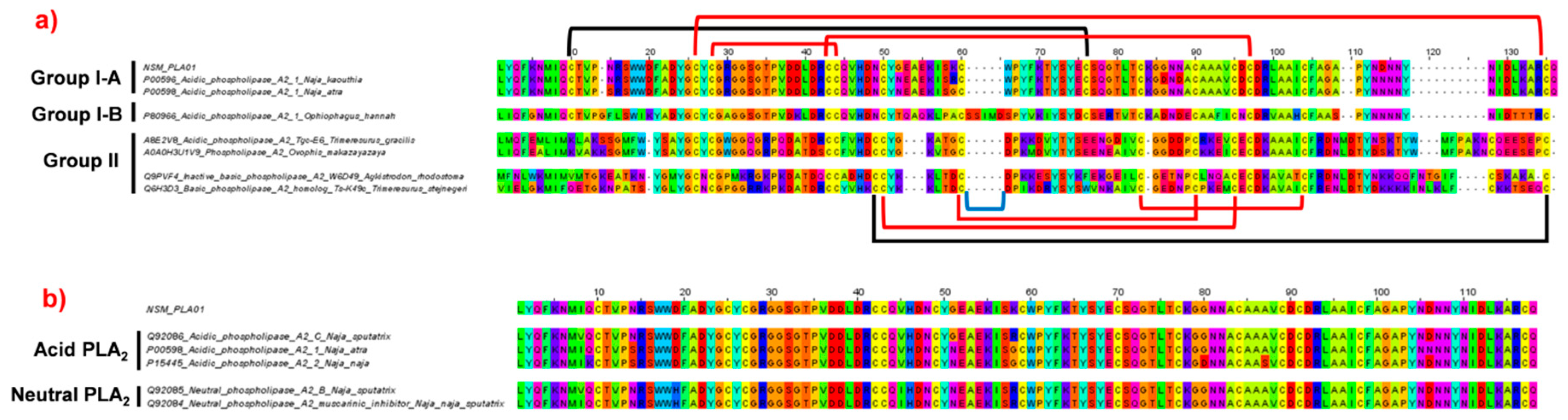
2.4.2. Phospholipase A2 (PLA2)
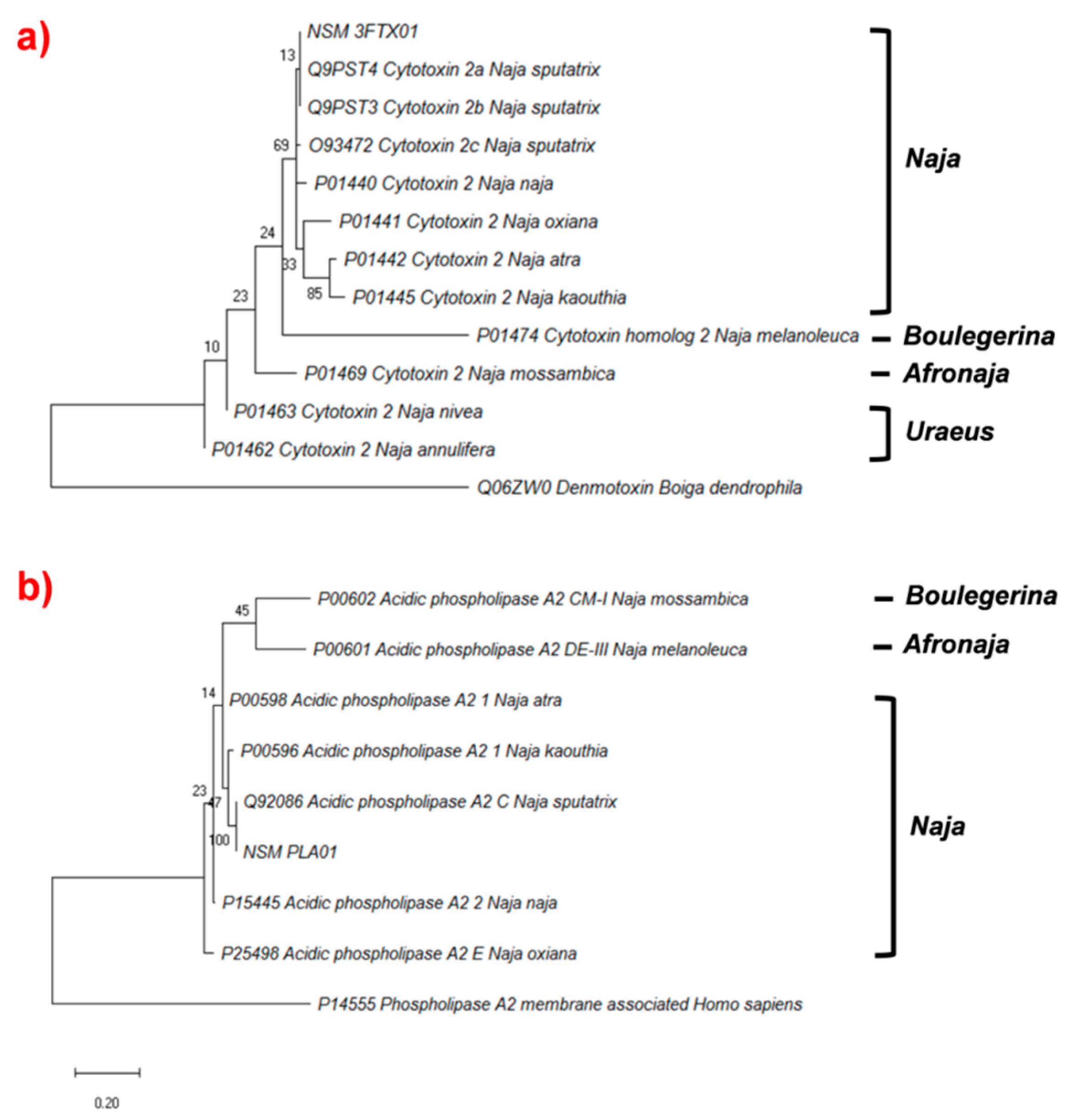
2.4.3. Phylogenetic Analysis of Cytotoxin and Phospholipase A2
2.5. Low Abundance Transcripts
2.5.1. Transcript Expression of Toxin Families Previously Reported in Venom Proteome
2.5.2. Transcript Expression of Toxin Families Not Reported in Venom Proteome of N. sumatrana
3. Conclusions
4. Materials and Methods
4.1. Preparation of Snake Venom-Gland Tissue
4.2. Extraction of RNA and Purification of mRNA
4.3. Construction of cDNA Library and Sequencing
4.4. Filtration of Raw Sequence Data
4.5. Assembly of De Novo Transcriptome
4.6. Clustering of Transcripts
4.7. Quantifying the Expression Annotation of Transcripts
4.8. Determination of Functional Annotation of Transcripts
4.9. Classification of Venom-Gland Transcripts Based on Toxinology
4.10. Multiple Sequence Alignment and Phylogenetic Tree Construction
4.11. Supporting Data
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sunagar, K.; Moran, Y. The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals. PLoS Genet 2015, 11, e1005596. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wuster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Daltry, J.C.; Wuster, W.; Thorpe, R.S. Diet and snake venom evolution. Nature 1996, 379, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wuster, W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. R. Soc. Lond. B Biol. Sci. 2009, 276, 2443–2449. [Google Scholar] [CrossRef] [PubMed]
- Augusto-de-Oliveira, C.; Stuginski, D.R.; Kitano, E.S.; Andrade-Silva, D.; Liberato, T.; Fukushima, I.; Serrano, S.M.; Zelanis, A. Dynamic Rearrangement in Snake Venom Gland Proteome: Insights into Bothrops jararaca Intraspecific Venom Variation. J. Proteome Res. 2016, 15, 3752–3762. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.Y.; Tan, C.H.; Chanhome, L.; Tan, N.H. Comparative venom gland transcriptomics of Naja kaouthia (monocled cobra) from Malaysia and Thailand: elucidating geographical venom variation and insights into sequence novelty. PeerJ 2017, 5, e3142. [Google Scholar] [CrossRef] [PubMed]
- Zelanis, A.; Tashima, A.K.; Rocha, M.M.; Furtado, M.F.; Camargo, A.C.; Ho, P.L.; Serrano, S.M. Analysis of the ontogenetic variation in the venom proteome/peptidome of Bothrops jararaca reveals different strategies to deal with prey. J. Proteome Res. 2010, 9, 2278–2291. [Google Scholar] [CrossRef]
- Tan, C.H.; Tan, K.Y.; Yap, M.K.; Tan, N.H. Venomics of Tropidolaemus wagleri, the sexually dimorphic temple pit viper: Unveiling a deeply conserved atypical toxin arsenal. Sci. Rep. 2017, 7, 43237. [Google Scholar] [CrossRef]
- Tan, K.Y.; Tan, C.H.; Fung, S.Y.; Tan, N.H. Venomics, lethality and neutralization of Naja kaouthia (monocled cobra) venoms from three different geographical regions of Southeast Asia. J. Proteom. 2015, 120, 105–125. [Google Scholar] [CrossRef]
- Oh, A.M.F.; Tan, C.H.; Ariaranee, G.C.; Quraishi, N.; Tan, N.H. Venomics of Bungarus caeruleus (Indian krait): Comparable venom profiles, variable immunoreactivities among specimens from Sri Lanka, India and Pakistan. J. Proteom. 2017, 164, 1–18. [Google Scholar] [CrossRef]
- Wong, K.Y.; Tan, C.H.; Tan, N.H. Venom and purified toxins of the spectacled cobra (Naja naja) from Pakistan: Insights into toxicity and antivenom neutralization. Am. J. Trop. Med. Hyg. 2016, 94, 1392–1399. [Google Scholar] [CrossRef] [PubMed]
- Faisal, T.; Tan, K.Y.; Sim, S.M.; Quraishi, N.; Tan, N.H.; Tan, C.H. Proteomics, functional characterization and antivenom neutralization of the venom of Pakistani Russell’s viper (Daboia russelii) from the wild. J. Proteom. 2018, 183, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Alirol, E.; Sharma, S.K.; Bawaskar, H.S.; Kuch, U.; Chappuis, F. Snake bite in South Asia: a review. PLoS Negl. Trop. Dis. 2010, 4, e603. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Liew, J.L.; Tan, N.H.; Khaldun, I.A.; Maharani, T.; Khomvilai, S.; Sitprija, V. Cross reactivity and lethality neutralization of venoms of Indonesian Trimeresurus complex species by Thai Green Pit Viper Antivenom. Toxicon 2017. [Google Scholar] [CrossRef] [PubMed]
- WHO. Guidelines for the Production Control and Regulation of Snake Antivenomimmunoglobulins; World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- Wuster, W. Taxonomic changes and toxinology: systematic revisions of the Asiatic cobras (Naja naja species complex). Toxicon 1996, 34, 399–406. [Google Scholar] [CrossRef]
- WHO. Guidelines for the Management of Snake Bites; Regional office for South-East Asia, World Health Organization: New Delhi, India, 2016. [Google Scholar]
- Chew, K.S.; Khor, H.W.; Ahmad, R.; Rahman, N.H. A five-year retrospective review of snakebite patients admitted to a tertiary university hospital in Malaysia. Int. J. Emerg. Med. 2011, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Reid, H. Cobra-bites. Br. Med. J. 1964, 2, 540. [Google Scholar] [CrossRef]
- Wong, K.Y.; Tan, C.H.; Tan, K.Y.; Quraishi, N.H.; Tan, N.H. Elucidating the biogeographical variation of the venom of Naja naja (spectacled cobra) from Pakistan through a venom-decomplexing proteomic study. J. Proteom. 2018, 175, 156–173. [Google Scholar] [CrossRef]
- Tan, N.H.; Wong, K.Y.; Tan, C.H. Venomics of Naja sputatrix, the Javan spitting cobra: A short neurotoxin-driven venom needing improved antivenom neutralization. J. Proteom. 2017, 157, 18–32. [Google Scholar] [CrossRef]
- Margres, M.J.; Walls, R.; Suntravat, M.; Lucena, S.; Sanchez, E.E.; Rokyta, D.R. Functional characterizations of venom phenotypes in the eastern diamondback rattlesnake (Crotalus adamanteus) and evidence for expression-driven divergence in toxic activities among populations. Toxicon 2016, 119, 28–38. [Google Scholar] [CrossRef]
- Sunagar, K.; Morgenstern, D.; Reitzel, A.M.; Moran, Y. Ecological venomics: How genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J. Proteom. 2016, 135, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Armugam, A.; Earnest, L.; Chung, M.; Gopalakrishnakone, P.; Tan, C.; Tan, N.; Jeyaseelan, K. Cloning and characterization of cDNAs encoding three isoforms of phospholipase A2 in Malayan spitting cobra (Naja naja sputatrix) venom. Toxicon 1997, 35, 27–37. [Google Scholar] [CrossRef]
- Afifiyan, F.; Armugam, A.; Tan, C.H.; Gopalakrishnakone, P.; Jeyaseelan, K. Postsynaptic α-neurotoxin gene of the spitting cobra, Naja naja sputatrix: structure, organization, and phylogenetic analysis. Genome Res. 1999, 9, 259–266. [Google Scholar] [PubMed]
- Jeyaseelan, K.; Armugam, A.; Lachumanan, R.; Tan, C.H.; Tan, N.H. Six isoforms of cardiotoxin in Malayan spitting cobra (Naja naja sputatrix) venom: cloning and characterization of cDNAs1. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 1998, 1380, 209–222. [Google Scholar] [CrossRef]
- Jeyaseelan, K.; Poh, S.L.; Nair, R.; Armugam, A. Structurally conserved α-neurotoxin genes encode functionally diverse proteins in the venom of Naja sputatrix. FEBS Lett. 2003, 553, 333–341. [Google Scholar] [CrossRef]
- Lay Poh, S.; Mourier, G.; Thai, R.; Armugam, A.; Molgó, J.; Servent, D.; Jeyaseelan, K.; Ménez, A. A synthetic weak neurotoxin binds with low affinity to Torpedo and chicken α7 nicotinic acetylcholine receptors. Eur. J. Biochem. 2002, 269, 4247–4256. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Y.; Lee, W.; Xu, X.; Zhang, Y.; Zhao, R.; Zhang, Y.; Wang, W. Venom gland transcriptomes of two elapid snakes (Bungarus multicinctus and Naja atra) and evolution of toxin genes. BMC Genom. 2011, 12, 1. [Google Scholar] [CrossRef]
- Tan, C.H.; Tan, K.Y.; Fung, S.Y.; Tan, N.H. Venom-gland transcriptome and venom proteome of the Malaysian king cobra (Ophiophagus hannah). BMC Genom. 2015, 16, 687. [Google Scholar] [CrossRef]
- Xu, N.; Zhao, H.Y.; Yin, Y.; Shen, S.S.; Shan, L.L.; Chen, C.X.; Zhang, Y.X.; Gao, J.F.; Ji, X. Combined venomics, antivenomics and venom gland transcriptome analysis of the monocoled cobra (Naja kaouthia) from China. J. Proteom. 2017, 159, 19–31. [Google Scholar] [CrossRef]
- Kini, R.M.; Doley, R. Structure, function and evolution of three-finger toxins: mini proteins with multiple targets. Toxicon 2010, 56, 855–867. [Google Scholar] [CrossRef]
- Antil, S.; Servent, D.; Menez, A. Variability among the sites by which curaremimetic toxins bind to torpedo acetylcholine receptor, as revealed by identification of the functional residues of alpha-cobratoxin. J. Biol. Chem. 1999, 274, 34851–34858. [Google Scholar] [CrossRef] [PubMed]
- Servent, D.; Antil-Delbeke, S.; Gaillard, C.; Corringer, P.J.; Changeux, J.P.; Menez, A. Molecular characterization of the specificity of interactions of various neurotoxins on two distinct nicotinic acetylcholine receptors. Eur. J. Pharmacol. 2000, 393, 197–204. [Google Scholar] [CrossRef]
- Antil-Delbeke, S.; Gaillard, C.; Tamiya, T.; Corringer, P.J.; Changeux, J.P.; Servent, D.; Menez, A. Molecular determinants by which a long chain toxin from snake venom interacts with the neuronal alpha 7-nicotinic acetylcholine receptor. J. Biol. Chem. 2000, 275, 29594–29601. [Google Scholar] [CrossRef] [PubMed]
- Yap, M.K.; Fung, S.Y.; Tan, K.Y.; Tan, N.H. Proteomic characterization of venom of the medically important Southeast Asian Naja sumatrana (Equatorial spitting cobra). Acta Trop. 2014, 133, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Gasanov, S.E.; Dagda, R.K.; Rael, E.D. Snake venom cytotoxins, phospholipase A2s, and Zn2+-dependent metalloproteinases: mechanisms of action and pharmacological relevance. J. Clin. Toxicol. 2014, 4, 1000181. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, N.H. Cytotoxicity of Snake Venoms and Toxins: Mechanisms and Applications. In Snake Venoms and Envenomation: Modern Trends and Future Prospects; Utkin, Y.N., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2016; pp. 215–254. [Google Scholar]
- Panagides, N.; Jackson, T.N.W.; Ikonomopoulou, M.P.; Arbuckle, K.; Pretzler, R.; Yang, D.C.; Ali, S.A.; Koludarov, I.; Dobson, J.; Sanker, B.; et al. How the Cobra Got Its Flesh-Eating Venom: Cytotoxicity as a Defensive Innovation and Its Co-Evolution with Hooding, Aposematic Marking, and Spitting. Toxins 2017, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Barber, C.M.; Isbister, G.K.; Hodgson, W.C. Alpha neurotoxins. Toxicon 2013, 66, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, N.H. Toxinology of Snake Venoms: The Malaysian Context. In Snake Venoms; Gopalakrishnakone, P., Inagaki, H., Mukherjee, A.K., Rahmy, T.R., Vogel, C.-W., Eds.; Toxinology Series; Springer: Dordrecht, Switzerland, 2015; pp. 1–37. [Google Scholar]
- Utkin, Y.N.; Kukhtina, V.V.; Kryukova, E.V.; Chiodini, F.; Bertrand, D.; Methfessel, C.; Tsetlin, V.I. “Weak toxin” from Naja kaouthia is a nontoxic antagonist of alpha 7 and muscle-type nicotinic acetylcholine receptors. J. Biol. Chem. 2001, 276, 15810–15815. [Google Scholar] [CrossRef] [PubMed]
- Leong, P.K.; Fung, S.Y.; Tan, C.H.; Sim, S.M.; Tan, N.H. Immunological cross-reactivity and neutralization of the principal toxins of Naja sumatrana and related cobra venoms by a Thai polyvalent antivenom (Neuro Polyvalent Snake Antivenom). Acta Trop. 2015, 149, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.Y.; Tan, C.H.; Fung, S.Y.; Tan, N.H. Neutralization of the Principal Toxins from the Venoms of Thai Naja kaouthia and Malaysian Hydrophis schistosus: Insights into Toxin-Specific Neutralization by Two Different Antivenoms. Toxins 2016, 8. [Google Scholar] [CrossRef]
- Vonk, F.J.; Casewell, N.R.; Henkel, C.V.; Heimberg, A.M.; Jansen, H.J.; McCleary, R.J.; Kerkkamp, H.M.; Vos, R.A.; Guerreiro, I.; Calvete, J.J.; et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA 2013, 110, 20651–20656. [Google Scholar] [CrossRef] [PubMed]
- Durban, J.; Juarez, P.; Angulo, Y.; Lomonte, B.; Flores-Diaz, M.; Alape-Giron, A.; Sasa, M.; Sanz, L.; Gutierrez, J.M.; Dopazo, J.; et al. Profiling the venom gland transcriptomes of Costa Rican snakes by 454 pyrosequencing. BMC Genom. 2011, 12, 259. [Google Scholar] [CrossRef] [PubMed]
- Aird, S.D.; Watanabe, Y.; Villar-Briones, A.; Roy, M.C.; Terada, K.; Mikheyev, A.S. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis). BMC Genom. 2013, 14, 790. [Google Scholar] [CrossRef] [PubMed]
- Rokyta, D.R.; Margres, M.J.; Calvin, K. Post-transcriptional Mechanisms Contribute Little to Phenotypic Variation in Snake Venoms. G3 Genes Genomes Genet. 2015, 5, 2375–2382. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Bickel, P.J.; Biggin, M.D. System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ 2014, 2, e270. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, K.Y.; Lim, S.E.; Tan, N.H. Venomics of the beaked sea snake, Hydrophis schistosus: A minimalist toxin arsenal and its cross-neutralization by heterologous antivenoms. J. Proteom. 2015, 126, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, K.Y.; Ng, T.S.; Sim, S.M.; Tan, N.H. Venom Proteome of Spine-Bellied Sea Snake (Hydrophis curtus) from Penang, Malaysia: Toxicity Correlation, Immunoprofiling and Cross-Neutralization by Sea Snake Antivenom. Toxins 2019, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Wong, K.Y.; Tan, K.Y.; Tan, N.H. Venom proteome of the yellow-lipped sea krait, Laticauda colubrina from Bali: Insights into subvenomic diversity, venom antigenicity and cross-neutralization by antivenom. J. Proteom. 2017, 166, 48–58. [Google Scholar] [CrossRef]
- Tan, N.-H.; Armugam, A. In vivo interactions between neurotoxin, cardiotoxin and phospholipases A2 isolated from Malayan cobra (Naja naja sputatrix) venom. Toxicon 1990, 28, 1193–1198. [Google Scholar] [CrossRef]
- Tan, C.H.; Wong, K.Y.; Tan, N.H.; Ng, T.S.; Tan, K.Y. Distinctive distribution of secretory phospholipases A2 in the venoms of Afro-Asian cobras (subgenus: Naja, Afronaja, Boulengerina and Uraeus. Toxins 2019, 11, 116. [Google Scholar] [CrossRef]
- Du, X.-Y.; Clemetson, K.J. Snake venom l-amino acid oxidases. Toxicon 2002, 40, 659–665. [Google Scholar] [CrossRef]
- Tan, N.H.; Fry, B.G.; Sunagar, K.; Jackson, T.N.W.; Reeks, T.; Fung, S.Y. L-amino acid oxidase enzymes. In Venomous Reptiles and Their Toxins: Evolution, Pathophysiology, and Biodiscovery; Fry, B.G., Ed.; Oxford University Press: New York, NY, USA, 2015; pp. 291–298. [Google Scholar]
- Tan, K.Y.; Tan, N.H.; Tan, C.H. Venom proteomics and antivenom neutralization for the Chinese eastern Russell’s viper, Daboia siamensis from Guangxi and Taiwan. Sci. Rep. 2018, 8, 8545. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.W.; Fritzinger, D.C. Cobra venom factor: Structure, function, and humanization for therapeutic complement depletion. Toxicon 2010, 56, 1198–1222. [Google Scholar] [CrossRef] [PubMed]
- Lavin, M.F.; Earl, S.; Birrel, G.; St Pierre, L.; Guddat, L.; de Jersey, J.; Masci, P. Snake venom nerve growth factors. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; Taylor and Francis Group, CRC Press: Boca Raton, FL, USA, 2009; pp. 377–391. [Google Scholar]
- Kostiza, T.; Meier, J. Nerve growth factors from snake venoms: chemical properties, mode of action and biological significance. Toxicon 1996, 34, 787–806. [Google Scholar] [CrossRef]
- Pung, Y.F.; Kumar, S.V.; Rajagopalan, N.; Fry, B.G.; Kumar, P.P.; Kini, R.M. Ohanin, a novel protein from king cobra venom: its cDNA and genomic organization. Gene 2006, 371, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Pung, Y.F.; Wong, P.T.; Kumar, P.P.; Hodgson, W.C.; Kini, R.M. Ohanin, a novel protein from king cobra venom, induces hypolocomotion and hyperalgesia in mice. J. Biol. Chem. 2005, 280, 13137–13147. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, K.Y.; Ng, T.S.; Quah, E.S.H.; Ismail, A.K.; Khomvilai, S.; Sitprija, V.; Tan, N.H. Venomics of Trimeresurus (Popeia) nebularis, the Cameron Highlands pit viper from Malaysia: Insights into venom proteome, toxicity and neutralization of antivenom. Toxins 2019, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Rotenberg, D.; Bamberger, E.S.; Kochva, E. Studies on ribonucleic acid synthesis in the venom glands of Vipera palaestinae (Ophidia, Reptilia). Biochem. J. 1971, 121, 609–612. [Google Scholar] [CrossRef][Green Version]
- Wery, M.; Descrimes, M.; Thermes, C.; Gautheret, D.; Morillon, A. Zinc-mediated RNA fragmentation allows robust transcript reassembly upon whole transcriptome RNA-Seq. Methods 2013, 63, 25–31. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]

| Parameter | Sequencing Output |
|---|---|
| Total raw reads | 47,494,560 |
| Total clean reads | 46,878,172 |
| Contigs created | 148,475 |
| Q20 percentage | 97.94% |
| N percentage | 0.00% |
| GC percentage | 44.16% |
| Unigenes/transcripts assembled | 75,387 |
| Number of transcripts (FPKM > 1) | 55,386 |
| Unidentified | Abundance |
| Number of transcripts | 35,449 |
| Clean reads | 123,432.1986 |
| Total FPKM percentage (%) | 7.95% |
| Non-toxin | Abundance |
| Number of transcripts | 19,877 |
| Clean reads | 199,393.4726 |
| Total FPKM percentage (%) | 12.84% |
| Toxin | Abundance |
| Number of transcripts | 60 |
| Clean reads | 1,230,548.6634 |
| Total FPKM percentage (%) | 79.22% |
| Protein Family/Protein ID | Annotated Accession | Species | Amino Acid Chain | Mature Chain of Accession ID | Coverage (Mature Chain) | Coverage Percentage (%) | |
|---|---|---|---|---|---|---|---|
| Three-finger toxin | |||||||
| NSM_FTX01 | Cytotoxin 2a | Q9PST4 | N. sputatrix | 81 | 81 | 1–81 | 100 |
| NSM_FTX02 | Neurotoxin homolog NL1 | Q9DEQ3 | N. atra | 81 a | 86 | 6–86 | 93 |
| NSM_FTX06 | Long neurotoxin 7 | O42257 | N. sputatrix | 89 | 90 | 2–90 | 98 |
| NSM_FTX07 | Alpha-neurotoxin NTX-4 | O57327 | N. sputatrix | 83 | 83 | 1–83 | 100 |
| Phospholipase A2 | |||||||
| NSM_PLA01 | Acidic phospholipase A2 C | Q92086 | N. sputatrix | 146 | 146 | 1–146 | 100 b |
| Cysteine-rich secretory protein | |||||||
| NSM_CRP01 | Cysteine-rich venom protein natrin-1 | Q7T1K6 | N. atra | 239 a | 239 | 1–239 | 100 b |
| Nerve growth factor | |||||||
| NSM_NGF01 | Venom nerve growth factor 2 | Q5YF89 | N. sputatrix | 246 | 241 | 1–241 | 100 b |
| Vespryn | |||||||
| NSM_VES01 | Ohanin | P83234 | O. hannah | 190 a | 190 | 1–190 | 100 b |
| C-type/lectin-like protein | |||||||
| NSM_SCL01 | C-type lectin BfL-1 | Q90WI8 | B. fasciatus | 158 a | 158 | 1–158 | 100 b |
| Snake venom metalloproteinase | |||||||
| NSM_SMP07 | Zinc metalloproteinase-disintegrin-like kaouthiagin-like | D3TTC1 | N. atra | 593 a | 593 | 1–593 | 100 b |
| NSM_SMP08 | Zinc metalloproteinase-disintegrin-like atrase-B | D6PXE8 | N. atra | 613 a | 593 | 1–593 | 100 b |
| 5’ nucleotidase | |||||||
| NSM_NUC01 | Ecto-5’-nucleotidase 1 | U3FYP9 | M. fulvius | 569 a | 574 | 1–569 | 99 |
| L-amino-acid oxidase | |||||||
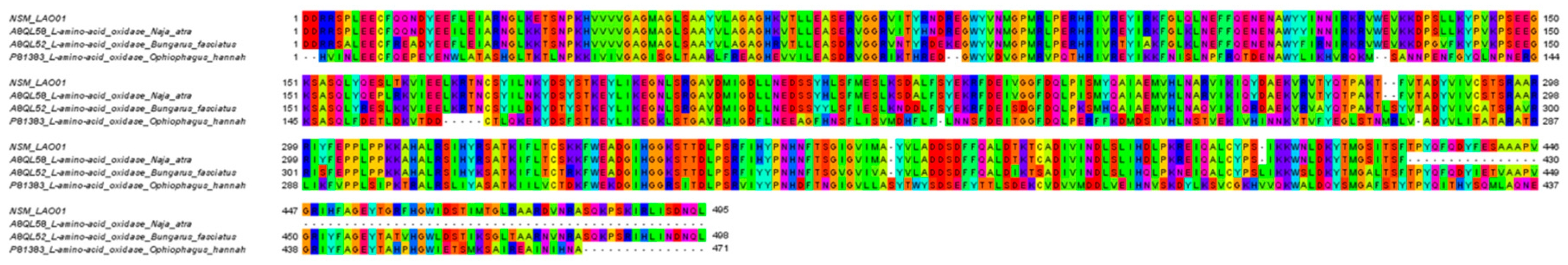
| NSM_LAO01 | L-amino-acid oxidase | A8QL58 | N. atra | 514 a | 449 | 1–449 | 100 b |
| Cobra venom factor | |||||||
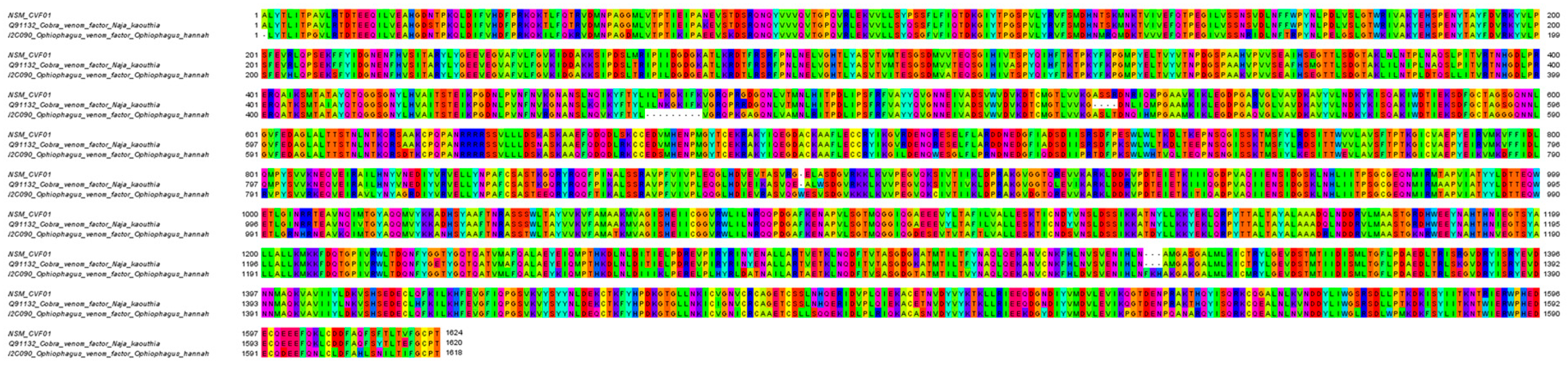
| NSM_CVF01 | Cobra venom factor | Q91132 | N. kaouthia | 1646 a | 1642 | 1–1642 | 100 b |
| Kunitz-type serine protease inhibitor | |||||||
| NSM_KPI01 | Putative Kunitz-type serine protease inhibitor | B2BS84 | A. labialis | 249 | 252 | 1–252 | 98 |
| NSM_KPI02 | Kunitz-type protease inhibitor | U3FZD6 | M. fulvius | 513 | 511 | 1–511 | 100 b |
| Aminopeptidase | |||||||
| NSM_AP01 | Aminopeptidase | U3FZS8 | M. fulvius | 1000 | 993 | 1–993 | 100 b |
| Phospholipase B | |||||||
| NSM_PLB01 | Phospholipase-B 81 | F8J2D3 | D. coronoides | 553 a | 553 | 1–553 | 100 b |
| Vascular endothelial growth factor | |||||||
| NSM_VGF01 | Vascular endothelial growth factor 2 | U3FAK1 | M. fulvius | 421 | 421 | 1–421 | 100 b |
| Dipeptidylpeptidase IV | |||||||
| NSM_DPP01 | Dipeptidyl peptidase 4 | V8P9G9 | O. hannah | 753 | 754 | 1–753 | 99 |
| Neprilysin | |||||||
| NSM_NP01 | Neprilysin | A0A0B8RU83 | B. irregularis | 750 | 750 | 1–750 | 100 b |
| Hyaluronidase | |||||||
| NSM_HY01 | Hyaluronidase | A0A194APD1 | M. tener | 449 | 447 | 1–447 | 100 b |
| NSM_HY02 | Hyaluronidase | A0A194APD1 | M. tener | 449 | 447 | 1–447 | 100 b |
| NSM_HY03 | Hyaluronidase | A0A194APD1 | M. tener | 449 | 447 | 1–447 | 100 b |
| NSM_HY04 | Hyaluronidase | A0A194APD1 | M. tener | 449 | 447 | 1–447 | 100 b |
| Phosphodiesterase | |||||||
| NSM_PDE01 | Snake venom phosphodiesterase | A0A2D0TC04 | N. atra | 850 | 830 | 1–830 | 100 b |
| NSM_PDE02 | Snake venom phosphodiesterase | A0A2D0TC04 | N. atra | 848 | 830 | 1–830 | 100 b |
| Snake venom serine protease | |||||||
| NSM_SSP01 | Serine protease HTRA1 | A0A0B8RTL3 | B. irregularis | 471 | 489 | 18–488 | 96 |
| NSM_SSP02 | Serine protease 23 | V8N8N4 | O. hannah | 365 | 372 | 8–372 | 98 |
| Protein Family/Protein Subtype | Accession/Species | Transcript Abundance % a |
|---|---|---|
| Three-finger toxin (3FTx) | 91.11 (10) | |
| S-3FTx | 73.14 (5) | |
| Cytotoxin 2a | Q9PST4 (N. sputatrix) | 72.83 (1) b |
| Neurotoxin homolog NL1 | Q9DEQ3 (N. atra) | 0.21 (1) b |
| Cardiotoxin 7 | Q91996 (N. atra) | 0.10 (2) |
| Muscarinic toxin-like protein 1 | P82462 (N. kaouthia) | 0.00 (1) |
| L-3FTx | 8.23 (4) | |
| Long neurotoxin 7 | O42257 (N. sputatrix) | 4.91 (1) b |
| Alpha-neurotoxin NTX-4 | O57327 (N. sputatrix) | 3.30 (1) b |
| Long neurotoxin-like OH-31 | Q53B55 (O. hannah) | 0.01 (2) |
| UC-3FTx | 9.74 (1) | |
| Weak neurotoxin 5 | O42255 (N. sputatrix) | 9.74 (1) |
| Phospholipase A2 (PLA2) | 7.42 (2) | |
| Acidic phospholipase A2 C | Q92086 (N. sputatrix) | 7.39 (1) b |
| Phospholipase A2 GL16-1 | Q8JFB2 (L. semifasciata) | 0.03 (1) |
| Cysteine-rich secretory protein (CRISP) | 0.33 (4) | |
| Cysteine-rich venom protein natrin-1 | Q7T1K6 (N. atra) | 0.32 (1) b |
| Cysteine-rich venom protein natrin-2 | Q7ZZN8 (N. atra) | 0.01 (2) |
| Cysteine-rich venom protein kaouthin-2 | P84808 (N. kaouthia) | 0.01 (1) |
| Nerve growth factor (NGF) | 0.31 (2) | |
| Venom nerve growth factor 2 | Q5YF89 (N. sputatrix) | 0.31 (1) b |
| Venom nerve growth factor 1 | Q5YF90 (N. sputatrix) | 0.00 (1) |
| Vespryn (VES) | 0.25 (1) | |
| Thaicobrin | P83234 (O. hannah) | 0.25 (1) b |
| Snake venom C-type/lectin-like protein (Snaclec) | 0.22 (2) | |
| C-type lectin BfL-1 | Q90WI8 (B. fasciatus) | 0.22 (1) b |
| C-type lectin BfL-2 | Q90WI7 (B. fasciatus) | 0.00 (1) |
| Snake venom metalloproteinase (SVMP) | 0.17 (10) | |
| Snake venom metalloproteinase-disintegrin-like morcarhagin | Q10749 (N. mossambica) | 0.03 (1) |
| Zinc metalloproteinase-disintegrin-like cobrin | Q9PVK7 (N. kaouthia) | 0.07 (3) |
| Carinatease-1 | B5KFV1 (Tr. carinatus) | 0.02 (1) |
| Zinc metalloproteinase-disintegrin-like atrase B | D6PXE8 (N. atra) | 0.03 (3) b |
| Zinc metalloproteinase-disintegrin-like kaouthiagin-like | D3TTC1 (N. atra) | 0.01 (1) b |
| Zinc metalloproteinase-disintegrin-like atrase A | D5LMJ3 (N. atra) | 0.01 (1) |
| 5’ nucleotidase (5’ NUC) | 0.05 (2) | |
| Ecto-5’-nucleotidase 1 | U3FYP9 (M. fulvius) | 0.05 (1) b |
| 5’ nucleotidase | A0A024AXW5 (M. ikaheca) | 0.00 (1) |
| Natriuretic peptide (NP) | 0.05 (2) | |
| Natriuretic peptide Na-NP | D9IX97 (N. atra) | 0.05 (2) |
| L-amino acid oxidase (LAAO) | 0.02 (1) | |
| L-amino-acid oxidase | A8QL58 (N. atra) | 0.02 (1) b |
| Cobra venom factor (CVF) | 0.02 (3) | |
| Cobra venom factor | Q91132 (N. kaouthia) | 0.02 (3) b |
| Kunitz-type serine protease inhibitor (KSPI) | 0.01 (2) | |
| Putative Kunitz-type serine protease inhibitor | B2BS84 (A. labialis) | 0.01 (1) b |
| Kunitz-type protease inhibitor | U3FZD6 (M. fulvius) | 0.00 (1) b |
| Aminopeptidase (AP) | 0.01 (1) | |
| Aminopeptidase | U3FZS8 (M. fulvius) | 0.01 (1) b |
| Phospholipase B (PLB) | 0.01 (1) | |
| Phospholipase-B 81 | F8J2D3 (D. coronoides) | 0.01 (1) b |
| Cystatin | 0.01 (5) | |
| Cystatin | V8NX38 (O. hannah) | 0.00 (1) |
| Cystatin | A0A098LYB6 (O. aestivus) | 0.00 (4) |
| Vascular endothelial growth factor (VEGF) | 0.01 (2) | |
| Vascular endothelial growth factor 2 | U3FAK1 (M. fulvius) | 0.01 (1) b |
| Vascular endothelial growth factor A | A0A098LYD7 (O. aestivus) | 0.00 (1) |
| Dipeptidylpeptidase IV (DPP IV) | 0.00 (1) | |
| Dipeptidyl peptidase 4 | V8P9G9 (O. hannah) | 0.00 (1) b |
| Neprilysin | 0.00 (1) | |
| Neprilysin | A0A0B8RU83 (B. irregularis) | 0.00 (1) b |
| Hyaluronidase (HY) | 0.00 (4) | |
| Hyaluronidase | A0A194APD1 (M. tener) | 0.00 (4) b |
| Phosphodiesterase (PDE) | 0.00 (2) | |
| Snake venom phosphodiesterase | A0A2D0TC04 (N. atra) | 0.00 (2) b |
| Snake venom serine protease (SVSP) | 0.00 (2) | |
| Serine protease HTRA1 | A0A0B8RTL3 (B. irregularis) | 0.00 (1) b |
| Serine protease 23 | V8N8N4 (O. hannah) | 0.00 (1) b |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chong, H.P.; Tan, K.Y.; Tan, N.H.; Tan, C.H. Exploring the Diversity and Novelty of Toxin Genes in Naja sumatrana, the Equatorial Spitting Cobra from Malaysia through De Novo Venom-Gland Transcriptomics. Toxins 2019, 11, 104. https://doi.org/10.3390/toxins11020104
Chong HP, Tan KY, Tan NH, Tan CH. Exploring the Diversity and Novelty of Toxin Genes in Naja sumatrana, the Equatorial Spitting Cobra from Malaysia through De Novo Venom-Gland Transcriptomics. Toxins. 2019; 11(2):104. https://doi.org/10.3390/toxins11020104
Chicago/Turabian StyleChong, Ho Phin, Kae Yi Tan, Nget Hong Tan, and Choo Hock Tan. 2019. "Exploring the Diversity and Novelty of Toxin Genes in Naja sumatrana, the Equatorial Spitting Cobra from Malaysia through De Novo Venom-Gland Transcriptomics" Toxins 11, no. 2: 104. https://doi.org/10.3390/toxins11020104
APA StyleChong, H. P., Tan, K. Y., Tan, N. H., & Tan, C. H. (2019). Exploring the Diversity and Novelty of Toxin Genes in Naja sumatrana, the Equatorial Spitting Cobra from Malaysia through De Novo Venom-Gland Transcriptomics. Toxins, 11(2), 104. https://doi.org/10.3390/toxins11020104






