Proteo-Transcriptomic Characterization of the Venom from the Endoparasitoid Wasp Pimpla turionellae with Aspects on Its Biology and Evolution
Abstract
1. Introduction
2. Results
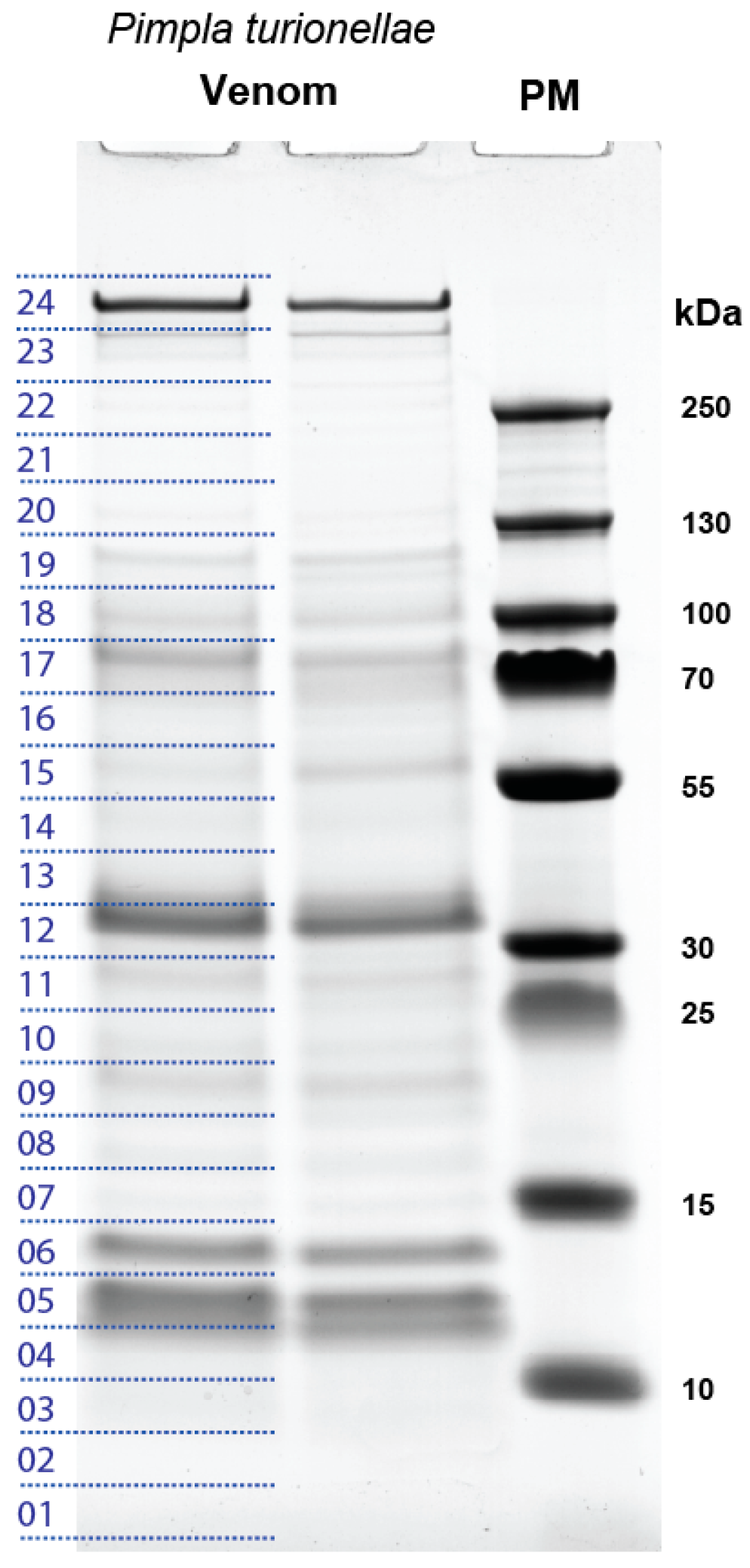
2.1. Proteo-Transcriptomic as a Strategy to Identify Coding Transcripts Based on Proteomics
2.2. General Overview of the Transcripts that are Supported by Proteomics
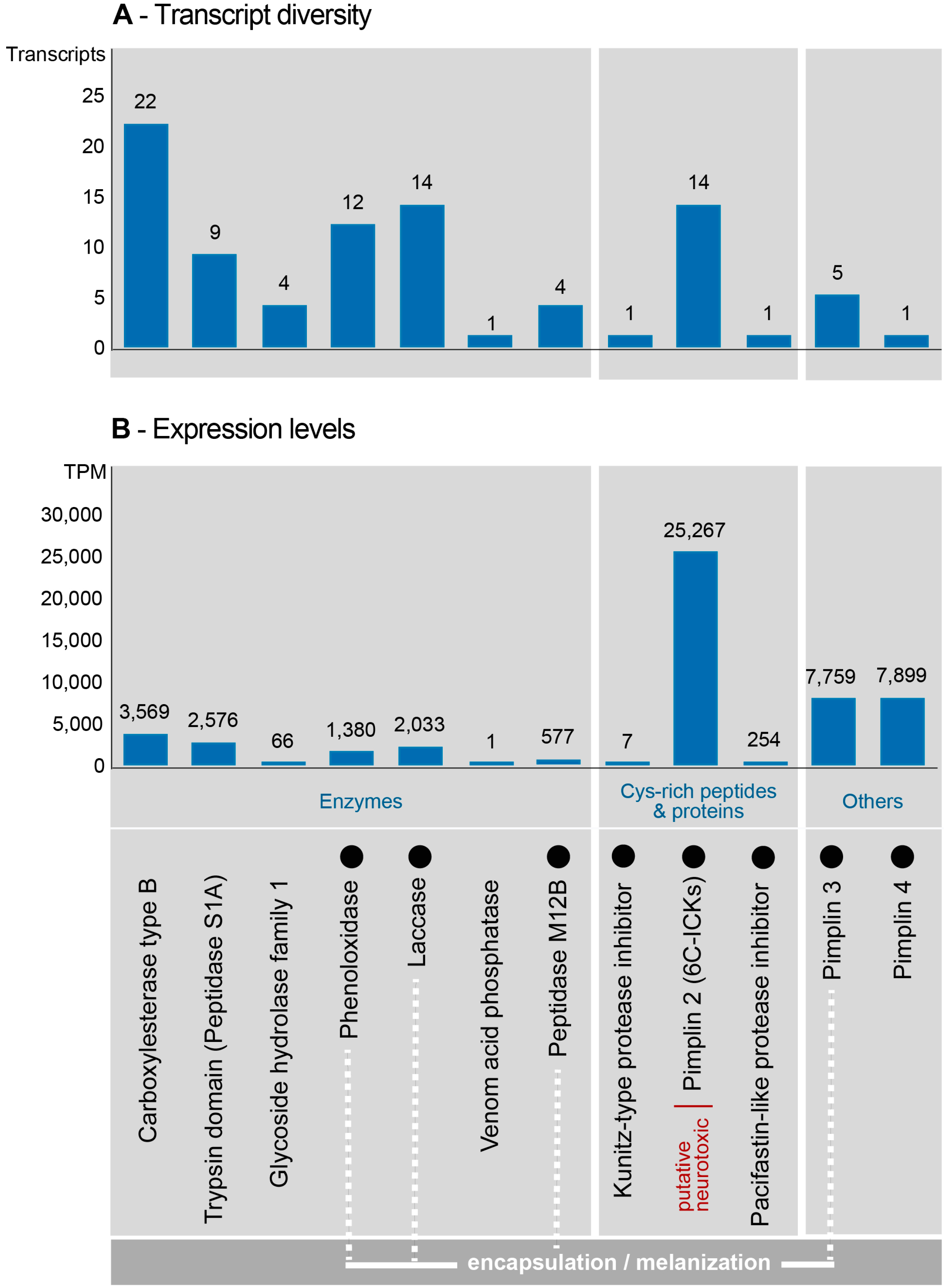
2.3. Composition and Expression of Genes from Known Venom Protein Classes
2.4. Novel, Uncharacterized Venom Peptides and Proteins
3. Discussion
3.1. Missing Evidence of the Paralytic Venom Component Pimplin Described for P. hypochondriaca
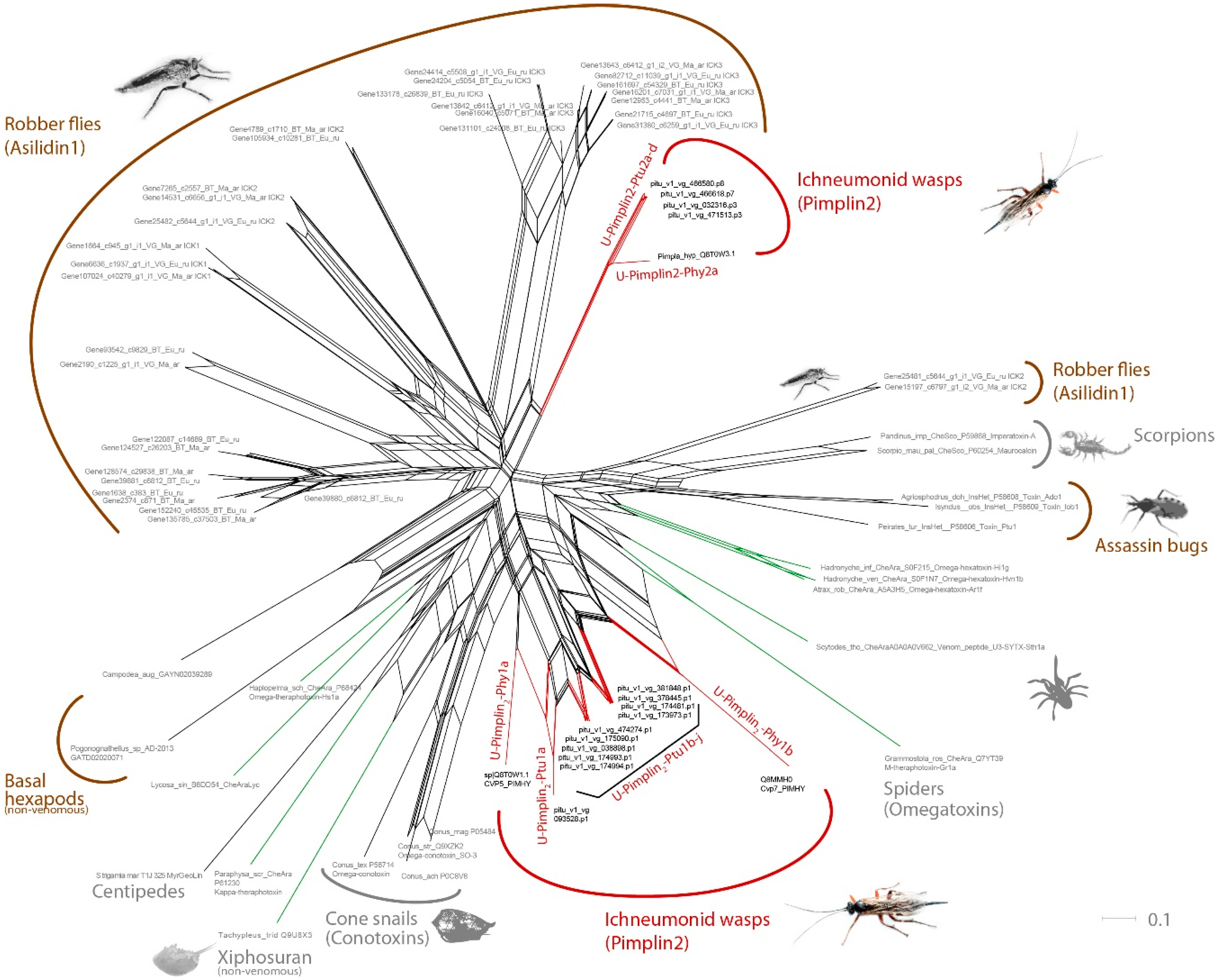
3.2. Pimplin2 (a New ICK Family) Might Act as Paralytic Factor in P. turionellae
3.3. Venom Components Linked to Encapsulation
3.4. Venom Components Involved in the Modulation of Melanization
3.5. Known and Novel Venom Components with Unknown Function
4. Conclusions
5. Materials and Methods
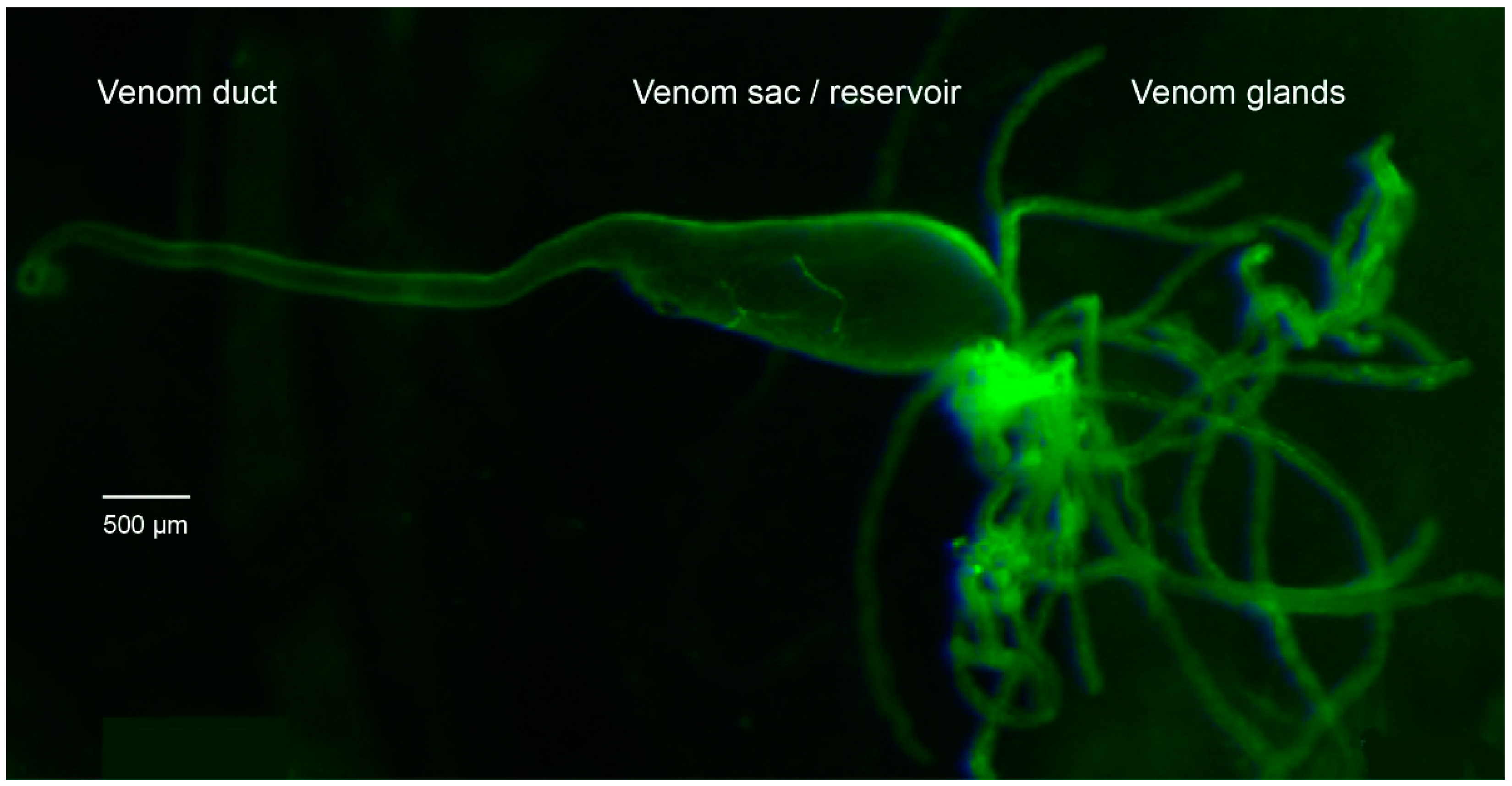
5.1. Rearing and Dissection of P. turionellae Specimens for Proteomics and Transcriptomics
5.2. RNA Isolation, Library Preparation, and Illumina Sequencing
5.3. Transcriptome Assembly, Read Mapping, and Identification of Venom Proteins
5.4. SDS-PAGE and LC-MS/MS Analysis of P. turionellae Venom Proteins
5.5. Matching Mass Spectrometry Data with Transcriptome Data
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aguilar, A.P.; Deans, A.R.; Engel, M.S.; Forshage, M.; Huber, J.T.; Jennings, J.T.; Johnson, N.F.; Lelej, A.S.; Longino, J.T.; Lohrmann, V.; et al. Order Hymenoptera. Zootaxa 2013, 3703, 51. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. (Amst) 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- von Reumont, B.M.; Blanke, A.; Richter, S.; Alvarez, F.; Bleidorn, C.; Jenner, R.A. The first venomous crustacean revealed by transcriptomics and functional morphology: Remipede venom glands express a unique toxin cocktail dominated by enzymes and a neurotoxin. Mol. Biol. Evol. 2014, 31, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.A.; Robinson, S.D.; Yeates, D.K.; Jin, J.; Baumann, K.; Dobson, J.; Fry, B.G.; King, G.F. Entomo-venomics: The evolution, biology and biochemistry of insect venoms. Toxicon 2018, 154, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Branstetter, M.G.; Danforth, B.N.; Pitts, J.P.; Faircloth, B.C.; Ward, P.S.; Buffington, M.L.; Gates, M.W.; Kula, R.R.; Brady, S.G. Phylogenomic insights into the evolution of stinging wasps and the origins of ants and bees. Curr. Biol. 2017, 27, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Peters, R.S.; Krogmann, L.; Mayer, C.; Donath, A.; Gunkel, S.; Meusemann, K.; Kozlov, A.; Podsiadlowski, L.; Petersen, M.; Lanfear, R.; et al. Evolutionary history of the Hymenoptera. Curr. Biol. 2017, 27, 1013–1018. [Google Scholar] [CrossRef]
- Piek, T. Venoms of the Hymenoptera: Biochemical, Pharmacological and Behavioural Aspects; Academic Press: London, UK, 1986; ISBN 9781483263700. [Google Scholar]
- Godfray, H.C.J. Parasitoids: Behavioral and Evolutionary Ecology; Princeton University Press Books: Princeton, NJ, USA, 1994; ISBN 9780691000473. [Google Scholar]
- Whitfield, J.B. Phylogeny and evolution of host-parasitoid interactions in hymenoptera. Annu. Rev. Entomol. 1998, 129–151. [Google Scholar] [CrossRef]
- Quicke, D.L.J. Biology, Systematics, Evolution and Ecology of Braconid and Ichneumonid Parasitoid Wasps; Wiley Blackwell: Hoboken, NJ, USA, 2014; ISBN 9781118907078. [Google Scholar]
- Pennacchio, F.; Strand, M.R. Evolution of developmental strategies in parasitic hymenoptera. Annu. Rev. Entomol. 2006, 51, 233–258. [Google Scholar] [CrossRef]
- Braud, S.; Bon, C.; Wisner, A. Snake venom proteins acting on hemostasis. Biochimie 2000, 82, 851–859. [Google Scholar] [CrossRef]
- Terlau, H.; Olivera, B.M. Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef]
- Baracchi, D.; Tragust, S. Evolution of Venomous Animals and Their Toxins. Venom as a Component of External Immune Defense in Hymenoptera; Springer: Dordrecht, The Netherlands, 2017; ISBN 978-94-007-6457-6. [Google Scholar]
- Lin, Z.; Cheng, Y.; Wang, R.J.; Du, J.; Volovych, O.; Li, J.C.; Hu, Y.; Lu, Z.Y.; Lu, Z.; Zou, Z. A metalloprotease homolog venom protein from a parasitoid wasp suppresses the toll pathway in host hemocytes. Front. Immunol. 2018, 9, 2301. [Google Scholar] [CrossRef] [PubMed]
- Graaf, D.C.; Aerts, M.; Brunain, M.; Desjardins, C.A.; Jacobs, F.J.; Werren, J.H.; Devreese, B. Insights into the venom composition of the ectoparasitoid wasp Nasonia vitripennis from bioinformatic and proteomic studies. Insect Mol. Biol. 2010, 19, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Moreau, S.J.M.; Asgari, S. Venom proteins from parasitoid wasps and their biological functions. Toxins 2015, 7, 2385–2412. [Google Scholar] [CrossRef] [PubMed]
- Laurino, S.; Grossi, G.; Pucci, P.; Flagiello, A.; Bufo, S.A.; Bianco, G.; Salvia, R.; Vinson, S.B.; Vogel, H.; Falabella, P. Identification of major Toxoneuron nigriceps venom proteins using an integrated transcriptomic/proteomic approach. Insect Biochem. Mol. Biol. 2016, 76, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Arvidson, R.; Kaiser, M.; Lee, S.S.; Urenda, J.P.; Dail, C.; Mohammed, H.; Nolan, C.; Pan, S.; Stajich, J.E.; Libersat, F.; et al. Parasitoid jewel wasp mounts multipronged neurochemical attack to hijack a host brain. Mol. Cell. Proteom. 2019, 18, 99–114. [Google Scholar] [CrossRef]
- Gauld, I.D. Evolutionary patterns of host utilization by ichneumonoid parasitoids (Hymenoptera: Ichneumonidae and Braconidae). Biol. J. Linn. Soc. 1988, 35, 351–377. [Google Scholar] [CrossRef]
- Poirié, M.; Carton, Y.; Dubuffet, A. Virulence strategies in parasitoid Hymenoptera as an example of adaptive diversity. C. R. Biol. 2009, 332, 311–320. [Google Scholar] [CrossRef]
- Poirié, M.; Colinet, D.; Gatti, J.L. Insights into function and evolution of parasitoid wasp venoms. Curr. Opin. Insect Sci. 2014, 6, 52–60. [Google Scholar] [CrossRef]
- Libersat, F.; Gal, R. What can parasitoid wasps teach us about decision-making in insects? J. Exp. Biol. 2013, 216, 47–55. [Google Scholar] [CrossRef]
- Desneux, N.; Barta, R.J.; Delebecque, C.J.; Heimpel, G.E. Transient host paralysis as a means of reducing self-superparasitism in koinobiont endoparasitoids. J. Insect Physiol. 2009, 55, 321–327. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Ye, G.Y.; Dong, S.Z.; Fang, Q.; Hu, C. Venom of Pteromalus puparum (Hymenoptera: Pteromalidae) induced endocrine changes in the hemolymph of its host, Pieris rapae (Lepidoptera: Pieridae). Arch. Insect Biochem. Physiol. 2009, 71, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.D.; Salvador, G.; Cônsoli, F.L. The parasitoid, Cotesia flavipes (Cameron) (Hymenoptera: Braconidae), influences food consumption and utilization by larval Diatraea saccharalis (F.) (Lepidoptera: Crambidae). Arch. Insect Biochem. Physiol. 2014, 87, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.M.A.; Luna-Santillana, E.J.; Rodríguez-Perez, M.A. Parasitism by the endoparasitoid, Cotesia flavipes induces cellular immunosuppression and enhances susceptibility of Parasitism by the endoparasitoid, Cotesia flavipes induces cellular immunosuppression and enhances susceptibility of the sugar cane borer, Diatraea saccharalis to Bacillus thuringiensis. J. Insect Sci. 2011, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Cavagnol, R.M. The pharmacological effects of Hymenoptera venoms. Annu. Rev. Pharmacol. Toxicol. 1977, 17, 479–498. [Google Scholar] [CrossRef] [PubMed]
- Teng, Z.W.; Xiong, S.J.; Xu, G.; Gan, S.Y.; Chen, X.; Stanley, D.; Yan, Z.C.; Ye, G.Y.; Fang, Q. Protein discovery: combined transcriptomic and proteomic analyses of venom from the endoparasitoid Cotesia chilonis (Hymenoptera: Braconidae). Toxins 2017, 9, 135. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Fang, Q.; Wang, L.; Hu, C.; Ye, G.Y. Proteomic analysis of the venom from the endoparasitoid wasp Pteromalus puparum (Hymenoptera: Pteromalidae). Arch. Insect Biochem. Physiol. 2010, 75, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Vincent, B.; Kaeslin, M.; Roth, T.; Heller, M.; Poulain, J.; Cousserans, F.; Schaller, J.; Poirié, M.; Lanzrein, B.; Drezen, J.M.; et al. The venom composition of the parasitic wasp Chelonus inanitus resolved by combined expressed sequence tags analysis and proteomic approach. BMC Genom. 2010, 11, 693. [Google Scholar] [CrossRef]
- Asgari, S.; Rivers, D.B. Venom proteins from endoparasitoid wasps and their role in host-parasite interactions. Annu. Rev. Entomol. 2011, 56, 313–335. [Google Scholar] [CrossRef]
- Yan, Z.; Fang, Q.; Liu, Y.; Xiao, S.; Yang, L.; Wang, F.; An, C.; Werren, J.H.; Ye, G. A venom serpin splicing isoform of the endoparasitoid wasp Pteromalus puparum suppresses host prophenoloxidase cascade by forming complexes with host hemolymph proteinases. J. Biol. Chem. 2017, 292, 1038–1051. [Google Scholar] [CrossRef]
- Parkinson, N.; Smith, I.; Audsley, N.; Edwards, J.P. Purification of pimplin, a paralytic heterodimeric polypeptide from venom of the parasitoid wasp Pimpla hypochondriaca, and cloning of the cDNA encoding one of the subunits. Insect Biochem. Mol. Biol. 2002, 32, 1769–1773. [Google Scholar] [CrossRef]
- Dani, M.P.; Richards, E.H. Identification, cloning and expression of a second gene (vpr1) from the venom of the endoparasitic wasp, Pimpla hypochondriaca that displays immunosuppressive activity. J. Insect Physiol. 2010, 56, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Dani, M.P.; Richards, E.H. Cloning and expression of the gene for an insect haemocyte anti-aggregation protein (VPr3), from the venom of the endoparasitic wasp, Pimpla hypochondriaca. Arch. Insect Biochem. Physiol. 2009, 71, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, N.M.; Weaver, R.J. Noxious components of venom from the pupa-specific parasitoid Pimpla hypochondriaca. J. Invertebr. Pathol. 1988, 73, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, N.; Smith, I.; Weaver, R.; Edwards, J.P. A new form of arthropod phenoloxidase is abundant in venom of the parasitoid wasp Pimpla hypochondriaca. Insect Biochem. Mol. Biol. 2001, 31, 57–63. [Google Scholar] [CrossRef]
- Parkinson, N.; Richards, E.H.; Conyers, C.; Smith, I.; Edwards, J.P. Analysis of venom constituents from the parasitoid wasp Pimpla hypochondriaca and cloning of a cDNA encoding a venom protein. Insect Biochem. Mol. Biol. 2002, 32, 729–735. [Google Scholar] [CrossRef]
- Parkinson, N.; Conyers, C.; Smith, I. A venom protein from the endoparasitoid wasp Pimpla hypochondriaca is similar to snake venom reprolysin-type metalloproteases. J. Invertebr. Pathol. 2002, 79, 129–131. [Google Scholar] [CrossRef]
- Parkinson, N.M.; Conyers, C.M.; Keen, J.N.; MacNicoll, A.D.; Smith, I.; Weaver, R.J. cDNAs encoding large venom proteins from the parasitoid wasp Pimpla hypochondriaca identified by random sequence analysis. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2003, 134, 513–520. [Google Scholar] [CrossRef]
- Parkinson, N.M.; Conyers, C.; Keen, J.; MacNicoll, A.; Smith, I.; Audsley, N.; Weaver, R. Towards a comprehensive view of the primary structure of venom proteins from the parasitoid wasp Pimpla hypochondriaca. Insect Biochem. Mol. Biol. 2004, 34, 565–571. [Google Scholar] [CrossRef]
- Richards, E.H.; Dani, M.P. Biochemical isolation of an insect haemocyte anti-aggregation protein from the venom of the endoparasitic wasp, Pimpla hypochondriaca, and identification of its gene. J. Insect Physiol. 2008, 54, 1041–1049. [Google Scholar] [CrossRef]
- Richards, E.H.; Bradish, H.; Dani, M.P.; Pietravalle, S.; Lawson, A. Recombinant immunosuppressive protein from Pimpla hypochondrica venom (rVPr1) increases the susceptibility of Mamestra brassicae larvae to the fungal biological control agent, Beauveria bassiana. Arch. Insect Biochem. Physiol. 2011, 78, 119–131. [Google Scholar] [CrossRef]
- Uçkan, F.; Sİnan, S.; Savaşçi, Ş.; Ergİn, E. Determination of venom components from the endoparasitoid wasp Pimpla turionellae L. (Hymenoptera: Ichneumonidae). Ann. Entomol. Soc. Am. 2004, 97, 775–780. [Google Scholar] [CrossRef]
- Smith, J.J.; Undheim, E.A.B. True lies: Using proteomics to assess the accuracy of transcriptome-based venomics in centipedes uncovers false positives and reveals startling intraspecific variation in Scolopendra subspinipes. Toxins 2018, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Von Reumont, B.M. Studying smaller and neglected organisms in modern evolutionary venomics implementing RNASeq (Transcriptomics) -A critical guide. Toxins 2018, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Holding, M.L.; Margres, M.J.; Mason, A.J.; Parkinson, C.L.; Rokyta, D.R. Evaluating the performance of de novo assembly methods for venom-gland transcriptomics. Toxins 2018, 10, 249. [Google Scholar] [CrossRef] [PubMed]
- Uçkan, F.; Er, A.; Ergin, E. Levels of encapsulation and melanization in Galleria mellonella (Lepidoptera: Pyralidae) parasitized and envenomated by Pimpla turionellae (Hymenoptera: Ichneumonidae). J. Appl. Entomol. 2010, 134, 718–726. [Google Scholar] [CrossRef]
- Er, A.; Uçkan, F.; Rivers, D.B.; Ergİn, E.; Sak, O. Effects of parasitization and envenomation by the endoparasitic wasp Pimpla turionellae (Hymenoptera: Ichneumonidae) on hemocyte numbers, morphology, and viability of its host Galleria mellonella (Lepidoptera: Pyralidae). Ann. Entomol. Soc. Am. 2010, 103, 273–282. [Google Scholar] [CrossRef]
- Er, A.; Uçkan, F.; Rivers, D.B.; Sak, O. Cytotoxic effects of parasitism and application of venom from the endoparasitoid Pimpla turionellae on hemocytes of the host Galleria mellonella. J. Appl. Entomol. 2011, 135, 225–236. [Google Scholar] [CrossRef]
- Quistad, G.; Nguyen, Q.; Bernasconi, P.; Leisy, D.J. Purification and characterization of insecticidal toxins from venom glands of the parasitic wasp, Bracon hebetor. Insect Biochem. Mol. Biol. 1994, 10, 955–961. [Google Scholar] [CrossRef]
- Undheim, E.A.B.; Mobli, M.; King, G.F. Toxin structures as evolutionary tools: Using conserved 3D folds to study the evolution of rapidly evolving peptides. Bioessays 2016, 38, 539–548. [Google Scholar] [CrossRef]
- Drukewitz, S.H.; Fuhrmann, N.; Undheim, E.A.B.; Blanke, A.; Giribaldi, J.; Mary, R.; Laconde, G.; Dutertre, S.; Reumont, B.M. von. A dipteran’s novel sucker punch: Evolution of arthropod atypical venom with a neurotoxic component in robber flies (Asilidae, Diptera). Toxins 2018, 10, 29. [Google Scholar] [CrossRef]
- Undheim, E.A.B.; Fry, B.G.; King, G.F. Centipede venom: Recent discoveries and current state of knowledge. Toxins 2015, 7, 679–704. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, A.; UlAbdin, Z.; Webb, B.A.; Arif, M.J.; Jamil, A. De novo sequencing and transcriptome analysis of female venom glands of ectoparasitoid Bracon hebetor (Say.) (Hymenoptera: Braconidae). Comp. Biochem. Physiol. Part D Genom. Proteom. 2016, 20, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Shaina, H.; UlAbdin, Z.; Webb, B.A.; Arif, M.J.; Jamil, A. De novo sequencing and transcriptome analysis of venom glands of endoparasitoid Aenasius arizonensis (Girault) (=Aenasius bambawalei Hayat) (Hymenoptera, Encyrtidae). Toxicon 2016, 121, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Price, D.R.; Bell, H.A.; Hinchliffe, G.; Fitches, E.; Weaver, R.; Gatehouse, J.A. Blackwell Publishing Ltd. A venom metalloproteinase from the parasitic wasp Eulophus pennicornis is toxic towards its host, tomato moth (Lacanobia oleracae). Insect Mol. Biol. 2009, 18, 195–202. [Google Scholar] [CrossRef]
- Zhang, G.; Schmidt, O.; Asgari, S. A calreticulin-like protein from endoparasitoid venom fluid is involved in host hemocyte inactivation. Dev. Comp. Immunol. 2006, 30, 756–764. [Google Scholar] [CrossRef]
- Wu, M.L.; Ye, G.Y.; Zhu, J.Y.; Chen, X.X.; Hu, C. Isolation and characterization of an immunosuppressive protein from venom of the pupa-specific endoparasitoid Pteromalus puparum. J. Invertebr. Pathol. 2008, 99, 186–191. [Google Scholar] [CrossRef]
- Labrosse, C.; Eslin, P.; Doury, G.; Drezen, J.M.; Poirié, M. Haemocyte changes in D. melanogaster in response to long gland components of the parasitoid wasp Leptopilina boulardi: A Rho-GAP protein as an important factor. J. Insect Physiol. 2005, 51, 161–170. [Google Scholar] [CrossRef]
- Labrosse, C.; Stasiak, K.; Lesobre, J.; Grangeia, A.; Huguet, E.; Drezen, J.M.; Poirie, M. A RhoGAP protein as a main immune suppressive factor in the Leptopilina boulardi (Hymenoptera, Figitidae)-Drosophila melanogaster interaction. Insect Biochem. Mol. Biol. 2005, 35, 93–103. [Google Scholar] [CrossRef]
- Takeda, S. ADAM and ADAMTS family proteins and snake venom metalloproteinases: A structural overview. Toxins 2016, 8, 155. [Google Scholar] [CrossRef]
- Griesch, J.; Vilcinskas, A. Proteases released by entomopathogenic fungi impair phagocytic activity, attachment and spreading of plasmatocytes isolated from haemolymph of the greater wax moth Galleria mellonella. Biocontrol Sci. Technol. 1998, 8, 517–531. [Google Scholar] [CrossRef]
- Liehl, P.; Blight, M.; Vodovar, N.; Boccard, F.; Lemaitre, B. Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathog. 2006, 2, e56. [Google Scholar] [CrossRef]
- Di Cera, E. Serine proteases. IUBMB Life 2009, 61, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhao, M.; Rotgans, B.A.; Ni, G.; Dean, J.F.D.; Nahrung, H.F.; Cummins, S.F. Proteomic analysis of the venom and venom sac of the woodwasp, Sirex noctilio— Towards understanding its biological impact. J. Proteom. 2016, 146, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Choo, Y.M.; Lee, K.S.; Yoon, H.J.; Kim, B.Y.; Sohn, M.R.; Roh, J.Y.; Je, Y.H.; Kim, N.J.; Kim, I.; Woo, S.D.; et al. Dual function of a bee venom serine protease: Prophenoloxidase-activating factor in arthropods and fibrin(ogen)olytic enzyme in mammals. PLoS ONE 2010, 5, e10393. [Google Scholar] [CrossRef]
- Han, J.; You, D.; Xu, X.; Han, W.; Lu, Y.; Lai, R.; Meng, Q. An anticoagulant serine protease from the wasp venom of Vespa magnifica. Toxicon 2008, 51, 914–922. [Google Scholar] [CrossRef]
- Czaikoski, P.G.; Menaldo, D.L.; Marcussi, S.; Baseggio, A.L.C.; Fuly, A.L.; Paula, R.C.; Quadros, A.U.; Romão, P.R.T.; Buschini, M.L.T.; Cunha, F.Q.; et al. Anticoagulant and fibrinogenolytic properties of the venom of Polybia occidentalis social wasp. Blood Coagul. Fibrinolysis 2010, 21, 653–659. [Google Scholar] [CrossRef]
- Ergin, E.; Uçkan, F.; Rivers, D.B.; Sak, O. In vivo and in vitro activity of venom from the endoparasitic wasp Pimpla turionellae (L.) (Hymenoptera: Ichneumonidae). Arch. Insect Biochem. Physiol. 2006, 61, 87–97. [Google Scholar] [CrossRef]
- Keenan, B.; Uçkan, F.; Ergin, E.; Rivers, D.B. Morphological and biochemical changes in cultured cells induced by venom from the Endoparasitoid, Pimpla turionellae. In Recent Advances in the Biochemistry, Toxicity, and mode of Action of Parasitic Wasp Venoms; Rivers, D., Jay, Y., Eds.; Research Signpost: Thiruvananthapuram, Kerala, India, 2007; pp. 75–92. ISBN 81-308-0161-2. [Google Scholar]
- Rivers, D.; Brogan, A. Venom glands from the ectoparasitoid Nasonia vitripennis (Walker) (Hymenoptera: Pteromalidae) produce a calreticulin-like protein that functions in developmental arrest and cell death in the flesh fly host, Sarcophaga bullata Parker (Diptera: Sarcophagidae). In Insect Physiology: New Research; Maes, R.P., Ed.; Nova Science Publishers: New York, NY, USA, 2008; pp. 259–278. [Google Scholar]
- Formesyn, E.M.; Heyninck, K.; de Graaf, D.C. The role of serine- and metalloproteases in Nasonia vitripennis venom in cell death related processes towards a Spodoptera frugiperda Sf21 cell line. J. Insect Physiol. 2013, 59, 795–803. [Google Scholar] [CrossRef]
- Dani, M.P.; Richards, E.H.; Edwards, J.P. Venom from the pupal endoparasitoid, Pimpla hypochondriaca, increases the susceptibility of larval Lacanobia oleracea to the entomopathogens Bacillus cereus and Beauveria bassiana. J. Invertebr. Pathol. 2004, 86, 19–25. [Google Scholar] [CrossRef]
- García-Estrada, C.; Cat, E.; Santamarta, I. Beauveria bassiana as biocontrol agent: Formulation and commercialization for pest management. In Agriculturally Important Microorganisms; Singh, H.B., Sarma, B.K., Keswani, C., Eds.; Springer: Singapore, 2016; pp. 81–96. ISBN 978-981-10-2575-4. [Google Scholar]
- Christensen, B.M.; Li, J.; Chen, C.C.; Nappi, A.J. Melanization immune responses in mosquito vectors. Trends Parasitol. 2005, 21, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Chen, C.S. Brugia pahangi: Effects of melanization on the uptake of nutrients by microfilariae in vitro. Exp. Parasitol. 1995, 81, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Nappi, A.J.; Christensen, B.M. Melanogenesis and associated cytotoxic reactions: Applications to insect innate immunity. Insect Biochem. Mol. Biol. 2005, 35, 443–459. [Google Scholar] [CrossRef] [PubMed]
- Abt, M.; Rivers, D.B. Characterization of phenoloxidase activity in venom from the ectoparasitoid Nasonia vitripennis (Walker) (Hymenoptera: Pteromalidae). J. Invertebr. Pathol. 2007, 94, 108–118. [Google Scholar] [CrossRef]
- Mason, H.S. Oxidases. Annu. Rev. Biochem. 1965, 34, 595–634. [Google Scholar] [CrossRef]
- Suderman, R.J.; Dittmer, N.T.; Kanost, M.R.; Kramer, K.J. Model reactions for insect cuticle sclerotization: Cross-linking of recombinant cuticular proteins upon their laccase-catalyzed oxidative conjugation with catechols. Insect Biochem. Mol. Biol. 2006, 36, 353–365. [Google Scholar] [CrossRef]
- Rivers, D.B.; Dani, M.P.; Richards, E.H. The mode of action of venom from the endoparasitic wasp Pimpla hypochondriaca (hymenoptera: Ichneumonidae) involves Ca+2-dependent cell death pathways. Arch. Insect Biochem. Physiol. 2009, 71, 173–190. [Google Scholar] [CrossRef]
- Kanost, M.R. Serine proteinase inhibitors in arthropod immunity. Dev. Comp. Immunol. 1999, 23, 291–301. [Google Scholar] [CrossRef]
- Colinet, D.; Dubuffet, A.; Cazes, D.; Moreau, S.; Drezen, J.M.; Poirié, M. A serpin from the parasitoid wasp Leptopilina boulardi targets the Drosophila phenoloxidase cascade. Dev. Comp. Immunol. 2009, 33, 681–689. [Google Scholar] [CrossRef]
- Perkin, L.C.; Friesen, K.S.; Flinn, P.W.; Oppert, B. Venom gland components of the ectoparasitoid wasp, Anisopteromalus calandrae. J. Venom. Res. 2015, 6, 19. [Google Scholar]
- Yuan, C.H.; He, Q.Y.; Peng, K.; Diao, J.B.; Jiang, L.P.; Tang, X.; Liang, S.P. Discovery of a distinct superfamily of Kunitz-type toxin (KTT) from tarantulas. PLoS ONE 2008, 3, e3414. [Google Scholar] [CrossRef]
- Grunwald, T.; Bockisch, B.; Spillner, E.; Ring, J.; Bredehorst, R.; Ollert, M.W. Molecular cloning and expression in insect cells of honeybee venom allergen acid phosphatase (Api m 3). J. Allergy Clin. Immunol. 2006, 117, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Dani, M.P.; Edwards, J.P.; Richards, E.H. Hydrolase activity in the venom of the pupal endoparasitic wasp, Pimpla hypochondriaca. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2005, 141, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Cui, F.; Qiao, C. Structure, function and applications of carboxylesterases from insects for insecticide resistance. Protein Pept. Lett. 2009, 16, 1181–1188. [Google Scholar] [CrossRef]
- Ross, M.K.; Streit, T.M.; Herring, K.L. Carboxylesterases: Dual roles in lipid and pesticide metabolism. J. Pestic. Sci. 2010, 35, 257–264. [Google Scholar] [CrossRef]
- Taylor, P.; Radic, Z. The cholinesterases: From genes to proteins. Annu. Rev. Pharmacol. Toxicol. 1994, 34, 281–320. [Google Scholar] [CrossRef]
- Oakeshott, J.G.; Claudianos, C.; Russell, R.J.; Robin, G.C. Carboxyl/cholinesterases: A case study of the evolution of a successful multigene family. Bioessays 1999, 21, 1031–1042. [Google Scholar] [CrossRef]
- Gilbert, L.I.; Granger, N.A.; Roe, R.M. The juvenile hormones: Historical facts and speculations on future research directions. Insect Biochem. Mol. Biol. 2000, 30, 617–644. [Google Scholar] [CrossRef]
- Mathé-Hubert, H.; Colinet, D.; Deleury, E.; Belghazi, M.; Ravallec, M.; Poulain, J.; Dossat, C.; Poirié, M.; Gatti, J.L. Comparative venomics of Psyttalia lounsburyi and P. concolor, two olive fruit fly parasitoids: A hypothetical role for a GH1 β-glucosidase. Sci. Rep. 2016, 6, 35873. [Google Scholar] [CrossRef]
- Davies, G.; Henrissat, B. Structures and mechanisms of glycosyl hydrolases. Structure 1995, 3, 853–859. [Google Scholar] [CrossRef]
- Berlemont, R.; Martiny, A.C. Glycoside hydrolases across environmental microbial communities. PLoS Comput. Biol. 2016, 12, e1005300. [Google Scholar] [CrossRef] [PubMed]
- Eyun, S.-I.; Wang, H.; Pauchet, Y.; Ffrench-Constant, R.H.; Benson, A.K.; Valencia-Jiménez, A.; Moriyama, E.N.; Siegfried, B.D. Molecular evolution of glycoside hydrolase genes in the Western corn rootworm (Diabrotica virgifera virgifera). PLoS ONE 2014, 9, e94052. [Google Scholar] [CrossRef] [PubMed]
- Koudounas, K.; Banilas, G.; Michaelidis, C.; Demoliou, C.; Rigas, S.; Hatzopoulos, P. A defence-related Olea europaea β-glucosidase hydrolyses and activates oleuropein into a potent protein cross-linking agent. J. Exp. Bot. 2015, 66, 2093–2106. [Google Scholar] [CrossRef] [PubMed]
- Mander, L.; Liu, H.W. Comprehensive Natural Products II. Chemistry and Biology; Elsevier: Oxford, UK, 2010; ISBN 978-0-08-045382-8. [Google Scholar]
- Cairo, F.; João, P.L.; Carazzolle, M.F.; Leonardo, F.C.; Mofatto, L.S.; Brenelli, L.B.; Gonçalves, T.A.; Uchima, C.A.; Domingues, R.R.; Alvarez, T.M.; et al. Expanding the knowledge on lignocellulolytic and redox enzymes of worker and soldier castes from the lower termite Coptotermes gestroi. Front. Microbiol. 2016, 7, 1518. [Google Scholar] [CrossRef]
- Martinson, E.O.; Kelkar, Y.D.; Chang, C.H.; Werren, J.H. The Evolution of venom by co-option of single-copy genes. Curr. Biol. 2017, 27, 2007–2013. [Google Scholar] [CrossRef] [PubMed]
- Uçkan, F.; Özbek, R.; Ergin, E. Effects of Indol-3-Acetic Acid on the biology of Galleria mellonella and its endoparasitoid Pimpla turionellae. Belg. J. Zool. 2015, 145, 49–58. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Babraham, UK, 2019; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 31 July 2019).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. rnaSPAdes: A de novo transcriptome assembler and its application to RNA-Seq data. Gigascience 2019, 8. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]

| Name | Structural Fold | Scaffold | Length(aa) | TPM |
|---|---|---|---|---|
| Pimplin * | Dimeric protein | Prolin scaffold | 143 | NA |
| Pimplin2 | ICK | X-CX7-[C-X6-C-X5–8-CC-X2–4-C-X6–9]-X | 63–115 | 25,267 |
| Pimplin3 | Protein | Potential P and C scaffold | 167–315 | 7759 |
| Pimplin4 | Short protein | No cysteine scaffold, 3 P residues | 70–78 | 7899 |
| Name | Transcript ID | Manual BLAST Match | Length(aa) | TPM | Signal Peptide |
|---|---|---|---|---|---|
| NovelP1 | pitu_v1_174267 | Inconclusive-non cytoplasmic domain | 126 | 7746 | Yes |
| NovelP2 | pitu_v1_002265 | Inconclusive-bacterial | 50 | 6141 | No |
| NovelP3 | pitu_v1_377983 | Inconclusive-bacterial | 17 | 1209 | No |
| NovelP4 | pitu_v1_378290 | Inconclusive-bacterial | 14 | 1159 | No |
| NovelP5 | pitu_v1_002208 | Inconclusive-non cytoplasmic domain | 73 | 288 | No |
| NovelP6 | pitu_v1_468063 | Inconclusive-non cytoplasmic domain | 70 | 239 | Yes |
| NovelP7 | pitu_v1_094627 | Inconclusive-non cytoplasmic domain | 167 | 219 | Yes |
| NovelP8 | pitu_v1_377800 | Inconclusive-non cytoplasmic domain | 214 | 208 | Yes |
| NovelP9 | pitu_v1_473891 | Inconclusive-bacterial | 11 | 180 | No |
| NovelP10 | pitu_v1_176834 | Inconclusive-Water bear-uncharacterized | 43 | 102 | No |
| NovelP11 | pitu_v1_172572 | Inconclusive-bacterial | 49 | 36 | No |
| NovelP12 | pitu_v1_285207 | Inconclusive-bacterial | 19 | 2 | No |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Özbek, R.; Wielsch, N.; Vogel, H.; Lochnit, G.; Foerster, F.; Vilcinskas, A.; von Reumont, B.M. Proteo-Transcriptomic Characterization of the Venom from the Endoparasitoid Wasp Pimpla turionellae with Aspects on Its Biology and Evolution. Toxins 2019, 11, 721. https://doi.org/10.3390/toxins11120721
Özbek R, Wielsch N, Vogel H, Lochnit G, Foerster F, Vilcinskas A, von Reumont BM. Proteo-Transcriptomic Characterization of the Venom from the Endoparasitoid Wasp Pimpla turionellae with Aspects on Its Biology and Evolution. Toxins. 2019; 11(12):721. https://doi.org/10.3390/toxins11120721
Chicago/Turabian StyleÖzbek, Rabia, Natalie Wielsch, Heiko Vogel, Günter Lochnit, Frank Foerster, Andreas Vilcinskas, and Björn Marcus von Reumont. 2019. "Proteo-Transcriptomic Characterization of the Venom from the Endoparasitoid Wasp Pimpla turionellae with Aspects on Its Biology and Evolution" Toxins 11, no. 12: 721. https://doi.org/10.3390/toxins11120721
APA StyleÖzbek, R., Wielsch, N., Vogel, H., Lochnit, G., Foerster, F., Vilcinskas, A., & von Reumont, B. M. (2019). Proteo-Transcriptomic Characterization of the Venom from the Endoparasitoid Wasp Pimpla turionellae with Aspects on Its Biology and Evolution. Toxins, 11(12), 721. https://doi.org/10.3390/toxins11120721






