Fusarium-Produced Mycotoxins in Plant-Pathogen Interactions
Abstract
1. Introduction
2. Fusarium Mycotoxins
2.1. Trichothecenes
2.2. Fumonisins
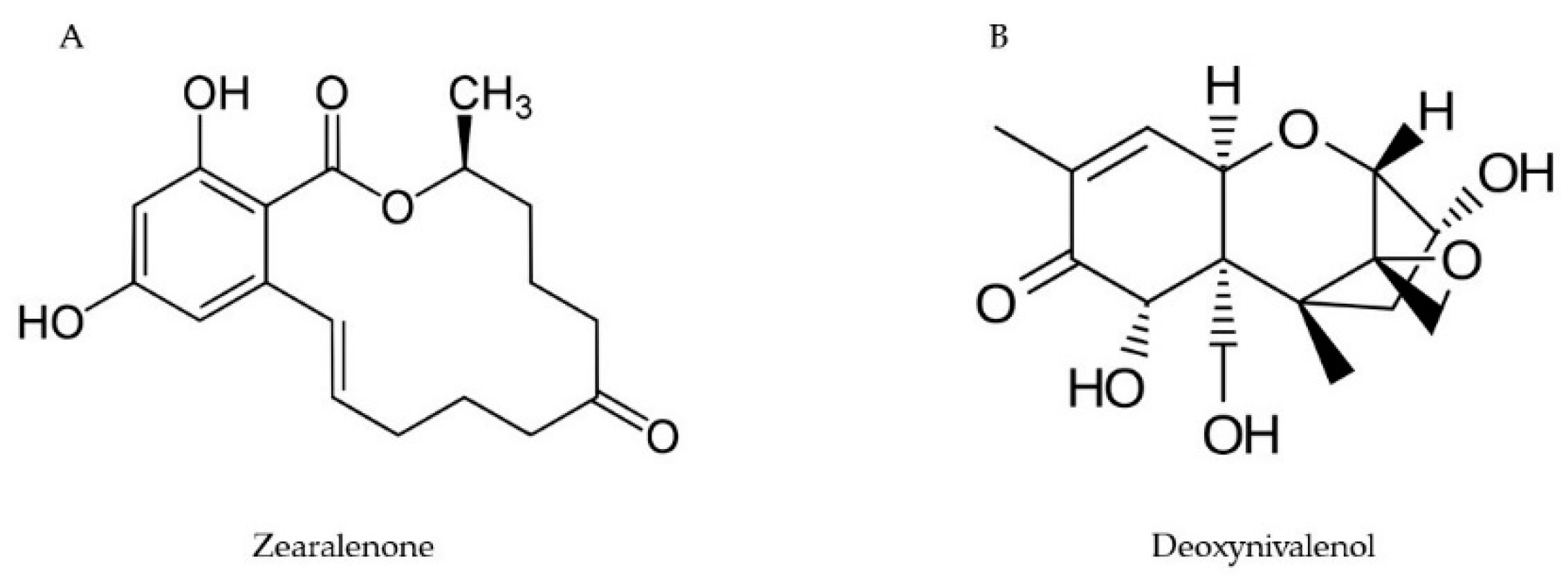
2.3. Zearalenone
2.4. Fusarins
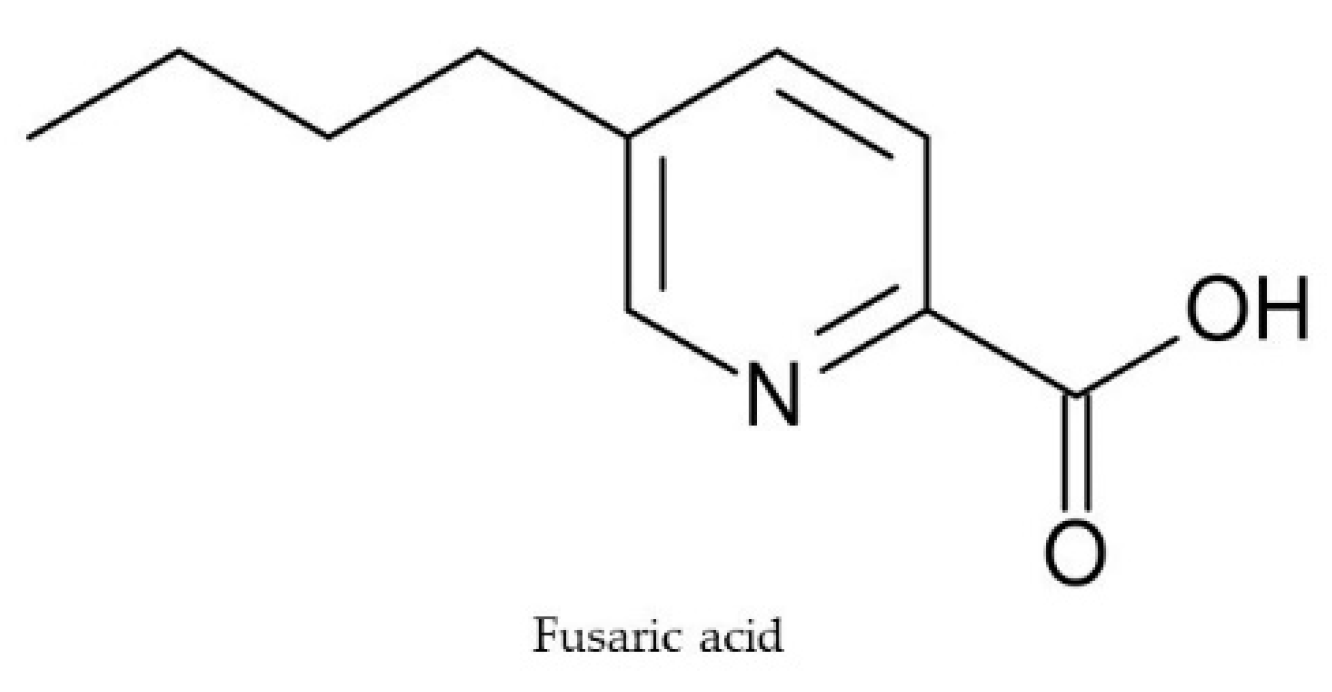
2.5. Fusaric Acid
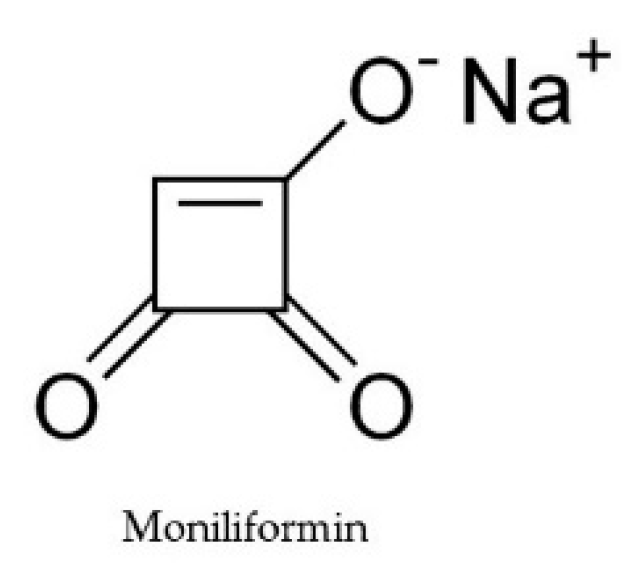
2.6. Moniliformin
2.7. Enniatins and Beauvericins
3. Effect of Climate Change/Environment Factors on Mycotoxin Biosynthesis—Overview
3.1. Temperature and Moisture Content
3.2. Effect of pH
3.3. Effect of Nitrogen Sources and Plant Extracts
4. Effect of Mycotoxins on Plant Secondary Metabolite Production During Infection
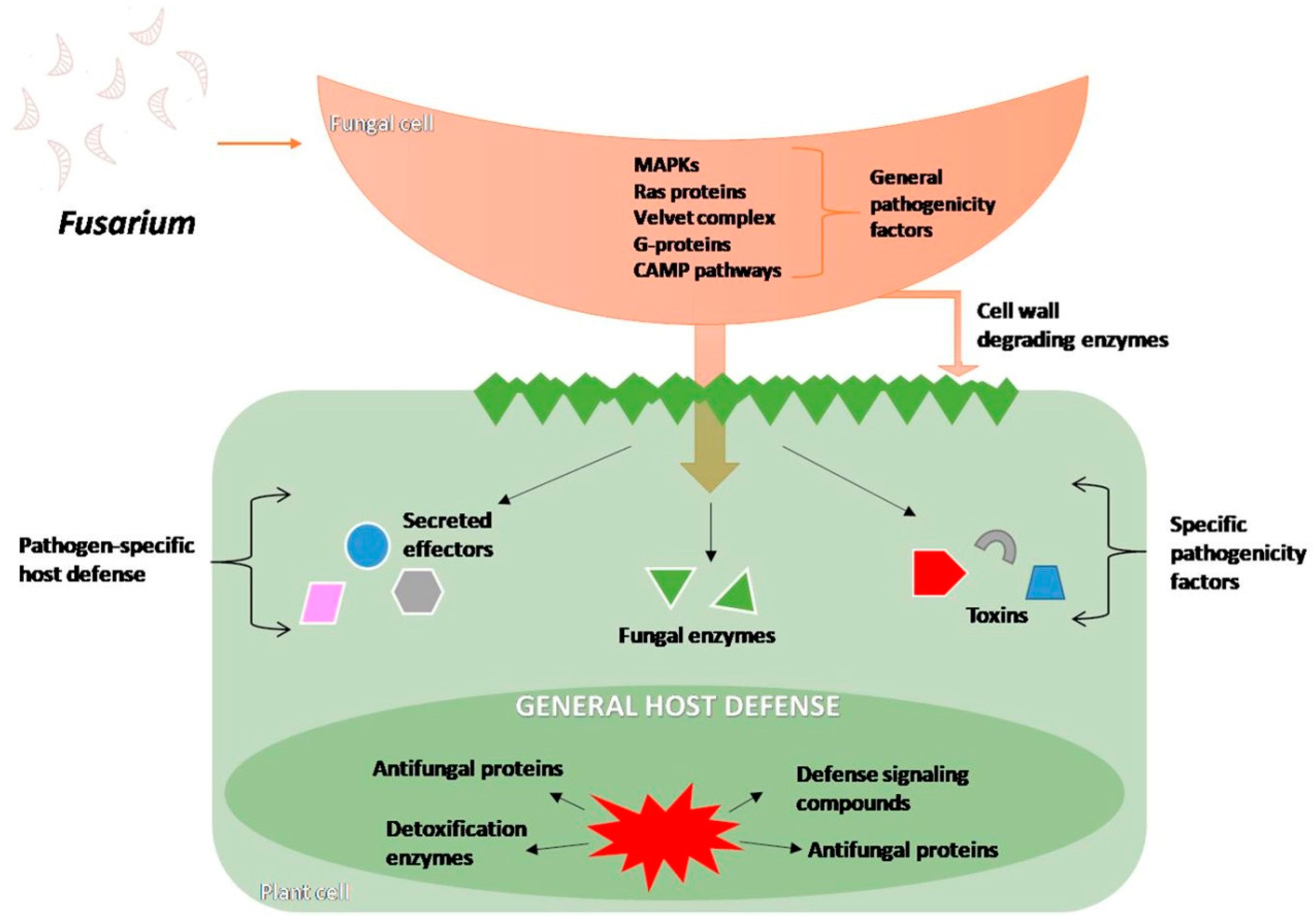
4.1. Infection Process and Changes Inside the Host Cells
4.2. The Signaling Crosstalk for Disease Resistance
4.3. Plant Strategies of Avoiding Fusarium Mycotoxins
4.4. Transgenic Plants Expressing Detoxification Genes
4.5. Secondary Metabolites Involved in Plant Resistance Against Fusarium
5. Conclusion and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Ploetz, R.C. Fusarium wilt of banana. Phytopathology 2015, 105, 1512–1521. [Google Scholar] [CrossRef] [PubMed]
- Nganje, W.E.; Bangsund, D.A.; Leistritz, F.L.; Wilson, W.W.; Tiapo, N.M. Regional economic impacts of Fusarium head blight in wheat and barley. Rev. Agric. Econ. 2004, 26, 332–347. [Google Scholar] [CrossRef]
- Jones, J.P.; Jones, J.B.; Miller, W. Fusarium wilt on tomato. Fla Dept Agric & Consumer Serv, Div of Plant Industry; Plant Pathology circular no. 237; 1982. Available online: https://www.fdacs.gov/content/download/11243/file/pp237.pdf (accessed on 14 November 2019).
- Ploetz, R.C. (Ed.) Diseases of Tropical Fruit Crops; CABI Publishing: Wallingford, UK, 2003. [Google Scholar]
- Chittem, K.; Mathew, F.M.; Gregoire, M.; Lamppa, R.S.; Chang, Y.W.; Markell, S.G.; Bradley, C.; Barasubiye, T.; Goswami, R.S. Identification and characterization of Fusarium spp. associated with root rots of field pea in North Dakota. Eur. J. Plant Pathol. 2015, 143, 641–649. [Google Scholar] [CrossRef]
- Tiwari, N.; Ahmed, S.; Kumar, S.; Sarker, A. Fusarium wilt: A killer disease of lentil. In Fusarium-Plant Diseases, Pathogen Diversity, Genetic Diversity, Resistance and Molecular Markers; Asku, T., Ed.; IntechOpen: Rijeka, Croatia, 2018. [Google Scholar]
- Karanyi, Z.; Holb, I.; Hornok, L.; Pocsi, I.; Miskei, M. FSRD: Fungal stress response database. Database 2013, 2013, bat037. [Google Scholar] [CrossRef]
- Reverberi, M.; Ricelli, A.; Zjalic, S.; Fabbri, A.A.; Fanelli, C. Natural functions of mycotoxins and control of their biosynthesis in fungi. Appl. Microbiol. Biotechnol. 2010, 87, 899–911. [Google Scholar] [CrossRef]
- Fanelli, C.; Ricelli, A.; Reverberi, M.; Fabbri, A.A.; Pandalai, S. Aflatoxins and ochratoxins in cereal grains: An open challenge. Recent Res. Dev. Crop Sci. 2004, 1, 295–317. [Google Scholar]
- Reverberi, M.; Fabbri, A.A.; Zjalic, S.; Ricelli, A.; Punelli, F.; Fanelli, C. Antioxidant enzymes stimulation in Aspergillus parasiticus by Lentinula edodes inhibits aflatoxin production. Appl. Microbiol. Biotechnol. 2005, 69, 207–215. [Google Scholar] [CrossRef]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant Microbe Interact. 1995, 8, 593–601. [Google Scholar] [CrossRef]
- Voigt, C.A.; Von Scheidt, B.; Gacser, A.; Kassner, H.; Lieberei, R.; Schäfer, W.; Salomon, S. Enhanced mycotoxin production of a lipase-deficient Fusarium graminearum mutant correlates to toxin-related gene expression. Eur. J. Plant Pathol. 2007, 117, 1–12. [Google Scholar] [CrossRef]
- Pusztahelyi, T.; Holb, I.J.; Pócsi, I. Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 2015, 6, 573. [Google Scholar] [CrossRef]
- Bakker, M.G.; Brown, D.W.; Kelly, A.C.; Kim, H.S.; Kurtzman, C.P.; Mccormick, S.P.; O’Donnell, K.L.; Proctor, R.H.; Vaughan, M.M.; Ward, T.J. Fusarium mycotoxins: A trans-disciplinary overview. Can. J. Plant Path. 2018, 40, 161–171. [Google Scholar] [CrossRef]
- Pasquali, M.; Beyer, M.; Logrieco, A.; Audenaert, K.; Balmas, V.; Basler, R.; González-Jaén, M.T. A European database of Fusarium graminearum and F. culmorum trichothecene genotypes. Front. Microbiol. 2016, 7, 406. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.H.; McCormick, S.P.; Alexander, N.J.; Desjardins, A.E. Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol. Microbiol. 2009, 74, 1128–1142. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Tokai, T.; Takahashi-Ando, N.; Ohsato, S.; Fujimura, M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: Pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 2007, 71, 2105–2123. [Google Scholar] [CrossRef] [PubMed]
- Alexander, N.J.; Proctor, R.H.; McCormick, S.P. Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 2009, 28, 198–215. [Google Scholar] [CrossRef]
- Cundliffe, E.; Cannon, M.; Davies, J. Mechanism of inhibition of eukaryotic protein synthesis by trichothecene fungal toxins. Proc. Natl. Acad. Sci. USA 1974, 71, 30–34. [Google Scholar] [CrossRef]
- Cundliffe, E.; Davies, J.E. Inhibition of initiation, elongation, and termination of eukaryotic protein synthesis by trichothecene fungal toxins. Antimicrob. Agents Chemother. 1977, 11, 491–499. [Google Scholar] [CrossRef]
- McLaughlin, C.S.; Vaughan, M.H.; Campbell, I.M.; Wei, C.M.; Stafford, M.E.; Hansen, B.S. Inhibition of protein synthesis by trichothecenes. Mycotoxins in human and animal health. In Mycotoxins in Human and Health; Rodericks, J.V., Hesseltine, C.W., Mehlman, M.A., Eds.; Pathotox: Park Forest, IL, USA, 1977; pp. 263–275. [Google Scholar]
- Ueno, Y.; Hsieh, D.P. The toxicology of mycotoxins. CRC Crit. Rev. Toxicol. 1985, 14, 99–132. [Google Scholar] [CrossRef]
- Eudes, F.; Comeau, A.; Rioux, S.; Collin, J. Phytotoxicité de huit mycotoxines associées à la fusariose de l’épi chez le blé. Can. J. Plant Path. 2000, 22, 286–292. [Google Scholar] [CrossRef]
- Desjardins, A.E.; McCormick, S.P.; Appell, M. Structure—Activity relationships of trichothecene toxins in an Arabidopsis thaliana leaf assay. J. Agric. Food Chem. 2007, 55, 6487–6492. [Google Scholar] [CrossRef]
- Wakuliński, W. Phytotoxicity of the secondary metabolites of fungi causing wheat head fusariosis (head blight). Acta Physiol. Plant. 1989, 11, 301–306. [Google Scholar]
- Terse, P.S.; Madhyastha, M.S.; Zurovac, O.; Stringfellow, D.; Marquardt, R.R.; Kemppainen, B.W. Comparison of in vitro and in vivo biological activity of mycotoxins. Toxicon 1993, 31, 913–919. [Google Scholar] [CrossRef]
- Masuda, D.; Ishida, M.; Yamaguchi, K.; Yamaguchi, I.; Kimura, M.; Nishiuchi, T. Phytotoxic effects of trichothecenes on the growth and morphology of Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.H.; Brown, D.W.; Plattner, R.D.; Desjardins, A.E. Co-expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genet. Biol. 2003, 38, 237–249. [Google Scholar] [CrossRef]
- Proctor, R.H.; Plattner, R.D.; Desjardins, A.E.; Busman, M.; Butchko, R.A. Fumonisin production in the maize pathogen Fusarium verticillioides: Genetic basis of naturally occurring chemical variation. J. Agric. Food Chem. 2006, 54, 2424–2430. [Google Scholar] [CrossRef]
- Proctor, R.H.; Busman, M.; Seo, J.A.; Lee, Y.W.; Plattner, R.D. A fumonisin biosynthetic gene cluster in Fusarium oxysporum strain O-1890 and the genetic basis for B versus C fumonisin production. Fungal Genet. Biol. 2008, 45, 1016–1026. [Google Scholar] [CrossRef]
- Stępień, Ł.; Koczyk, G.; Waśkiewicz, A. FUM cluster divergence in fumonisins-producing Fusarium species. Fungal Biol. 2011, 115, 112–123. [Google Scholar] [CrossRef]
- Smith, G.W. Fumonisins. In Veterinary Toxicology, 3rd ed.; Gupta, R., Ed.; Elsevier: New York, NY, USA, 2018; pp. 1003–1018. [Google Scholar]
- Pekkarinen, A.; Mannonen, L.; Jones, B.L.; Niku-Paavola, M.L. Production of proteases by Fusarium species grown on barley grains and in media containing cereal proteins. J. Cereal Sci. 2000, 31, 253–261. [Google Scholar] [CrossRef]
- Abbas, H.K.; Duke, S.O.; Tanaka, T. Phytotoxicity of Fumonisins and Related Compounds. J. Toxicol. Toxin Rev. 1993, 12, 225–251. [Google Scholar] [CrossRef]
- Merrill, A.H., Jr.; Sullards, M.C.; Wang, E.; Voss, K.A.; Riley, R.T. Sphingolipid metabolism: Roles in signal transduction and disruption by fumonisins. Environ. Health Perspect. 2001, 109, 283–289. [Google Scholar]
- Doehlert, D.C.; Knutson, C.A.; Vesonder, R.F. Phytotoxic effects of fumonisin B 1 on maize seedling growth. Mycopathologia 1994, 127, 117–121. [Google Scholar] [CrossRef]
- Koen, J.S.; Smith, H.C. An unusual case of genital involvement in swine, associated with eating moldy com. Vet. Med. 1946, 40, 131–133. [Google Scholar]
- Vianello, A.; Macri, F. Inhibition of plant cell membrane transport phenomena induced by zearalenone (F-2). Planta 1978, 143, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Gelderblom, W.C.; Marasas, W.F.; Steyn, P.S.; Thiel, P.G.; van der Merwe, K.J.; van Rooyen, P.H.; Vleggaar, R.; Wessels, P.L. Structure elucidation of fusarin C, a mutagen produced by Fusarium moniliforme. J. Chem. Soc. Chem. Commun. 1984, 2, 122–124. [Google Scholar] [CrossRef]
- Niehaus, E.M.; Díaz-Sánchez, V.; von Bargen, K.W.; Kleigrewe, K.; Humpf, H.U.; Limón, M.C.; Tudzynski, B. Fusarins and fusaric acid in fusaria. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Springer: New York, NY, USA, 2014; pp. 239–262. [Google Scholar]
- Wiebe, L.A.; Bjeldanes, L.F. Fusarin C, a mutagen from Fusarium moniliforme grown on corn. J. Food Sci. 1981, 46, 1424–1426. [Google Scholar] [CrossRef]
- Cheng, S.J.; Jiang, Y.Z.; Li, M.H.; Lo, H.Z. A mutagenic metabolite produced by Fusarium moniliforme isolated from Linxian County, China. Carcinogenesis 1985, 6, 903–905. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Proctor, R.H. Molecular biology of Fusarium mycotoxins. Int. J. Food Microbiol. 2007, 119, 47–50. [Google Scholar] [CrossRef]
- Li, M.X. Fusarin C induced esophageal and forestomach carcinoma in mice and rats. Chin. J. Oncol. 1992, 14, 27–29. [Google Scholar]
- Selim, M.E.; El-Gammal, N.A. Role of fusaric acid mycotoxin in pathogensis process of tomato wilt disease caused by Fusarium oxysporum. J. Bioprocess Biotech. 2015, 5, 1. [Google Scholar] [CrossRef]
- López-Díaz, C.; Rahjoo, V.; Sulyok, M.; Ghionna, V.; Martín-Vicente, A.; Capilla, J.; Di Pietro, A.; López-Berges, M.S. Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Plant Pathol. 2018, 19, 440–453. [Google Scholar] [CrossRef]
- Singh, V.K.; Upadhyay, R.S. Fusaric acid induced cell death and changes in oxidative metabolism of Solanum lycopersicum L. Bot. Stud. 2014, 55, 66. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Singh, H.B.; Upadhyay, R.S. Role of fusaric acid in the development of ‘Fusarium wilt’ symptoms in tomato: Physiological, biochemical and proteomic perspectives. Plant Physiol. Biochem. 2017, 118, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Bullerman, L.B. Mycotoxins Classifications. In Encyclopedia of Food Sciences and Nutrition, 2nd ed.; Caballero, B., Ed.; Academic Press: Cambridge, UK, 2003; pp. 4080–4089. [Google Scholar]
- Vesonder, R.F.; Labeda, D.P.; Peterson, R.E. Phytotoxic activity of selected water-soluble metabolites of Fusarium against Lemna minor L. (duckweed). Mycopathologia 1992, 118, 185–189. [Google Scholar] [CrossRef]
- Urbaniak, M.; Stępień, Ł.; Uhlig, S. Evidence for Naturally Produced Beauvericins Containing N-Methyl-Tyrosine in Hypocreales Fungi. Toxins 2019, 11, 182. [Google Scholar] [CrossRef]
- Mallebrera, B.; Prosperini, A.; Font, G.; Ruiz, M.J. In vitro mechanisms of Beauvericin toxicity: A review. Food Chem. Toxicol. 2018, 111, 537–545. [Google Scholar] [CrossRef]
- Paciolla, C.; Dipierro, N.; Mule, G.; Logrieco, A.; Dipierro, S. The mycotoxins beauvericin and T-2 induce cell death and alteration to the ascorbate metabolism in tomato protoplasts. Physiol. Mol. Plant Pathol. 2004, 65, 49–56. [Google Scholar] [CrossRef]
- Llorens, A.; Mateo, R.; Hinojo, M.J.; Valle-Algarra, F.M.; Jiménez, M. Influence of environmental factors on the biosynthesis of type B trichothecenes by isolates of Fusarium spp. from Spanish crops. Int. J. Food Microbiol. 2004, 94, 43–54. [Google Scholar] [CrossRef]
- Hope, R.; Aldred, D.; Magan, N. Comparison of environmental profiles for growth and deoxynivalenol production by Fusarium culmorum and F. graminearum on wheat grain. Lett. Appl. Microbiol. 2005, 40, 295–300. [Google Scholar] [CrossRef]
- Rybecky, A.I.; Chulze, S.N.; Chiotta, M.L. Effect of water activity and temperature on growth and trichothecene production by Fusarium meridionale. Int. J. Food Microbiol. 2018, 285, 69–73. [Google Scholar] [CrossRef]
- Schmidt-Heydt, M.; Parra, R.; Geisen, R.; Magan, N. Modelling the relationship between environmental factors, transcriptional genes and deoxynivalenol mycotoxin production by strains of two Fusarium species. J. R. Soc. Interface 2010, 8, 117–126. [Google Scholar] [CrossRef]
- Aliakbari, F.; Mirabolfathy, M.; Emami, M.; Mazhar, S.F.; Karami-Osboo, R. Natural occurrence of Fusarium species in maize kernels at Gholestan province in northern Iran. Asian J. Plant Sci. 2007, 8, 1276–1281. [Google Scholar]
- Cavaglieri, L.R.; Keller, K.M.; Pereyra, C.M.; Pereyra, M.G.; Alonso, V.A.; Rojo, F.G.; Rosa, C.A.R. Fungi and natural incidence of selected mycotoxins in barley rootlets. J. Stored Products Res. 2009, 45, 147–150. [Google Scholar] [CrossRef]
- Munkvold, G.P. Epidemiology of Fusarium diseases and their mycotoxins in maize ears. Eur. J. Plant Pathol. 2003, 109, 705–713. [Google Scholar] [CrossRef]
- Cendoya, E.; del Pilar Monge, M.; Chiacchiera, S.M.; Farnochi, M.C.; Ramirez, M.L. Influence of water activity and temperature on growth and fumonisin production by Fusarium proliferatum strains on irradiated wheat grains. Int. J. Food Microbiol. 2018, 266, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, F.; Schmidt-Heydt, M.; Haidukowski, M.; Geisen, R.; Logrieco, A.; Mulè, G. Influence of light on growth, fumonisin biosynthesis and FUM1 gene expression by Fusarium proliferatum. Int. J. Food Microbiol. 2012, 153, 148–153. [Google Scholar] [CrossRef]
- Seo, J.A.; Proctor, R.H.; Plattner, R.D. Characterization of four clustered and coregulated genes associated with fumonisin biosynthesis in Fusarium verticillioides. Fungal Genet. Biol. 2001, 34, 155–165. [Google Scholar] [CrossRef]
- Jurado, M.; Marín, P.; Magan, N.; González-Jaén, M.T. Relationship between solute and matric potential stress, temperature, growth, and FUM1 gene expression in two Fusarium verticillioides strains from Spain. Appl. Environ. Microbiol. 2008, 74, 2032–2036. [Google Scholar] [CrossRef]
- Tag, A.G.; Garifullina, G.F.; Peplow, A.W.; Ake, C.; Phillips, T.D.; Hohn, T.M.; Beremand, M.N. A novel regulatory gene, Tri10, controls trichothecene toxin production and gene expression. Appl. Environ. Microbiol. 2001, 67, 5294–5302. [Google Scholar] [CrossRef]
- Seong, K.Y.; Pasquali, M.; Zhou, X.; Song, J.; Hilburn, K.; McCormick, S.; Dong, Y.; Xu, J.R.; Kistler, H.C. Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 2009, 72, 354–367. [Google Scholar] [CrossRef]
- Marín, P.; Magan, N.; Vázquez, C.; González-Jaén, M.T. Differential effect of environmental conditions on the growth and regulation of the fumonisin biosynthetic gene FUM1 in the maize pathogens and fumonisin producers Fusarium verticillioides and Fusarium proliferatum. FEMS Microbiol. Ecol. 2010, 73, 303–311. [Google Scholar] [CrossRef]
- Zong, Y.; Li, B.; Tian, S. Effects of carbon, nitrogen and ambient pH on patulin production and related gene expression in Penicillium expansum. Int. J. Food Microbiol. 2015, 206, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Liu, F.; Wang, Q.; Selvaraj, J.N.; Xing, F.; Zhao, Y.; Liu, Y. Ochratoxin A producing fungi, biosynthetic pathway and regulatory mechanisms. Toxins 2016, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Sanzani, S.M.; Reverberi, M.; Geisen, R. Mycotoxins in harvested fruits and vegetables: Insights in producing fungi, biological role, conducive conditions, and tools to manage postharvest contamination. Postharvest Biol. Technol. 2016, 122, 95–105. [Google Scholar] [CrossRef]
- Sandoval-Contreras, T.; Villarruel-López, A.; Sierra-Beltrán, A.P.; Torres-Vitela, R.; Ascencio, F. Effect of pH and temperature in production of mycotoxins and antibiotics by phytopathogenic moulds for Persian lime (Citrus latifolia T.) in a complex lime pericarp-base medium. Emirates J. Food Agric. 2017, 29, 751–759. [Google Scholar] [CrossRef]
- Li, T.; Gong, L.; Wang, Y.; Chen, F.; Gupta, V.K.; Jian, Q.; Duan, X.; Jiang, Y. Proteomics analysis of Fusarium proliferatum under various initial pH during fumonisin production. J. Proteom. 2017, 164, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, D.M.; Osborne, S.; Kazan, K.; Manners, J.M. Low pH regulates the production of deoxynivalenol by Fusarium graminearum. Microbiology 2009, 155, 3149–3156. [Google Scholar] [CrossRef]
- Espeso, E.A.; Tilburn, J.; Sánchez-Pulido, L.; Brown, C.V.; Valencia, A.; Arst, H.N.; Peñalva, M.A. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J. Mol. Biol. 1997, 274, 466–480. [Google Scholar] [CrossRef]
- Negrete-Urtasun, S.; Reiter, W.; Diez, E.; Denison, S.H.; Tilburn, J.; Espeso, E.A.; Peñalva, M.A.; Arst, H.N., Jr. Ambient pH signal transduction in Aspergillus: Completion of gene characterization. Mol. Microbiol. 1999, 33, 994–1003. [Google Scholar] [CrossRef]
- Caracuel, Z.; Roncero, M.I.G.; Espeso, E.A.; González-Verdejo, C.I.; García-Maceira, F.I.; Di Pietro, A. The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum. Mol. Microbiol. 2003, 48, 765–779. [Google Scholar] [CrossRef]
- Merhej, J.; Richard-Forget, F.; Barreau, C. The pH regulatory factor Pac1 regulates Tri gene expression and trichothecene production in Fusarium graminearum. Fungal Genet. Biol. 2011, 48, 275–284. [Google Scholar] [CrossRef]
- Flaherty, J.E.; Pirttilä, A.M.; Bluhm, B.H.; Woloshuk, C.P. PAC1, a pH-regulatory gene from Fusarium verticillioides. Appl. Environ. Microbiol. 2003, 69, 5222–5227. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.E.; Sullivan, T.M.; Chirtel, S. Factors affecting the growth of Fusarium proliferatum and the production of fumonisin B1: Oxygen and pH. J. Ind. Microbiol. Biotechnol. 1997, 19, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Anthony, S.; Abeywickrama, K.; Dayananda, R.; Wijeratnam, S.; Arambewela, L. Fungal pathogens associated with banana fruit in Sri Lanka, and their treatment with essential oils. Mycopathologia 2004, 157, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Zhan, R.L.; Yang, S.J.; Ho, H.H.; Liu, F.; Zhao, Y.L.; Chang, J.M.; He, Y.B. Mango malformation disease in south China caused by Fusarium proliferatum. J. Phytopathol. 2010, 158, 721–725. [Google Scholar] [CrossRef]
- Wu, H.S.; Yin, X.M.; Liu, D.Y.; Ling, N.; Bao, W.; Ying, R.R.; Zhu, Y.Y.; Guo, S.W.; Shen, Q.R. Effect of fungal fusaric acid on the root and leaf physiology of watermelon (Citrullus lanatus) seedlings. Plant Soil 2008, 308, 255. [Google Scholar] [CrossRef]
- Li, J.; Jiang, G.; Yang, B.; Dong, X.; Feng, L.; Lin, S.; Chen, F.; Ashraf, M.; Jiang, Y. A luminescent bacterium assay of fusaric acid produced by Fusarium proliferatum from banana. Anal. Bioanal. Chem. 2012, 402, 1347–1354. [Google Scholar] [CrossRef]
- Skrinjar, M.; Dimić, G. Ochratoxigenicity of Aspergillus ochraceus group and Penicillium verrucosum var. cyclopium strains on various media. Acta Microbiol. Hung. 1992, 39, 257–261. [Google Scholar]
- Shim, W.B.; Woloshuk, C.P. Nitrogen repression of fumonisin B1 biosynthesis in Gibberella fujikuroi. FEMS Microbiol. Lett. 1999, 177, 109–116. [Google Scholar] [CrossRef]
- Stępień, Ł.; Waśkiewicz, A.; Wilman, K. Host extract modulates metabolism and fumonisin biosynthesis by the plant-pathogenic fungus Fusarium proliferatum. Int. J. Food Microbiol. 2015, 193, 74–81. [Google Scholar] [CrossRef]
- Marı́n, S.; Velluti, A.; Ramos, A.J.; Sanchis, V. Effect of essential oils on zearalenone and deoxynivalenol production by Fusarium graminearum in non-sterilized maize grain. Food Microbiol. 2004, 21, 313–318. [Google Scholar] [CrossRef]
- Marín, S.; Velluti, A.; Muñoz, A.; Ramos, A.J.; Sanchis, V. Control of fumonisin B1 accumulation in naturally contaminated maize inoculated with Fusarium verticillioides and Fusarium proliferatum, by cinnamon, clove, lemongrass, oregano and palmarosa essential oils. Eur. Food Res. Technol. 2003, 217, 332–337. [Google Scholar] [CrossRef]
- Handa, A.K.; Fatima, T.; Mattoo, A.K. Polyamines: Bio-molecules with diverse functions in plant and human health and disease. Front. Chem. 2018, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Watson, M.B.; Malmberg, R.L. Regulation of Arabidopsis thaliana (L.) Heynh arginine decarboxylase by potassium deficiency stress. Plant Physiol. 1996, 111, 1077–1083. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, A.; Nakajima, T.; Hirayae, K. Effects of carbon sources and amines on induction of trichothecene production by Fusarium asiaticum in liquid culture. FEMS Microbiol. Lett. 2014, 352, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Bluhm, B.H.; Woloshuk, C.P. Amylopectin induces fumonisin B1 production by Fusarium verticillioides during colonization of maize kernels. Mol. Plant-Microbe Interact. 2005, 18, 1333–1339. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Chevalier, P.M.; Rupp, R.A. Storage and remobilization of soluble carbohydrates after heading in different plant parts of a winter wheat cultivar. Plant Prod. Sci. 2001, 4, 160–165. [Google Scholar] [CrossRef][Green Version]
- Ponts, N.; Pinson-Gadais, L.; Verdal-Bonnin, M.N.; Barreau, C.; Richard-Forget, F. Accumulation of deoxynivalenol and its 15-acetylated form is significantly modulated by oxidative stress in liquid cultures of Fusarium graminearum. FEMS Microbiol. Lett. 2006, 258, 102–107. [Google Scholar] [CrossRef]
- Ponts, N.; Pinson-Gadais, L.; Barreau, C.; Richard-Forget, F.; Ouellet, T. Exogenous H2O2 and catalase treatments interfere with Tri genes expression in liquid cultures of Fusarium graminearum. FEBS Lett. 2007, 581, 443–447. [Google Scholar] [CrossRef]
- Ma, L.J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef]
- Parry, D.W.; Jenkinson, P.; McLeod, L. Fusarium ear blight (scab) in small grain cereals—A review. Plant Pathol. 1995, 44, 207–238. [Google Scholar] [CrossRef]
- Bottalico, A.; Perrone, G. Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. In Mycotoxins in Plant Disease; Springer: Dordrecht, The Netherlands, 2002; pp. 611–624. [Google Scholar]
- Rana, A.; Sahgal, M.; Johri, B.N. Fusarium oxysporum: Genomics, Diversity and Plant–Host Interaction. In Developments in Fungal Biology and Applied Mycology; Springer: Singapore, 2017; pp. 159–199. [Google Scholar]
- Jansen, C.; Von Wettstein, D.; Schäfer, W.; Kogel, K.H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Buchenauer, H. Ultrastructural and cytochemical studies on cellulose, xylan and pectin degradation in wheat spikes infected by Fusarium culmorum. J. Phytopathol. 2000, 148, 263–275. [Google Scholar] [CrossRef]
- Kang, Z.; Zingen-Sell, I.; Buchenauer, H. Infection of wheat spikes by Fusarium avenaceum and alterations of cell wall components in the infected tissue. Eur. J. Plant Pathol. 2005, 111, 19–28. [Google Scholar] [CrossRef]
- Kang, Z.; Buchenauer, H. Studies on the infection process of Fusarium culmorum in wheat spikes: Degradation of host cell wall components and localization of trichothecene toxins in infected tissue. Eur. J. Plant Pathol. 2002, 108, 653–660. [Google Scholar] [CrossRef]
- Spassieva, S.D.; Markham, J.E.; Hille, J. The plant disease resistance gene Asc-1 prevents disruption of sphingolipid metabolism during AAL-toxin-induced programmed cell death. Plant J. 2002, 32, 561–572. [Google Scholar] [CrossRef]
- Williams, L.D.; Glenn, A.E.; Zimeri, A.M.; Bacon, C.W.; Smith, M.A.; Riley, R.T. Fumonisin disruption of ceramide biosynthesis in maize roots and the effects on plant development and Fusarium verticillioides-induced seedling disease. J. Agric. Food Chem. 2007, 55, 2937–2946. [Google Scholar] [CrossRef]
- Jiménez-Fernández, D.; Landa, B.B.; Kang, S.; Jiménez-Díaz, R.M.; Navas-Cortés, J.A. Quantitative and microscopic assessment of compatible and incompatible interactions between chickpea cultivars and Fusarium oxysporum f. sp. ciceris races. PLoS ONE 2013, 8, e61360. [Google Scholar] [CrossRef]
- Stephens, A.E.; Gardiner, D.M.; White, R.G.; Munn, A.L.; Manners, J.M. Phases of infection and gene expression of Fusarium graminearum during crown rot disease of wheat. Mol. Plant Microbe Interact. 2008, 21, 1571–1581. [Google Scholar] [CrossRef]
- Boddu, J.; Cho, S.; Muehlbauer, G.J. Transcriptome analysis of trichothecene-induced gene expression in barley. Mol. Plant Microbe Interact. 2007, 20, 1364–1375. [Google Scholar] [CrossRef]
- Packa, D. Cytogenetic effects of Fusarium mycotoxins on root tip cells of rye (Secale cereale L.), wheat (Triticum aestivum L.) and field bean (Vicia faba L. var. minor). J. Appl. Genet. 1997, 3, 259–272. [Google Scholar]
- Packa, D.; Śliwińska, E. Trichothecene fusarial toxins perturb the cell cycle in meristem-atic cells of Secale cereale L., Triticum aestivum L. and Vicia faba L. Caryologia 2005, 58, 86–93. [Google Scholar]
- Bushnell, W.R.; Perkins-Veazie, P.; Russo, V.M.; Collins, J.; Seeland, T.M. Effects of deoxynivalenol on content of chloroplast pigments in barley leaf tissues. Phytopathology 2010, 100, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Cossette, F.; Miller, J.D. Phytotoxic effect of deoxynivalenol and gibberella ear rot resistance of com. Nat. Toxins 1995, 3, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Uetsuka, K. Mechanisms of mycotoxin-induced neurotoxicity through oxidative stress-associated pathways. Int. J. Mol. Sci. 2011, 12, 5213–5237. [Google Scholar]
- Damann, K.E., Jr.; Gardner, J.M.; Scheffer, R.P. An Assay for Helminthosporium victoriae Toxin Based on Induced Leakage of Electrolytes from Oat. Phytopathology 1974, 64, 652–654. [Google Scholar] [CrossRef]
- Bronson, C.R.; Scheffer, R.P. Heat-and aging-induced tolerance of sorghum and oat tissues to host-selective toxins. Phytopathology 1977, 67, 1232–1238. [Google Scholar] [CrossRef]
- Mansoori, B.; Smith, C.J. Verticillium-toxins: Their Role in Pathogenesis. J. Agric. Sci. Technol. 2005, 7, 103–114. [Google Scholar]
- Maier, F.J.; Miedaner, T.; Hadeler, B.; Felk, A.; Salomon, S.; Lemmens, M.; Kassner, H.; Schafer, W. Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 2006, 7, 449–461. [Google Scholar] [CrossRef]
- Nishiuchi, T.; Masuda, D.; Nakashita, H.; Ichimura, K.; Shinozaki, K.; Yoshida, S.; Kimura, M.; Yamaguchi, I.; Yamaguchi, K. Fusarium phytotoxin trichothecenes have an elicitor-like activity in Arabidopsis thaliana, but the activity differed significantly among their molecular species. Mol. Plant Microbe Interact. 2006, 19, 512–520. [Google Scholar] [CrossRef]
- Desmond, O.J.; Manners, J.M.; Stephens, A.E.; Maclean, D.J.; Schenk, P.M.; Gardiner, D.M.; Munn, A.L.; Kazan, K. The Fusarium mycotoxin deoxynivalenol elicits hydrogen peroxide production, programmed cell death and defence responses in wheat. Mol. Plant Pathol. 2008, 9, 435–445. [Google Scholar] [CrossRef]
- Orzaez, D.; de Jong, A.J.; Woltering, E.J. A tomato homologue of the human protein PIRIN is induced during programmed cell death. Plant Mol. Biol. 2001, 46, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Mesterházy, Á.; Bartók, T.; Mirocha, C.G.; Komoroczy, R. Nature of wheat resistance to Fusarium head blight and the role of deoxynivalenol for breeding. Plant Breed. 1999, 118, 97–110. [Google Scholar] [CrossRef]
- Clinesmith, M.A.; Fritz, A.K.; Lemes da Silva, C.; Bockus, W.W.; Poland, J.A.; Dowell, F.E.; Peiris, K.H. QTL Mapping of Fusarium Head Blight Resistance in Winter Wheat Cultivars ‘Art’and ‘Everest’. Crop Sci. 2019, 59, 911–924. [Google Scholar] [CrossRef]
- Wang, Y.Z.; Miller, J.D. Effects of Fusarium graminearum metabolites on wheat tissue in relation to Fusarium head blight resistance. J. Phytopathol. 1988, 122, 118–125. [Google Scholar] [CrossRef]
- Maschietto, V.; Colombi, C.; Pirona, R.; Pea, G.; Strozzi, F.; Marocco, A.; Lanubile, A. QTL mapping and candidate genes for resistance to Fusarium ear rot and fumonisin contamination in maize. BMC Plant Biol. 2017, 17, 20. [Google Scholar] [CrossRef]
- Kluger, B.; Bueschl, C.; Lemmens, M.; Michlmayr, H.; Malachova, A.; Koutnik, A.; Schuhmacher, R. Biotransformation of the mycotoxin deoxynivalenol in Fusarium resistant and susceptible near isogenic wheat lines. PLoS ONE 2015, 10, e0119656. [Google Scholar] [CrossRef]
- Ma, J.; Li, H.B.; Zhang, C.Y.; Yang, X.M.; Liu, Y.X.; Yan, G.J.; Liu, C.J. Identification and validation of a major QTL conferring crown rot resistance in hexaploid wheat. Theor. Appl. Genet. 2010, 120, 1119–1128. [Google Scholar] [CrossRef]
- Poole, G.J.; Smiley, R.W.; Paulitz, T.C.; Walker, C.A.; Carter, A.H.; See, D.R.; Garland-Campbell, K. Identification of quantitative trait loci (QTL) for resistance to Fusarium crown rot (Fusarium pseudograminearum) in multiple assay environments in the Pacific Northwestern US. Theor. Appl. Genet. 2012, 125, 91–107. [Google Scholar] [CrossRef]
- Xiao, J.; Jin, X.; Jia, X.; Wang, H.; Cao, A.; Zhao, W.; Pei, H.; Xue, Z.; He, L.; Chen, Q.; et al. Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genom. 2013, 14, 197. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. Jasmonate signaling: Toward an integrated view. Plant Physiol. 2008, 146, 1459–1468. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, L.F.; Manners, J.M.; Kazan, K. Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. 2009, 58, 927–939. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, Q.; Cui, S.; Ren, J.; Qian, W.; Yang, Y.; He, S.; Chu, J.; Sun, X.; Yan, C.; et al. Hijacking of the jasmonate pathway by the mycotoxin fumonisin B1 (FB1) to initiate programmed cell death in Arabidopsis is modulated by RGLG3 and RGLG4. J. Exp. Bot. 2015, 66, 2709–2721. [Google Scholar] [CrossRef] [PubMed]
- Poppenberger, B.; Berthiller, F.; Lucyshyn, D.; Sieberer, T.; Schuhmacher, R.; Krska, R.; Kuchler, K.; Glössl, J.; Luschnig, C.; Adam, G. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 47905–47914. [Google Scholar] [CrossRef] [PubMed]
- Walter, S.; Brennan, J.M.; Arunachalam, C.; Ansari, K.I.; Hu, X.; Khan, M.R.; Trognitz, F.; Trognitz, B.; Leonard, G.; Egan, D.; et al. Components of the gene network associated with genotype-dependent response of wheat to the Fusarium mycotoxin deoxynivalenol. Funct. Integr. Genom. 2008, 8, 421–427. [Google Scholar] [CrossRef]
- Ross, J.; Li, Y.; Lim, E.K.; Bowles, D.J. Higher plant glycosyltransferases. Genome Boil. Rev. 2001, 2, reviews3004.1. [Google Scholar]
- Schweiger, W.; Boddu, J.; Shin, S.; Poppenberger, B.; Berthiller, F.; Lemmens, M.; Muehlbauer, G.J.; Adam, G. Validation of a candidate deoxynivalenol-inactivating UDP-glucosyltransferase from barley by heterologous expression in yeast. Mol. Plant Microbe Interact. 2010, 23, 977–986. [Google Scholar] [CrossRef]
- Schweiger, W.; Pasquet, J.C.; Nussbaumer, T.; Paris, M.P.K.; Wiesenberger, G.; Macadré, C.; Ametz, C.; Berthiller, F.; Lemmens, M.; Saindrenan, P.; et al. Functional characterization of two clusters of Brachypodium distachyon UDP-glycosyltransferases encoding putative deoxynivalenol detoxification genes. Mol. Plant Microbe Interact. 2013, 26, 781–792. [Google Scholar] [CrossRef]
- Michlmayr, H.; Malachová, A.; Varga, E.; Kleinová, J.; Lemmens, M.; Newmister, S.; Rayment, I.; Berthiller, F.; Adam, G. Biochemical Characterization of a Recombinant UDP-glucosyltransferase from Rice and Enzymatic Production of Deoxynivalenol-3-O-β-D-glucoside. Toxins 2015, 7, 2685–2700. [Google Scholar] [CrossRef]
- Huang, J.; Pang, C.; Fan, S.; Song, M.; Yu, J.; Wei, H.; Yu, S. Genome-wide analysis of the family 1 glycosyltransferases in cotton. Mol. Genet. Genom. 2015, 290, 1805–1818. [Google Scholar] [CrossRef]
- Warth, B.; Fruhmann, P.; Wiesenberger, G.; Kluger, B.; Sarkanj, B.; Lemmens, M.; Schuhmacher, R. Deoxynivalenol-sulfates: Identification and quantification of novel conjugated (masked) mycotoxins in wheat. Anal. Bioanal. Chem. 2015, 407, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, M.; Ducos, E.; Martinoia, E.; Boutry, M. The ATP-binding cassette transporters: Structure, function, and gene family comparison between rice and Arabidopsis. Plant Physiol. 2003, 131, 1169–1177. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, N.A. Move it on out with MATEs. Plant Cell 2001, 13, 1477–1480. [Google Scholar] [CrossRef]
- Brown, M.H.; Paulsen, I.T.; Skurray, R.A. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 1999, 31, 394–395. [Google Scholar] [CrossRef] [PubMed]
- Kreuz, K.; Tommasini, R.; Martinoia, E. Old enzymes for a new job (herbicide detoxification in plants). Plant Physiol. 1996, 111, 349–353. [Google Scholar] [CrossRef]
- Frangne, N.; Eggmann, T.; Koblischke, C.; Weissenböck, G.; Martinoia, E.; Klein, M. Flavone glucoside uptake into barley mesophyll and Arabidopsis cell culture vacuoles. Energization occurs by H+-antiport and ATP-binding cassette-type mechanisms. Plant Physiol. 2002, 128, 726–733. [Google Scholar] [CrossRef]
- Van Schie, C.C.; Takken, F.L. Susceptibility genes 101: How to be a good host. Annu. Rev. Phytopathol. 2014, 52, 551–581. [Google Scholar] [CrossRef]
- Beeton, S.; Bull, A.T. Biotransformation and detoxification of T-2 toxin by soil and freshwater bacteria. Appl. Environ. Microbiol. 1989, 55, 190–197. [Google Scholar]
- Berthiller, F.; Werner, U.; Sulyok, M.; Krska, R.; Hauser, M.T.; Schuhmacher, R. Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) determination of phase II metabolites of the mycotoxin zearalenone in the model plant Arabidopsis thaliana. Food Addit. Contam. 2006, 23, 1194–1200. [Google Scholar] [CrossRef]
- Muhitch, M.J.; McCormick, S.P.; Alexander, N.J.; Hohn, T.M. Transgenic expression of the TRI101 or PDR5 gene increases resistance of tobacco to the phytotoxic effects of the trichothecene 4, 15-diacetoxyscirpenol. Plant Sci. 2000, 157, 201–207. [Google Scholar] [CrossRef]
- Kimura, M.; Kaneko, I.; Komiyama, M.; Takatsuki, A.; Koshino, H.; Yoneyama, K.; Yamaguchi, I. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins cloning and characterization of Tri101. J. Biol. Chem. 1998, 273, 1654–1661. [Google Scholar] [CrossRef] [PubMed]
- Ohsato, S.; Ochiai-Fukuda, T.; Nishiuchi, T.; Takahashi-Ando, N.; Koizumi, S.; Hamamoto, H.; Kimura, M. Transgenic rice plants expressing trichothecene 3-O-acetyltransferase show resistance to the Fusarium phytotoxin deoxynivalenol. Plant Cell Rep. 2007, 26, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shin, S.; Heinen, S.; Dill-Macky, R.; Berthiller, F.; Nersesian, N.; Muehlbauer, G.J. Transgenic wheat expressing a barley UDP-glucosyltransferase detoxifies deoxynivalenol and provides high levels of resistance to Fusarium graminearum. Mol. Plant Microbe Interact. 2015, 28, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.J.; Gleddie, S.C. A modified Rpl3 gene from rice confers tolerance of the Fusarium graminearum mycotoxin deoxynivalenol to transgenic tobacco. Physiol. Mol. Plant Pathol. 2001, 58, 173–181. [Google Scholar] [CrossRef]
- Perochon, A.; Váry, Z.; Malla, K.B.; Halford, N.G.; Paul, M.J.; Doohan, F.M. The wheat SnRK1α family and its contribution to Fusarium toxin tolerance. Plant Sci. 2019, 288, 110217. [Google Scholar] [CrossRef]
- Fuchs, E.; Binder, E.M.; Heidler, D.; Krska, R. Structural characterization of metabolites after the microbial degradation of type A trichothecenes by the bacterial strain BBSH 797. Food Addit. Contam. 2002, 19, 379–386. [Google Scholar] [CrossRef]
- Atanasova-Penichon, V.; Barreau, C.; Richard-Forget, F. Antioxidant secondary metabolites in cereals: Potential involvement in resistance to Fusarium and mycotoxin accumulation. Front. Microbiol. 2016, 7, 566. [Google Scholar] [CrossRef]
- Balmer, D.; Flors, V.; Glauser, G.; Mauch-Mani, B. Metabolomics of cereals under biotic stress: Current knowledge and techniques. Front. Plant Sci. 2013, 4, 82. [Google Scholar] [CrossRef]
- Boutigny, A.L. Etude de l’effet de composés du grain de blé dur sur la régulation de la voie de biosynthèse des trichothécènes B: Purification de composés inhibiteurs, analyse des mécanismes impliqués. Ph.D. Thesis, University of Bordeaux–Talence Campus, Bordeaux, France, 2007. [Google Scholar]
- Picot, A.; Atanasova-Pénichon, V.; Pons, S.; Marchegay, G.; Barreau, C.; Pinson-Gadais, L.; Roucolle, J.; Daveau, F.; Caron, D.; Richard-Forget, F. Maize kernel antioxidants and their potential involvement in Fusarium ear rot resistance. J. Agric. Food Chem. 2013, 61, 3389–3395. [Google Scholar] [CrossRef]
- Etzerodt, T.; Gislum, R.; Laursen, B.B.; Heinrichson, K.; Gregersen, P.L.; Jørgensen, L.N.; Fomsgaard, I.S. Correlation of deoxynivalenol accumulation in Fusarium-infected winter and spring wheat cultivars with secondary metabolites at different growth stages. J. Agric. Food Chem. 2016, 64, 4545–4555. [Google Scholar] [CrossRef]
- Kumaraswamy, G.K.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S. Differential metabolic response of barley genotypes, varying in resistance, to trichothecene-producing and-nonproducing (tri5−) isolates of Fusarium graminearum. Plant Pathol. 2012, 61, 509–521. [Google Scholar] [CrossRef]
- Ferruz, E.; Atanasova-Pénichon, V.; Bonnin-Verdal, M.N.; Marchegay, G.; Pinson-Gadais, L.; Ducos, C.; Lorán, S.; Ariño, A.; Barreau, C.; Richard-Forget, F. Effects of Phenolic Acids on the Growth and Production of T-2 and HT-2 Toxins by Fusarium langsethiae and F. sporotrichioides. Molecules 2016, 21, 449. [Google Scholar] [CrossRef] [PubMed]
- Sicker, D.; Frey, M.; Schulz, M.; Gierl, A. Role of natural benzoxazinones in the survival strategy of plants. In International Review of Cytology; Academic Press: Cambridge, MA, USA, 2000; Volume 198, pp. 319–346. [Google Scholar]
- Do, H.; Kim, I.S.; Jeon, B.W.; Lee, C.W.; Park, A.K.; Wi, A.R.; Shin, S.C.; Park, H.; Kim, Y.S.; Yoon, H.S.; et al. Structural understanding of the recycling of oxidized ascorbate by dehydroascorbate reductase (OsDHAR) from Oryza sativa L. japonica. Sci. Rep. 2016, 6, 19498. [Google Scholar] [CrossRef] [PubMed]
- Stępień, Ł.; Lalak-Kańczugowska, J.; Witaszak, N.; Urbaniak, M. Fusarium Secondary Metabolism Biosynthetic Pathways: So Close but So Far Away. In Co-Evolution of Secondary Metabolites; Series in Phytochemistry; Mérillon, J.-M., Ramawat, K.G., Eds.; Springer: Cham, Switzerland, 2019; pp. 1–37. [Google Scholar]
- Allwood, J.W.; Ellis, D.I.; Goodacre, R. Metabolomic technologies and their application to the study of plants and plant-host interactions. Physiol. Plant. 2008, 132, 117–135. [Google Scholar] [CrossRef] [PubMed]
- Mehrabi, R.; Bahkali, A.H.; Abd-Elsalam, K.A.; Moslem, M.; Ben M’barek, S.; Gohari, A.M.; Jashni, M.K.; Stergiopoulos, I.; Kema, G.H.; de Wit, P.J. Horizontal gene and chromosome transfer in plant pathogenic fungi affecting host range. FEMS Microbiol. Rev. 2011, 35, 542–554. [Google Scholar] [CrossRef]
- Fitzpatrick, D.A. Horizontal gene transfer in fungi. FEMS Microbiol. Lett. 2012, 329, 1–8. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perincherry, L.; Lalak-Kańczugowska, J.; Stępień, Ł. Fusarium-Produced Mycotoxins in Plant-Pathogen Interactions. Toxins 2019, 11, 664. https://doi.org/10.3390/toxins11110664
Perincherry L, Lalak-Kańczugowska J, Stępień Ł. Fusarium-Produced Mycotoxins in Plant-Pathogen Interactions. Toxins. 2019; 11(11):664. https://doi.org/10.3390/toxins11110664
Chicago/Turabian StylePerincherry, Lakshmipriya, Justyna Lalak-Kańczugowska, and Łukasz Stępień. 2019. "Fusarium-Produced Mycotoxins in Plant-Pathogen Interactions" Toxins 11, no. 11: 664. https://doi.org/10.3390/toxins11110664
APA StylePerincherry, L., Lalak-Kańczugowska, J., & Stępień, Ł. (2019). Fusarium-Produced Mycotoxins in Plant-Pathogen Interactions. Toxins, 11(11), 664. https://doi.org/10.3390/toxins11110664





