Venom Diversity and Evolution in the Most Divergent Cone Snail Genus Profundiconus
Abstract
:1. Introduction
2. Results
2.1. Analysis of Profundiconus Transcriptomes
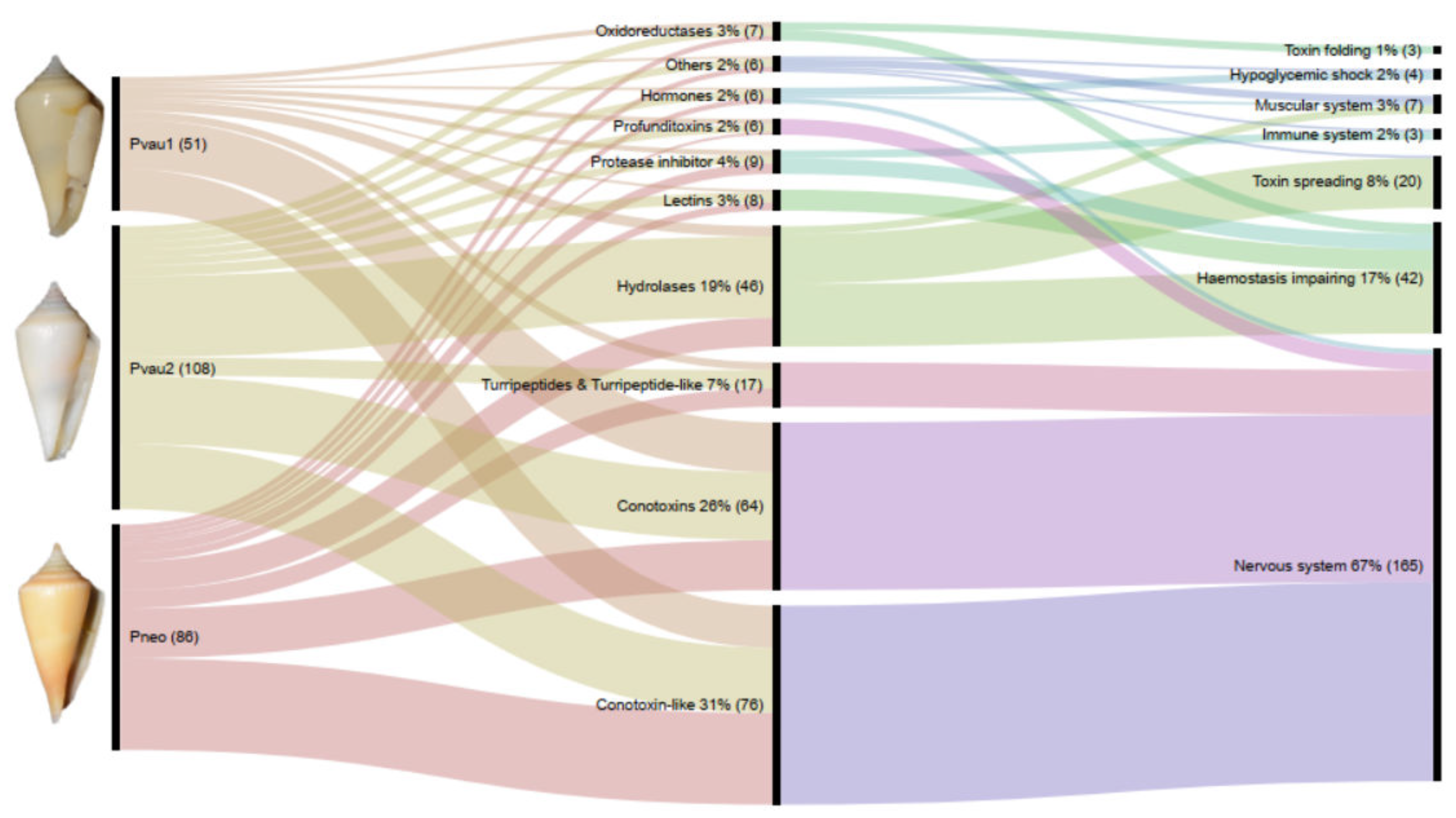
2.2. Molecular Types and Putative Targets of Profundiconus Venom Components
2.3. Analysis of Putative Conotoxins and Profunditoxins Transcripts
2.4. Analysis of Putative Turripeptide Trancripts
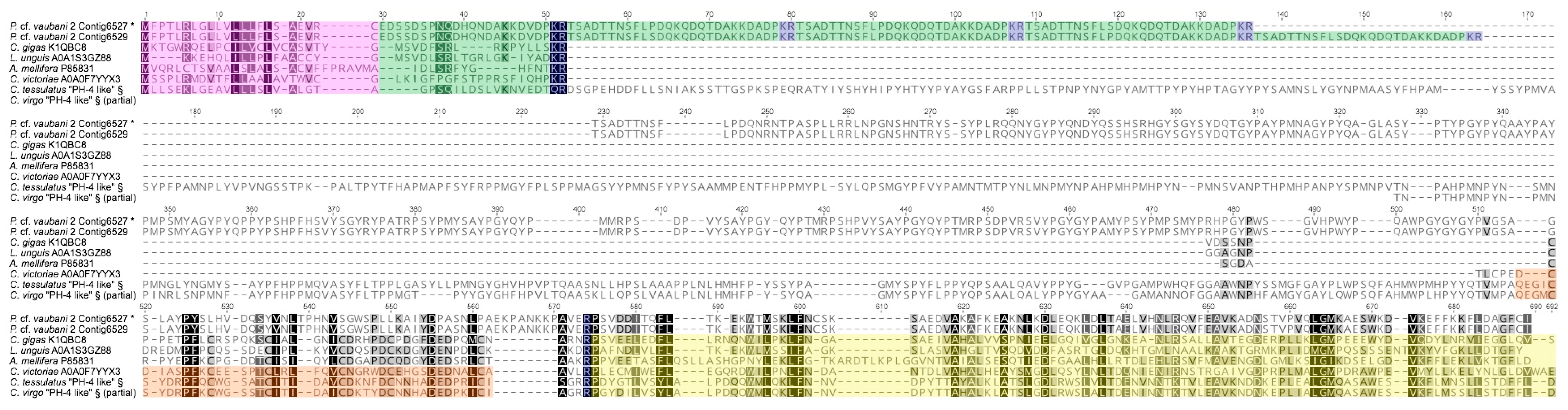
2.5. Analysis of Prohormone-4 Transcripts
2.6. Analysis of Insulin Transcripts
2.7. Analysis of Lectin Transcripts
2.8. Analysis of Chymotrypsin Transcripts
3. Discussion
3.1. P. cf. Vaubani Displayed a High Diversity of Nonneuropeptide Venom Components
3.2. Limited Conotoxin Diversity May Indicate a Narrow Worm or Molluscan Diet
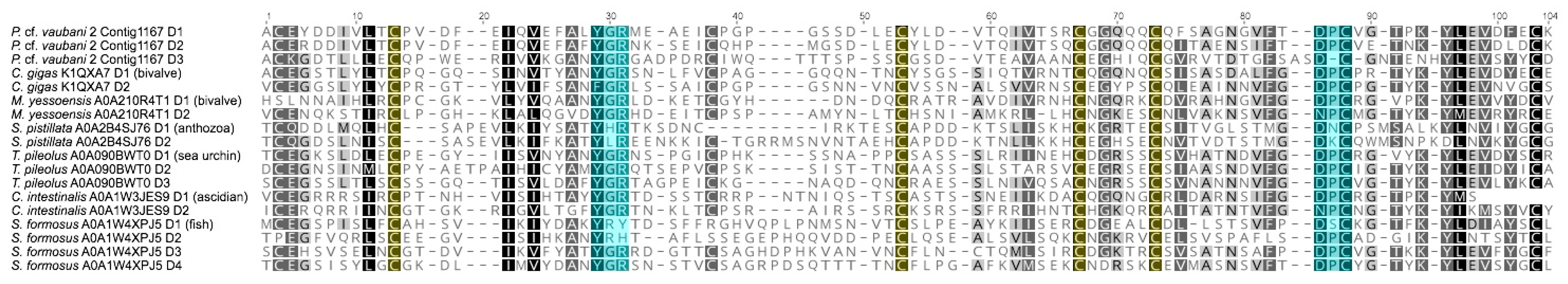
3.3. Characterization of the First Putative Profunditoxin Gene Superfamilies Indicate They Are Divergent
3.4. Turripeptides Retained in Profundiconus Venom
4. Conclusions
5. Materials and Methods
5.1. Sample Collection and Identification
5.2. RNA Extraction, Sequencing, and De Novo Assembly
5.3. Transcriptome Annotation and Differential Expression Analysis
5.4. Venom Component Identification and Diversity
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Paracelsus. Opera Omnia Medico-Chemico-Chirurgica, Tribus Voluminibus Comprehensa. Editio Novissima et Emendatissima ad Germanica & Latina Exemplaria Accuratissime Collata; Sumptibus Joan. Antonii & Samuelis De Tournes: Geneva, Switzerland, 1658. [Google Scholar]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.A.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef]
- Puillandre, N.; Kantor, Y.I.; Sysoev, A.; Couloux, A.; Meyer, C.; Rawlings, T.; Todd, J.A.; Bouchet, P. The dragon tamed? A molecular phylogeny of the Conoidea (Gastropoda). J. Molluscan Stud. 2011, 77, 259–272. [Google Scholar] [CrossRef]
- Abdelkrim, J.; Aznar-Cormano, L.; Buge, B.; Buge, B.; Kantor, Y.; Zaharias, P.; Puillandre, N. Delimiting species of marine gastropods (Turridae, Conoidea) using RAD sequencing in an integrative taxonomy framework. Mol. Ecol. 2018, 27, 4591–4611. [Google Scholar] [CrossRef]
- Phuong, M.A.; Alfaro, M.E.; Mahardika, G.N.; Marwoto, R.M.; Prabowo, R.E.; von Rintelen, T.; Vogt, P.W.H.; Hendricks, J.R.; Puillandre, N. Lack of signal for the impact of conotoxin gene diversity on speciation rates in cone snails. Syst. Biol. 2019, syz016. [Google Scholar] [CrossRef]
- Olivera, B.M.; Seger, J.; Horvath, M.P.; Fedosov, A.E. Prey-Capture Strategies of Fish-Hunting Cone Snails: Behavior, Neurobiology and Evolution. Brain Behav. Evol. 2015, 86, 58–74. [Google Scholar] [CrossRef] [Green Version]
- Puillandre, N.; Koua, D.; Favreau, P.; Olivera, B.M.; Stocklin, R. Molecular Phylogeny, Classification and Evolution of Conopeptides. J. Mol. Evol. 2012, 74, 297–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaas, Q.; Westermann, J.; Craik, D.J. Conopeptide characterization and classifications: An analysis using ConoServer. Toxicon 2010, 55, 1491–1509. [Google Scholar] [CrossRef] [PubMed]
- Rivera-ortiz, J.A.; Cano, H.; Marí, F. Intraspecies variability and conopeptide profiling of the injected venom of Conus ermineus. Peptides 2011, 32, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.M.; Dutertre, S.; Lewis, R.J.; Marí, F. Intraspecific variations in Conus purpurascens injected venom using LC/MALDI-TOF-MS and LC-ESI-TripleTOF-MS. Anal. Bioanal. Chem. 2015, 407, 6105–6116. [Google Scholar] [CrossRef] [PubMed]
- Prator, C.A.; Murayama, K.M.; Schulz, J.R. Venom Variation during Prey Capture by the Cone Snail, Conus textile. PLoS ONE 2014, 9, e98991. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Jin, A.-H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J.; et al. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Bandyopadhyay, P.K.; Olivera, B.M.; Yandell, M. Characterization of the Conus bullatus genome and its venom-duct transcriptome. BMC Genomics 2011, 12, 60–75. [Google Scholar] [CrossRef] [PubMed]
- Terrat, Y.; Biass, D.; Dutertre, S.; Favreau, P.; Remm, M.; Stöcklin, R.; Piquemal, D.; Ducancel, F. High-resolution picture of a venom gland transcriptome: Case study with the marine snail Conus consors. Toxicon 2012, 59, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Violette, A.; Leonardi, A.; Piquemal, D.; Terrat, Y.; Biass, D. Recruitment of Glycosyl Hydrolase Proteins in a Cone Snail Venomous Arsenal: Further Insights into Biomolecular Features of Conus Venoms. Mar. Drugs 2012, 10, 258–280. [Google Scholar] [CrossRef]
- Safavi-Hemami, H.; Hu, H.; Gorasia, D.G.; Bandyopadhyay, P.K.; Veith, P.D.; Young, N.D.; Reynolds, E.C.; Yandell, M.; Olivera, B.M.; Purcell, A.W. Combined Proteomic and Transcriptomic Interrogation of the Venom Gland of Conus geographus Uncovers Novel Components and Functional Compartmentalization. Mol. Cell. Proteomics 2014, 13, 938–953. [Google Scholar] [CrossRef]
- Figueroa-Montiel, A.; Ramos, M.A.; Mares, R.E.; Duenas, S.; Pimienta, G.; Ortiz, E.; Possani, L.D.; LIcea-Navarro, A.F. In Silico Identification of Protein Disulfide Isomerase Gene Families in the De Novo Assembled Transcriptomes of Four Different Species of the Genus Conus. PLoS ONE 2016, 11, e0148390. [Google Scholar] [CrossRef]
- Safavi-Hemami, H.; Li, Q.; Jackson, R.L.; Song, A.S.; Boomsma, W. Rapid expansion of the protein disulfide isomerase gene family facilitates the folding of venom peptides. Proc. Natl. Acad. Sci. USA 2016, 113, 3227–3232. [Google Scholar] [CrossRef] [Green Version]
- Safavi-Hemami, H.; Lu, A.; Li, Q.; Fedosov, A.E.; Biggs, J.; Corneli, P.S.; Seger, J.; Yandell, M.; Olivera, B.M. Venom Insulins of Cone Snails Diversify Rapidly and Track Prey Taxa. Mol. Biol. Evol. 2016, 33, 2924–2934. [Google Scholar] [CrossRef]
- Uribe, J.E.; Puillandre, N.; Zardoya, R. Beyond Conus: Phylogenetic relationships of Conidae based on complete mitochondrial genomes. Mol. Phylogenet. Evol. 2017, 107, 142–151. [Google Scholar] [CrossRef]
- Abdelkrim, J.; Aznar-cormano, L.; Fedosov, A.E.; Kantor, Y.I.; Lozouet, P.; Phuong, M.A.; Zaharias, P.; Puillandre, N. Exon-Capture-Based Phylogeny and Diversification of the Venomous Gastropods (Neogastropoda, Conoidea). Mol. Biol. Evol. 2018, 35, 2355–2374. [Google Scholar] [CrossRef]
- MolluscaBase Profundiconus Kuroda, 1956. Accessed Through: World Register of Marine Species. 2019. Available online: http://www.marinespecies.org/aphia.php?p=taxdetails&id=428962 on (accessed on 4 September 2019).
- Tenorio, M.J.; Castelin, M. Genus Profundiconus Kuroda, 1956 (Gastropoda, Conoidea): Morphological and molecular studies, with the description of five new species from the Solomon Islands and New Caledonia. Eur. J. Taxon. 2016, 173, 1–45. [Google Scholar] [CrossRef]
- Marshall, B.A. New records of Conidae (Mollusca: Gastropoda) from the New Zealand region. New Zeal. J. Zool. 2012, 8, 493–501. [Google Scholar] [CrossRef]
- Mauri, M.; Elli, T.; Caviglia, G.; Uboldi, G.; Azzi, M. RAWGraphs: A Visualisation Platform to Create Open Outputs open output. In Proceedings of the 12th Biannual Conference on Italian SIGCHI Chapter ACM, New York, NY, USA, 18–20 September 2017; pp. 28:1–28:5. [Google Scholar]
- Elliger, C.A.; Richmond, T.A.; Lebaric, Z.N.; Pierce, N.T.; Sweedler, J.V.; Gilly, W.F. Diversity of conotoxin types from Conus californicus reflects a diversity of prey types and a novel evolutionary history. Toxicon 2011, 57, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.D.; Li, Q.; Bandyopadhyay, P.K.; Gajewiak, J.; Yandell, M.; Papenfuss, A.T.; Purcell, A.W.; Norton, R.S.; Safavi-hemami, H. General and Comparative Endocrinology Hormone-like peptides in the venoms of marine cone snails. Gen. Comp. Endocrinol. 2017, 244, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Quinton, L.; Gilles, N.; De Pauw, E. TxXIIIA, an atypical homodimeric conotoxin found in the Conus textile venom. J. Proteomics 2009, 72, 219–226. [Google Scholar] [CrossRef]
- Degueldre, M.; Verdenaud, M.; Legarda, G.; Minambres, R.; Zuniga, S.; Leblanc, M.; Gilles, N.; Ducancel, F.; De Pauw, E.; Quinton, L. Diversity in sequences, post-translational modifications and expected pharmacological activities of toxins from four Conus species revealed by the combination of cutting-edge proteomics, transcriptomics and bioinformatics. Toxicon 2017, 130, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, D.T.T.; Saloma, C.P. A bioinformatics survey for conotoxin-like sequences in three turrid snail venom duct transcriptomes. Toxicon 2014, 92, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Heralde, F.M.; Imperial, J.; Bandyopadhyay, P.K.; Olivera, B.M.; Concepcion, G.P.; Santos, A.D. A rapidly diverging superfamily of peptide toxins in venomous Gemmula species. Toxicon 2008, 51, 890–897. [Google Scholar] [CrossRef]
- Olivera, B.M. Conus Venom Peptides: Reflections from the Biology of Clades and Species. Annu. Rev. Ecol. Syst. 2002, 33, 25–47. [Google Scholar] [CrossRef]
- Watkins, M.; Hillyard, D.R.; Olivera, B.M. Genes Expressed in a Turrid Venom Duct: Divergence and Similarity to Conotoxins. J. Mol. Evol. 2006, 62, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Olivera, B.M.; Watkins, M.; Bandyopadhyay, P.; Imperial, J.S.; de la Cotera, E.P.H.; Aguilar, M.B.; Lopez Vera, E.; Concepcion, G.P.; Lluisma, A. Adaptive radiation of venomous marine snail lineages and the accelerated evolution of venom peptide genes. Ann. N. Y. Acad. Sci. 2012, 1267, 61–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brockmann, A.; Annangudi, S.P.; Richmond, T.A.; Ament, S.A.; Xie, F.; Southey, B.R.; Rodriguez-zas, S.R.; Robinson, G.E.; Sweedler, J. V Quantitative peptidomics reveal brain peptide signatures of behavior. Proc. Natl. Acad. Sci. USA 2009, 106, 2383–2388. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Cummins, S.F.; Fitzgibbon, Q.; Battaglene, S.; Elizur, A. Analysis of the Central Nervous System Transcriptome of the Eastern Rock Lobster Sagmariasus verreauxi Reveals Its Putative Neuropeptidome. PLoS ONE 2014, 9, e97323. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.; van Kesteren, R.; Li, K.; van Minnen, J.; Spijker, S.; van Heerikhuizen, H.; Geraerts, W. Toward Understanding the Role of Insulin in the Brain: Lessons from Insulin-Related Signaling Systems in the Invertebrate Brain. Prog. Neurobiol. 1998, 54, 35–54. [Google Scholar] [CrossRef]
- Safavi-Hemami, H.; Gajewiak, J.; Karanth, S.; Robinson, S.D.; Ueberheide, B.; Douglass, A.D.; Schlegel, A.; Imperial, J.S.; Watkins, M.; Bandyopadhyay, P.K.; et al. Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc. Natl. Acad. Sci. USA 2015, 112, 1–6. [Google Scholar] [CrossRef]
- Ogawa, T.; Watanabe, M.; Naganuma, T.; Muramoto, K. Diversified Carbohydrate-Binding Lectins from Marine Resources. J. Amino Acids 2011, 2011, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, H.; Tanigawa, T.; Tomita, K.; Tomihara, Y.; Araki, Y.; Tachikawa, E. Recent Studies on the Pathological Effects of Purified Sea Urchin Toxins. J. Toxicol. 2003, 22, 633–649. [Google Scholar] [CrossRef]
- Tateno, H.; Saneyoshi, A.; Ogawa, T.; Muramoto, K.; Kamiya, H.; Saneyoshi, M. Isolation and Characterization of Rhamnose-binding Lectins from Eggs of Steelhead Trout (Oncorhynchus mykiss) Homologous to Low Density Lipoprotein Receptor Superfamily. J. Biol. Chem. 1998, 273, 19190–19197. [Google Scholar] [CrossRef]
- Hosono, M.; Ishikawa, K.; Mineki, R.; Murayama, K.; Numata, C.; Ogawa, Y.; Takayanagi, Y.; Nitta, K. Tandem repeat structure of rhamnose-binding lectin from catfish (Silurus asotus) eggs. Biochim. Biophys. Acta 1999, 1472, 668–675. [Google Scholar] [CrossRef]
- Gasparini, F.; Franchi, N.; Spolaore, B.; Loriano, B. Novel rhamnose-binding lectins from the colonial ascidian Botryllus schlosseri. Dev. Comp. Immunol. 2008, 32, 1177–1191. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, T.; Ogawa, T.; Hirabayashi, J.; Kasai, K.; Muramoto, K. Isolation, characterization and molecular evolution of a novel pearl shell lectin from a marine bivalve, Pteria penguin. Mol. Divers. 2006, 10, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Edo, K.; Sakai, H.; Nakagawa, H.; Hashimoto, T.; Shinohara, M.; Ohura, K. Immunomodulatory activity of a pedicellarial venom lectin from the toxopneustid sea urchin, Toxopneustes pileolus. Toxin Rev. 2012, 31, 54–60. [Google Scholar] [CrossRef]
- Takei, M.; Nakagawa, H. A sea urchin lectin, SUL-1, from the Toxopneustid sea urchin induces DC maturation from human monocyte and drives Th1 polarization in vitro. Toxicol. Appl. Pharmacol. 2006, 213, 27–36. [Google Scholar] [CrossRef]
- Hatakeyama, T.; Ichise, A.; Unno, H.; Goda, S.; Oda, T.; Tateno, H.; Hirabayashi, J.; Sakai, H.; Nakagawa, H. Carbohydrate recognition by the rhamnose-binding lectin SUL-I with a novel three-domain structure isolated from the venom of globiferous pedicellariae of the flower sea urchin Toxopneustes pileolus. Protein Sci. 2017, 26, 1574–1583. [Google Scholar] [CrossRef]
- Ekici, O.D.; Paetzel, M.; Dalbey, R.E. Unconventional serine proteases: Variations on the catalytic Ser/His/Asp triad configuration. Protein Sci. 2008, 17, 2023–2037. [Google Scholar] [CrossRef] [Green Version]
- Wlodawer, A.; Li, M.; Dauter, Z.; Gustchina, A.; Uchida, K.; Oyama, H.; Dunn, B.M.; Oda, K. Carboxyl proteinase from Pseudomonas defines a novel family of subtilisin-like enzymes. Nat. Struct. Biol. 2001, 8, 442–446. [Google Scholar] [CrossRef]
- Siigur, E.; Tõnismägi, K.; Trummal, K.; Samel, M.; Vija, H.; Aaspõllu, A.; Rönnholm, G.; Subbi, J.; Kalkkinen, N.; Siigur, J. A new tyrosine-specific chymotrypsin-like and angiotensin-degrading serine proteinase from Vipera lebetina snake venom. Biochimie 2011, 93, 321–330. [Google Scholar] [CrossRef]
- Li, Q.; Barghi, N.; Lu, A.; Fedosov, A.E.; Bandyopadhyay, P.K.; Lluisma, A.O.; Concepcion, G.P.; Yandell, M.; Olivera, B.M.; Safavi-hemami, H. Divergence of the Venom Exogene Repertoire in Two Sister Species of Turriconus. Genome Biol. Evol. 2017, 9, 2211–2225. [Google Scholar] [CrossRef]
- Phuong, M.A.; Mahardika, G.N.; Alfaro, M.E. Dietary breadth is positively correlated with venom complexity in cone snails. BMC Genom. 2016, 17, 401. [Google Scholar] [CrossRef]
- Abalde, S.; Tenorio, M.J.; Afonso, C.M.L.; Zardoya, R. Conotoxin Diversity in Chelyconus ermineus (Born, 1778) and the Convergent Origin of Piscivory in the Atlantic and Indo-Pacific Cones. Genome Biol. Evol. 2018, 10, 2643–2662. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Bandyopadhyay, P.K.; Olivera, B.M.; Yandell, M. Elucidation of the molecular envenomation strategy of the cone snail Conus geographus through transcriptome sequencing of its venom duct. BMC Genom. 2012, 13, 284. [Google Scholar] [CrossRef] [PubMed]
- Barghi, N.; Concepcion, G.P.; Olivera, B.M.; Lluisma, A.O. Comparison of the Venom Peptides and Their Expression in Closely Related Conus Species: Insights into Adaptive Post-speciation Evolution of Conus Exogenomes. Genome Biol. Evol. 2015, 7, 1797–1814. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.; Kaas, Q.; Lavergne, V.; Kubala, P.; Lewis, R.J.; Alewood, P.F. Transcriptomic Messiness in the Venom Duct of Conus miles Contributes to Conotoxin Diversity. Mol. Cell. Proteomics 2013, 12, 3824–3833. [Google Scholar] [CrossRef] [PubMed]
- Lavergne, V.; Harliwong, I.; Jones, A.; Miller, D.; Taft, R.J.; Alewood, P.F. Optimized deep-targeted proteotranscriptomic profiling reveals unexplored Conus toxin diversity and novel cysteine frameworks. Proc. Natl. Acad. Sci. USA 2015, 112, E3782–E3791. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Duda, T.F., Jr. Age-related association of venom gene expression and diet of predatory gastropods. BMC Evol. Biol. 2016, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kaas, Q.; Westermann, J.; Halai, R.; Wang, C.K.L.; Craik, D.J. ConoServer, a database for conopeptide sequences and structures. Bioinform. Appl. Note 2008, 24, 445–446. [Google Scholar] [CrossRef]
- Biggs, J.S.; Watkins, M.; Puillandre, N.; Ownby, J.; Lopez-vera, E.; Christensen, S.; Juarez, K.; Bernaldez, J.; Licea-navarro, A.; Showers, P.; et al. Molecular Phylogenetics and Evolution Evolution of Conus peptide toxins: Analysis of Conus californicus Reeve, 1844. Mol. Phylogenet. Evol. 2010, 56, 1–12. [Google Scholar] [CrossRef]
- Barghi, N.; Concepcion, G.P.; Olivera, B.M.; Lluisma, A.O. High Conopeptide Diversity in Conus tribblei Revealed Through Analysis of Venom Duct Transcriptome Using Two High-Throughput Sequencing Platforms. Mar. Biotechnol. 2015, 17, 81–98. [Google Scholar] [CrossRef]
- Modica, M.V.; Lombardo, F.; Franchini, P.; Oliverio, M. The venomous cocktail of the vampire snail Colubraria reticulata (Mollusca, Gastropoda). BMC Genom. 2015, 16, 441. [Google Scholar] [CrossRef]
- Von Reumont, B.M.; Campbell, L.I.; Richter, S.; Hering, L.; Sykes, D.; Hetmank, J.; Jenner, R.A.; Bleidorn, C. A Polychaete’s Powerful Punch: Venom Gland Transcriptomics of Glycera Reveals a Complex Cocktail of Toxin Homologs. Genome Biol. Evol. 2014, 6, 2406–2423. [Google Scholar] [CrossRef]
- Verdes, A.; Simpson, D.; Holford, M. Are Fireworms Venomous? Evidence for the Convergent Evolution of Toxin Homologs in Three Species of Fireworms (Annelida, Amphinomidae). Genome Biol. Evol. 2017, 10, 249–268. [Google Scholar] [CrossRef]
- Gerdol, M.; Puillandre, N.; De Moro, G.; Guarnaccia, C.; Lucafo, M.; Benincasa, M.; Zlatev, V.; Manfrin, C.; Torboli, V.; Giulianini, P.G.; et al. Identification and Characterization of a Novel Family of Cysteine-Rich Peptides (MgCRP-I) from Mytilus galloprovincialis. Genome Biol. Evol. 2015, 7, 2203–2219. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinforma. Appl. Note 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Gorson, J.; Ramrattan, G.; Verdes, A.; Wright, M.; Kantor, Y.I.; Srinivasan, R.; Musunuri, R.; Packer, D.; Albano, G.; Qiu, W.; et al. Molecular Diversity and Gene Evolution of the Venom Arsenal of Terebridae Predatory Marine Snails. Genome Biol. Evol. 2015, 7, 1761–1778. [Google Scholar] [CrossRef] [Green Version]
- Altschup, S.F.; Gish, W.; Pennsylvania, T.; Park, U.; Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kaas, Q.; Yu, R.; Jin, A.; Dutertre, S.; Craik, D.J. ConoServer: Updated content, knowledge, and discovery tools in the conopeptide database. Nucl. Acids Res. 2012, 40, 325–330. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucl. Acids Res. 2019, 47, 506–515. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.L.; Eddy, S.R.; Durbin, R. Pfam: A Comprehensive Database of Protein Domain Families Based on Seed Alignments. Proteins Struct. Funct. Genet. 1997, 28, 405–420. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucl. Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Tarazona, S.; Garcia-Alcade, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef] [Green Version]
- Armenteros, J.A.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucl. Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation. Nucl. Acids Res. 2018, 46, 493–496. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucl. Acids Res. 2018, 46, 296–303. [Google Scholar] [CrossRef]
- Lopez-Vera, E.; Heimer de la Cotera, E.P.; Maillo, M.; Riesgo-Escovar, J.R.; Olivera, B.M.; Aguilar, M.B. A novel structural class of toxins: The methionine-rich peptides from the venoms of turrid marine snails (Mollusca, Conoidea). Toxicon 2004, 43, 365–374. [Google Scholar] [CrossRef]
- Puillandre, N.; Holford, M. The Terebridae and teretoxins: Combining phylogeny and anatomy for concerted discovery of bioactive compounds. BMC Chem. Biol. 2010, 10, 7. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability Article Fast Track. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef]
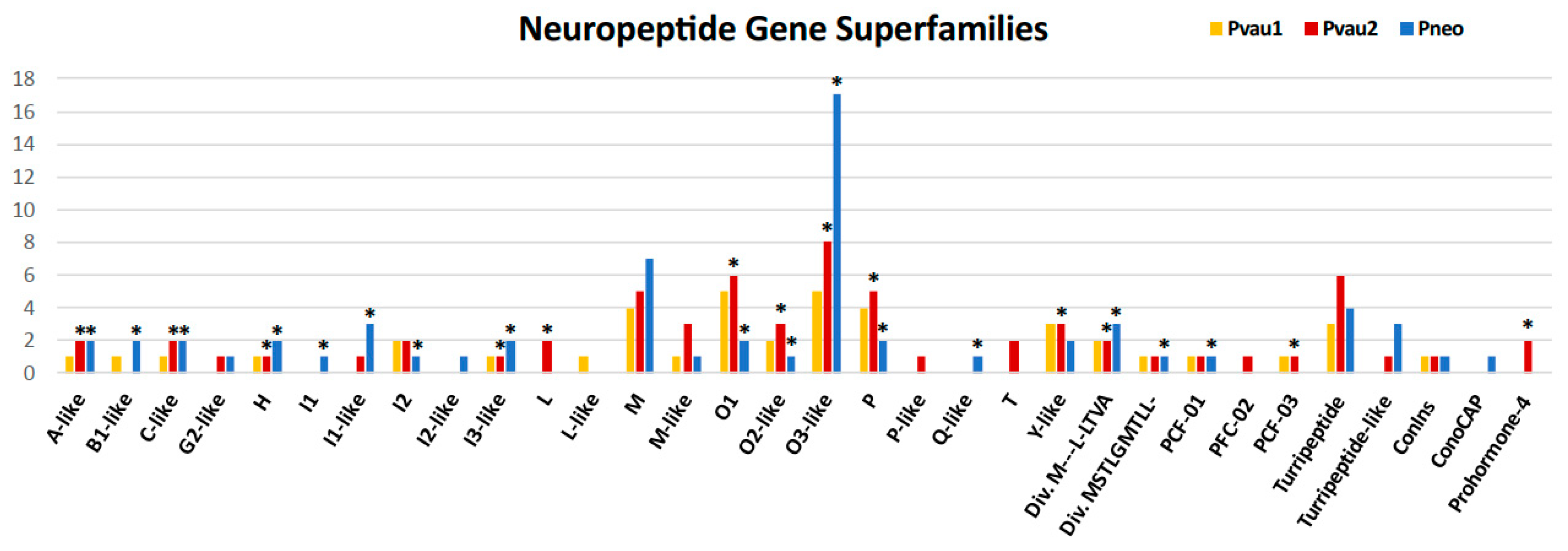
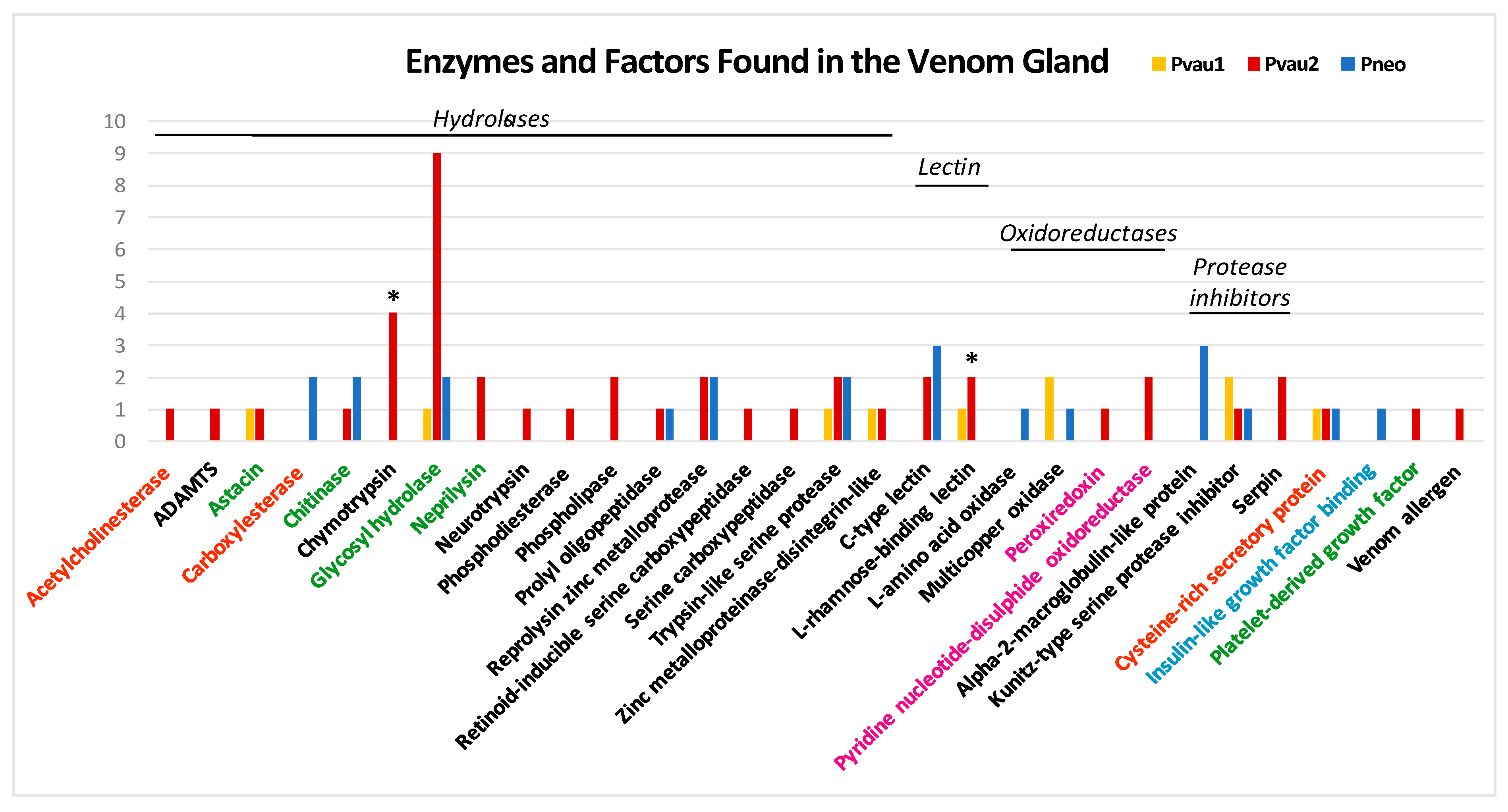
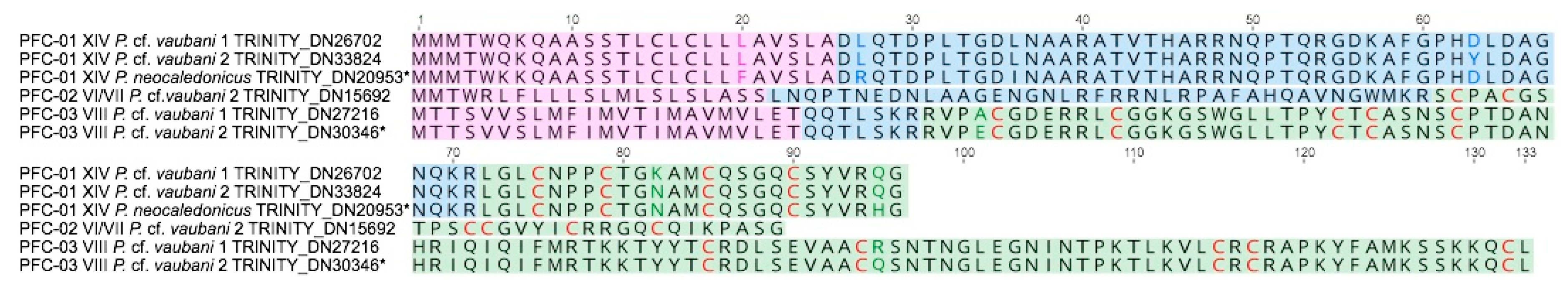
| Gene Superfamily | Pvau1 | Pvau2 | Pneo | Cysteine Framework |
|---|---|---|---|---|
| A-like | 1 | 2 (1) | 2 (1) | VI/VII, IX 1 |
| B1-like | 1 | 0 | 2 (1) | 0 |
| C-like | 1 | 2 (1) | 2 (1) | 0 |
| G2-like | 0 | 1 | 1 | VI/VII 1 |
| H | 1 | 1 (1) | 2 (1) | VIII 1, C-C 1 |
| I1 | 0 | 0 | 1 (1) | XI |
| I1-like | 0 | 1 | 3 (1) | XI, 0 1 |
| I2 | 2 | 2 | 1 (1) | VI/VII1, XI, C-C-C-CC-CC 1, C-C-C-CC-CC-C 1 |
| I2-like | 0 | 0 | 1 | XI |
| I3-like | 1 | 1 (1) | 2 (1) | XI, XV 1 |
| L | 0 | 2 (1) | 0 | XIV |
| L-like | 1 | 0 | 0 | XIV |
| M | 4 | 5 | 7 | 0, VI/VII, XIII 1, XXII 1, C-C 1, C-C-C 1, C-CC-C-CC-C-C 1 |
| M-like | 1 | 3 | 1 | 0, VI/VII, XV 1 |
| O1 | 5 | 6 (2) | 2 (2) | 0 1, VI/VII, XVI |
| O2-like | 2 | 3 (2) | 1 (1) | VI/VII, XV |
| O3-like | 5 | 8 (2) | 17 (6) | 0, VI/VII, XI 1, XIV 1, C1, C-C-C-C-C 1 |
| P | 4 | 5 (2) | 2 (1) | 0 1, XXV 1, IX 1, C-C-C-C-C 1 |
| P-like | 0 | 1 | 0 | IX |
| Q-like | 0 | 0 | 1 (1) | XVI |
| T | 0 | 2 | 0 | 0 1 |
| Y-like | 3 | 3 (1) | 2 | VI/VII 1 |
| Div.M---L-LTVA | 2 | 2 (1) | 3 (1) | VI/VII, IX |
| Div.MSTLGMTLL- | 1 | 1 | 1 (1) | XV 1, C-C-CC-CC-C-C-C 1 |
| PFC-01 | 1 | 1 | 1 (1) | XIV |
| PFC-02 | 0 | 1 | 0 | VI/VII |
| PFC-03 | 1 | 1 (1) | 0 | VIII |
| Turripeptide | 3 | 6 | 4 | 0, XVI, IX, C |
| Turripeptide-like | 0 | 1 | 3 | IX |
| Intragroup | Pneo | Pvau | |||||
|---|---|---|---|---|---|---|---|
| Insulin | Conus prey | % Id. | % Sim. | % Id. | % Sim. | % Id. | % Sim. |
| Chain A | Fish | 43–95 | 62–100 | 38–43 | 52–67 | 26–32 | 52–64 |
| Mollusc | 81–100 | 90–100 | 68–81 1 | 76–90 1,2 | 41–50 | 65–76 | |
| Worm | 44–100 | 72–100 | 52–76 | 68–86 | 38–56 5 | 71–85 4,6 | |
| Chain B | Fish | 59–100 | 91–100 | 23–22 | 48–64 | 38–43 | 52–67 |
| Mollusc | 50–97 | 79–97 | 41–50 1 | 62–74 | 69–81 1 | 88–90 | |
| Worm | 32–100 | 53–100 | 41–50 3,4 | 62–82 4 | 52–77 | 81–92 7 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fassio, G.; Modica, M.V.; Mary, L.; Zaharias, P.; Fedosov, A.E.; Gorson, J.; Kantor, Y.I.; Holford, M.; Puillandre, N. Venom Diversity and Evolution in the Most Divergent Cone Snail Genus Profundiconus. Toxins 2019, 11, 623. https://doi.org/10.3390/toxins11110623
Fassio G, Modica MV, Mary L, Zaharias P, Fedosov AE, Gorson J, Kantor YI, Holford M, Puillandre N. Venom Diversity and Evolution in the Most Divergent Cone Snail Genus Profundiconus. Toxins. 2019; 11(11):623. https://doi.org/10.3390/toxins11110623
Chicago/Turabian StyleFassio, Giulia, Maria Vittoria Modica, Lou Mary, Paul Zaharias, Alexander E. Fedosov, Juliette Gorson, Yuri I. Kantor, Mandё Holford, and Nicolas Puillandre. 2019. "Venom Diversity and Evolution in the Most Divergent Cone Snail Genus Profundiconus" Toxins 11, no. 11: 623. https://doi.org/10.3390/toxins11110623
APA StyleFassio, G., Modica, M. V., Mary, L., Zaharias, P., Fedosov, A. E., Gorson, J., Kantor, Y. I., Holford, M., & Puillandre, N. (2019). Venom Diversity and Evolution in the Most Divergent Cone Snail Genus Profundiconus. Toxins, 11(11), 623. https://doi.org/10.3390/toxins11110623






