Spider Venom: Components, Modes of Action, and Novel Strategies in Transcriptomic and Proteomic Analyses
Abstract
1. Introduction
2. Spider Venom Composition
2.1. Spider Venom
2.2. Small Molecular Mass Compounds
2.3. Antimicrobial Peptides
2.4. Cysteine-Rich Peptides
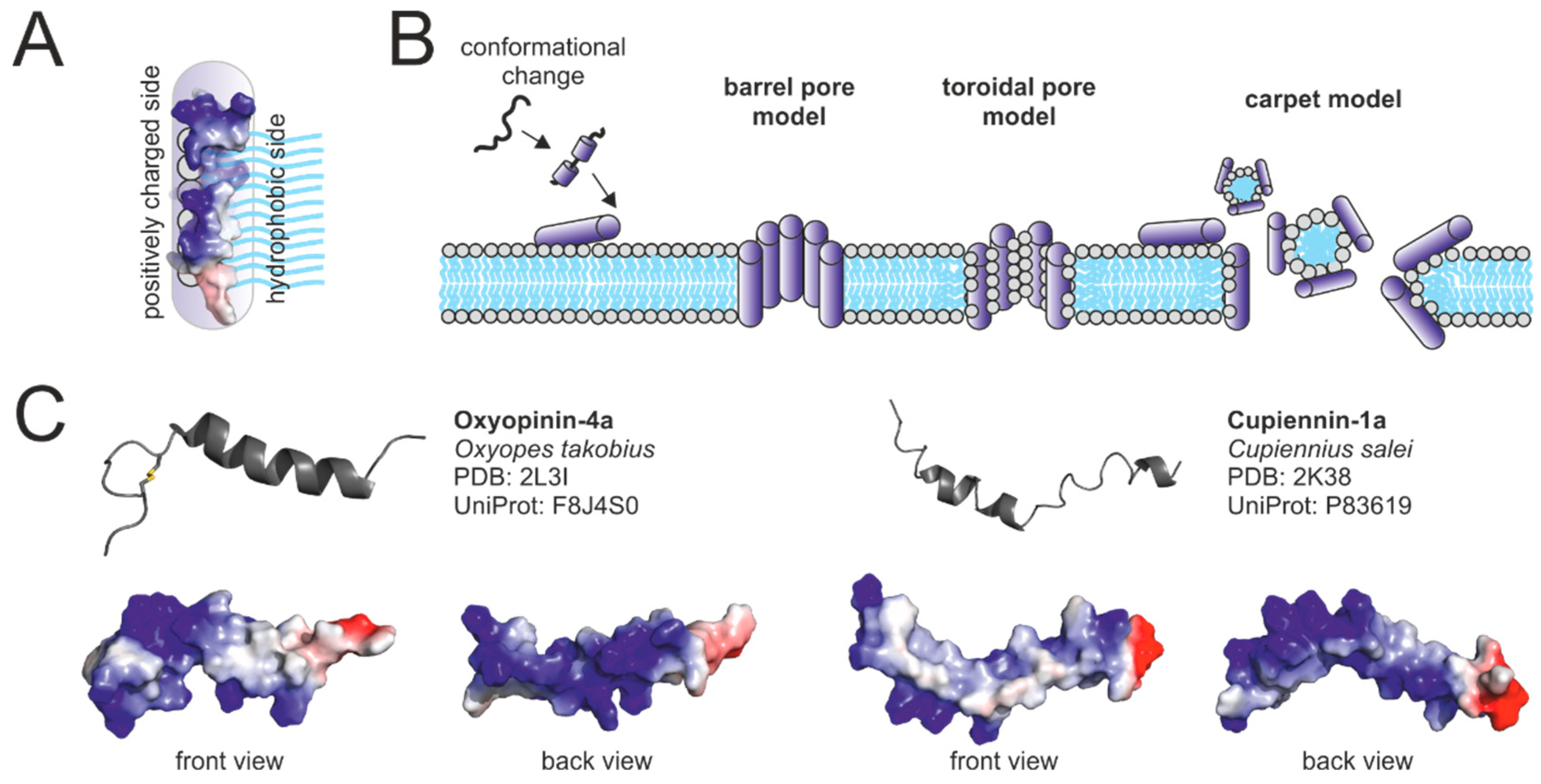
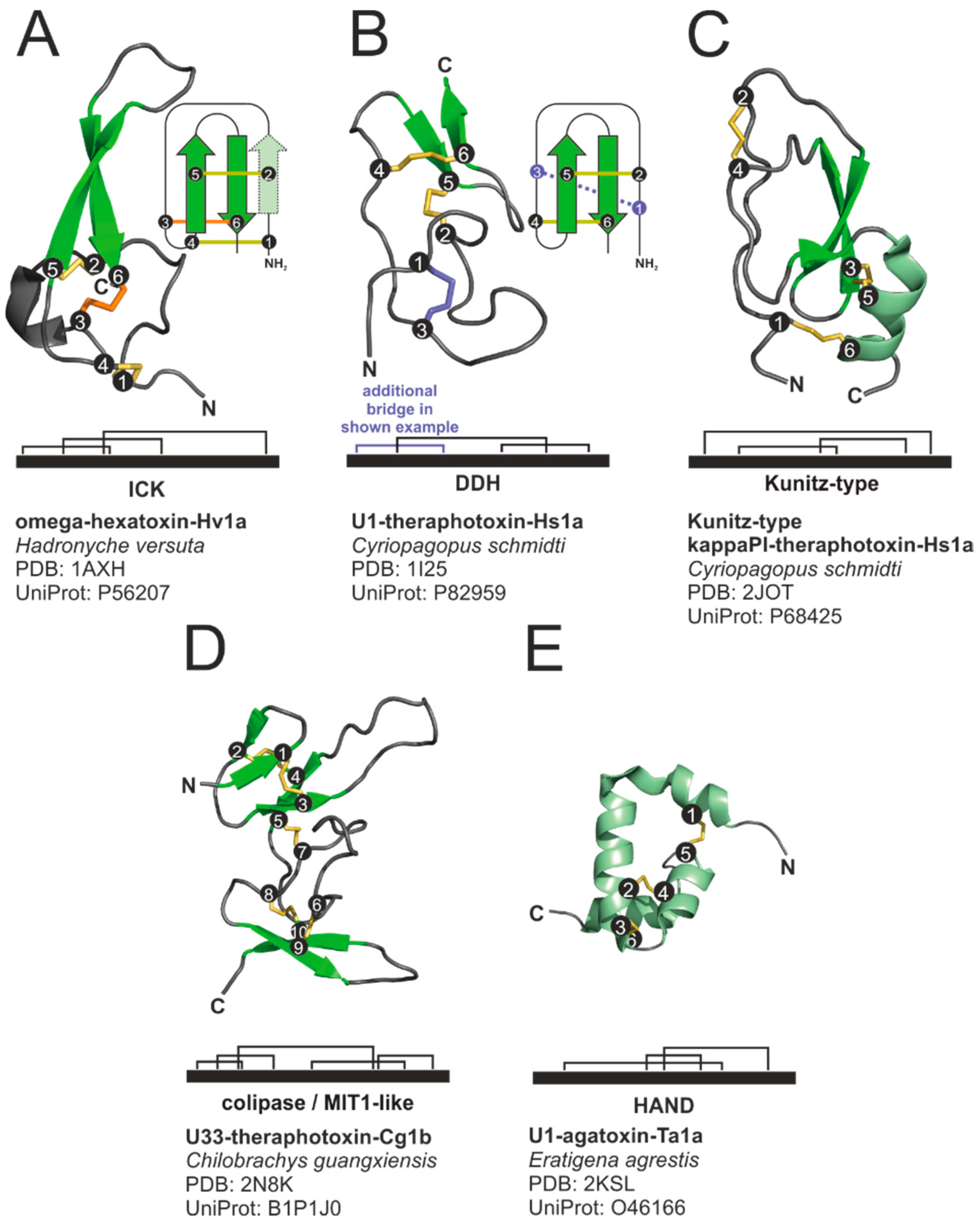
2.4.1. Structural Motifs
2.4.2. Targets of Neurotoxic Peptides
2.4.3. Nomenclature of Toxins
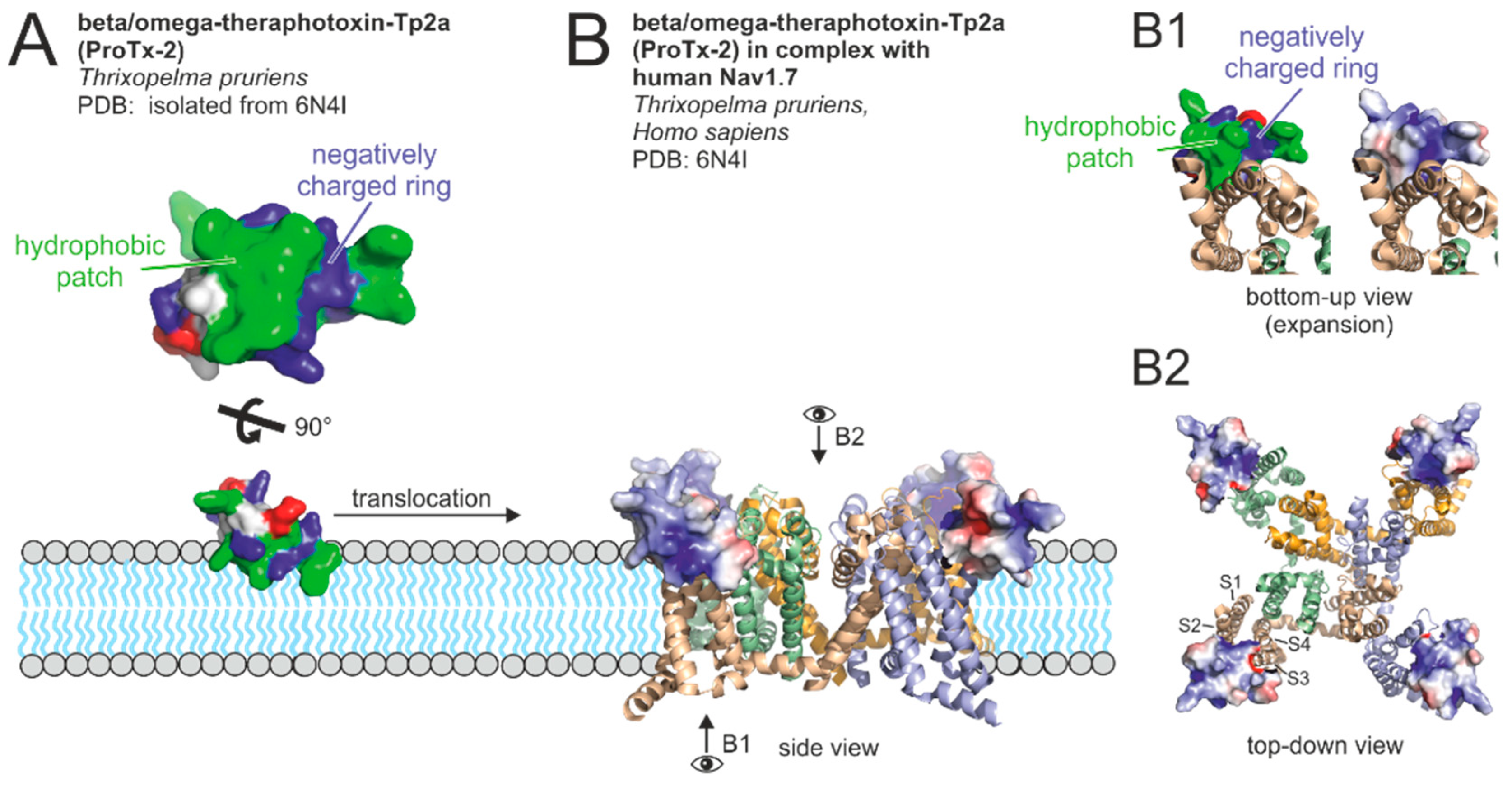
2.4.4. Molecular Binding of Neurotoxins
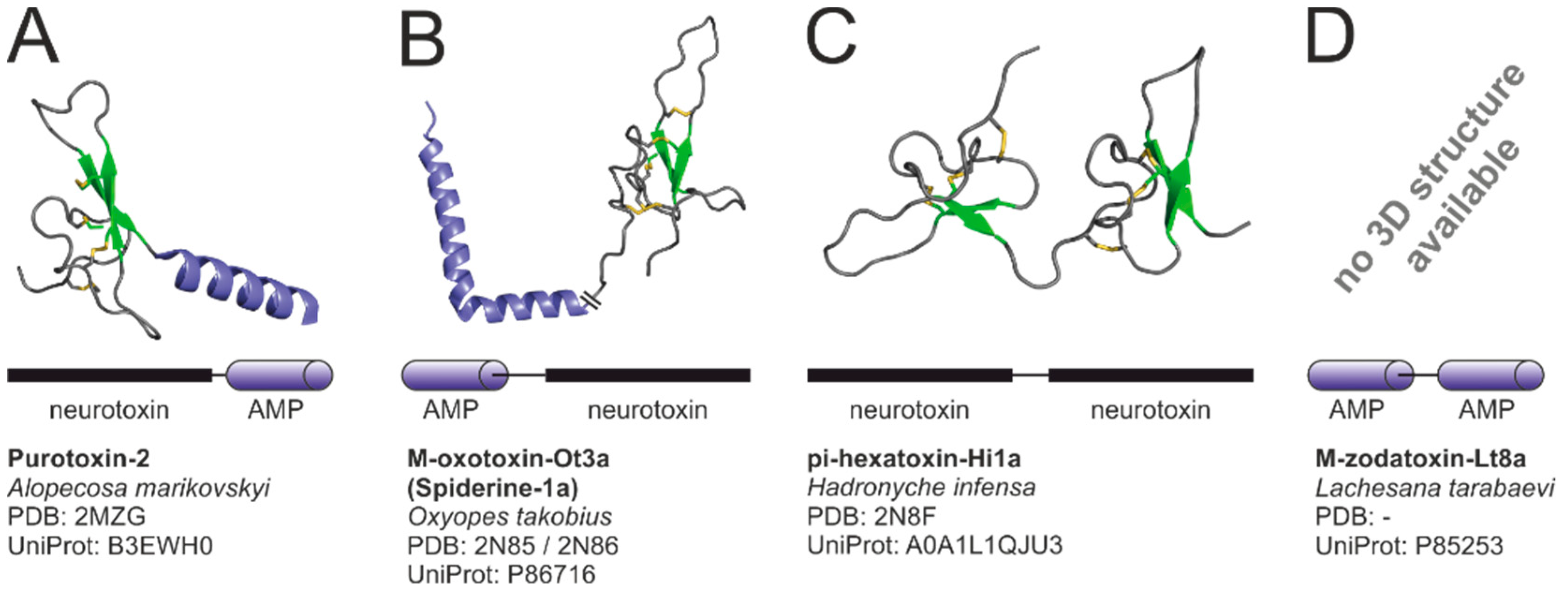
2.4.5. Modular Toxins—Combination of Toxin Structures
2.5. Enzymes and Proteins
2.6. Spider Venom Peptide Precursors and Their Maturing
2.6.1. Comparison of Protease Cleavage Motifs to Motifs of Other Organisms
2.6.2. Cleavage for Heterodimerization of Spider Toxins
2.7. Concluding Remarks on Venom Components and Their Toxic Action
3. Analysis of Venom Components
3.1. Venom Gland Transcriptome Analysis
3.1.1. Identification of Toxin-Like Transcripts
3.1.2. Recent Developments in Sequencing Techniques
3.1.3. Cross Contamination among Multiplexed Samples in Illumina Sequencing
3.2. High Throughput Mass Spectrometric Analysis of the Venom Proteoms
3.2.1. Bottom-Up Proteomics
3.2.2. Top-Down Proteomics
3.3. Annotation of Spider Venom Toxins
3.4. Concluding Remarks on the Analysis of Venom Components
Author Contributions
Funding
Conflicts of Interest
References
- Calvete, J.J. Venomics: Integrative venom proteomics and beyond. Biochem. J. 2017, 474, 611–634. [Google Scholar] [CrossRef]
- Peigneur, S.; Tytgat, J. Toxins in Drug Discovery and Pharmacology. Toxins 2018, 10, 126. [Google Scholar] [CrossRef]
- Soares, S.C.; Lindstrom, B.; Esteves, F.; Ohman, A. The Hidden Snake in the Grass: Superior Detection of Snakes in Challenging Attentional Conditions. PLoS ONE 2014, 9, e114724. [Google Scholar] [CrossRef]
- Isbell, L.A. Snakes as agents of evolutionary change in primate brains. J. Hum. Evol. 2006, 51, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wuster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.E. Poison Arrows: North American Indian Hunting and Warfare; University of Texas Press: Austin, TX, USA, 2007. [Google Scholar] [CrossRef]
- Gomes, A.; Bhattacharya, S.; Chakraborty, M.; Bhattacharjee, P.; Mishra, R.; Gomes, A. Anti-arthritic activity of Indian monocellate cobra (Naja kaouthia) venom on adjuvant induced arthritis. Toxicon 2010, 55, 670–673. [Google Scholar] [CrossRef] [PubMed]
- Norton, R.S. Enhancing the therapeutic potential of peptide toxins. Expert Opin. Drug Discov. 2017, 12, 611–623. [Google Scholar] [CrossRef]
- Wie, C.S.; Derian, A. Ziconotide. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2019. [Google Scholar]
- Opie, L.H.; Kowolik, H. The discovery of captopril: From large animals to small molecules. Cardiovasc. Res. 1995, 30, 18–25. [Google Scholar] [CrossRef]
- Dutertre, S.; Lewis, R.J. Use of venom peptides to probe ion channel structure and function. J. Biol. Chem. 2010, 285, 13315–13320. [Google Scholar] [CrossRef]
- Harrison, R.A.; Cook, D.A.; Renjifo, C.; Casewell, N.R.; Currier, R.B.; Wagstaff, S.C. Research strategies to improve snakebite treatment: Challenges and progress. J. Proteom. 2011, 74, 1768–1780. [Google Scholar] [CrossRef]
- Smith, J.J.; Herzig, V.; King, G.F.; Alewood, P.F. The insecticidal potential of venom peptides. Cell. Mol. Life Sci. 2013, 70, 3665–3693. [Google Scholar] [CrossRef] [PubMed]
- Kuhn-Nentwig, L.; Stöcklin, R.; Nentwig, W. Venom Composition and Strategies in Spiders: Is Everything Possible? Adv. Insect Physiol. 2011, 40, 1–86. [Google Scholar] [CrossRef]
- Escoubas, P.; Sollod, B.; King, G.F. Venom landscapes: Mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon 2006, 47, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Satake, H.; Villegas, E.; Oshiro, N.; Terada, K.; Shinada, T.; Corzo, G. Rapid and efficient identification of cysteine-rich peptides by random screening of a venom gland cDNA library from the hexathelid spider Macrothele gigas. Toxicon 2004, 44, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S.; Malyavka, A.; McCutchen, B.; Lu, A.; Schepers, E.; Herrmann, R.; Grishin, E. A novel strategy for the identification of toxinlike structures in spider venom. Proteins 2005, 59, 131–140. [Google Scholar] [CrossRef]
- Fernandes-Pedrosa Mde, F.; Junqueira-de-Azevedo Ide, L.; Goncalves-de-Andrade, R.M.; Kobashi, L.S.; Almeida, D.D.; Ho, P.L.; Tambourgi, D.V. Transcriptome analysis of Loxosceles laeta (Araneae, Sicariidae) spider venomous gland using expressed sequence tags. BMC Genom. 2008, 9, 279. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, L.; Jiang, L.; Meng, E.; Zhang, Y.; Xiong, X.; Liang, S. Transcriptome analysis revealed novel possible venom components and cellular processes of the tarantula Chilobrachys jingzhao venom gland. Toxicon 2008, 52, 794–806. [Google Scholar] [CrossRef]
- Kuhn-Nentwig, L.; Fedorova, I.M.; Lüscher, B.P.; Kopp, L.S.; Trachsel, C.; Schaller, J.; Vu, X.L.; Seebeck, T.; Streitberger, K.; Nentwig, W.; et al. A venom-derived neurotoxin, CsTx-1, from the spider Cupiennius salei exhibits cytolytic activities. J. Biol. Chem. 2012, 287, 25640–25649. [Google Scholar] [CrossRef]
- Undheim, E.A.; Sunagar, K.; Herzig, V.; Kely, L.; Low, D.H.; Jackson, T.N.; Jones, A.; Kurniawan, N.; King, G.F.; Ali, S.A.; et al. A proteomics and transcriptomics investigation of the venom from the barychelid spider Trittame loki (brush-foot trapdoor). Toxins 2013, 5, 2488–2503. [Google Scholar] [CrossRef]
- Haney, R.A.; Ayoub, N.A.; Clarke, T.H.; Hayashi, C.Y.; Garb, J.E. Dramatic expansion of the black widow toxin arsenal uncovered by multi-tissue transcriptomics and venom proteomics. BMC Genom. 2014, 15, 366. [Google Scholar] [CrossRef]
- Haney, R.A.; Matte, T.; Forsyth, F.S.; Garb, J.E. Alternative Transcription at Venom Genes and Its Role as a Complementary Mechanism for the Generation of Venom Complexity in the Common House Spider. Front. Ecol. Evol. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Duan, Z.; Yu, Y.; Liu, Z.; Liu, Z.; Liang, S. The venom gland transcriptome of Latrodectus tredecimguttatus revealed by deep sequencing and cDNA library analysis. PLoS ONE 2013, 8, e81357. [Google Scholar] [CrossRef] [PubMed]
- Penney, D. Spider Research in the 21st Century: Trends and Perspectives; Siri Scientific Press: Manchester, UK, 2013. [Google Scholar]
- Foelix, R.; Erb, B. Mesothelae have venom glands. J. Arachnol. 2010, 38, 596–598. [Google Scholar] [CrossRef]
- Foelix, R. Biology of Spiders, 3rd ed.; Oxford University Press: New York, NY, USA, 2011. [Google Scholar]
- Weng, J.L.; Barrantes, G.; Eberhard, W.G. Feeding by Philoponella vicina (Araneae, Uloboridae) and how uloborid spiders lost their venom glands. Can. J. Zool. 2006, 84, 1752–1762. [Google Scholar] [CrossRef]
- Zobel-Thropp, P.A.; Correa, S.M.; Garb, J.E.; Binford, G.J. Spit and venom from Scytodes spiders: A diverse and distinct cocktail. J. Proteome Res. 2014, 13, 817–835. [Google Scholar] [CrossRef] [PubMed]
- Pekar, S.; Toft, S.; Hruskova, M.; Mayntz, D. Dietary and prey-capture adaptations by which Zodarion germanicum, an ant-eating spider (Araneae: Zodariidae), specialises on the Formicinae. Naturwissenschaften 2008, 95, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Dippenaar-Schoeman, A.S.; Meyer, M.K.P. On the species of the African genus Ammoxenus (Araneae: Ammoxenidae), with decriptions of two new species. J. Entomol. Soc. South. Afr. 1980, 43, 41–49. [Google Scholar]
- Gemeno, C.; Yeargan, K.V.; Haynes, K.F. Aggressive Chemical Mimicry by the Bolas Spider Mastophora hutchinsoni: Identification and Quantification of a Major Prey’s Sex Pheromone Components in the Spider’s Volatile Emissions. J. Chem. Ecol. 2000, 26, 1235–1243. [Google Scholar] [CrossRef]
- Meehan, C.J.; Olson, E.J.; Reudink, M.W.; Kyser, T.K.; Curry, R.L. Herbivory in a spider through exploitation of an ant-plant mutualism. Curr. Biol. 2009, 19, R892–R893. [Google Scholar] [CrossRef]
- Escoubas, P.; Quinton, L.; Nicholson, G.M. Venomics: Unravelling the complexity of animal venoms with mass spectrometry. J. Mass Spectrom. 2008, 43, 279–295. [Google Scholar] [CrossRef]
- Zobel-Thropp, P.A.; Bulger, E.A.; Cordes, M.H.J.; Binford, G.J.; Gillespie, R.G.; Brewer, M.S. Sexually dimorphic venom proteins in long-jawed orb-weaving spiders (Tetragnatha) comprise novel gene families. PeerJ 2018, 6, e4691. [Google Scholar] [CrossRef] [PubMed]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wuster, W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. Biol. Sci. 2009, 276, 2443–2449. [Google Scholar] [CrossRef] [PubMed]
- Jorge da Silva, N., Jr.; Aird, S.D. Prey specificity, comparative lethality and compositional differences of coral snake venoms. Comp. Biochem. Physiol. Toxicol. Pharmacol. 2001, 128, 425–456. [Google Scholar] [CrossRef]
- Gibbs, H.L.; Mackessy, S.P. Functional basis of a molecular adaptation: Prey-specific toxic effects of venom from Sistrurus rattlesnakes. Toxicon 2009, 53, 672–679. [Google Scholar] [CrossRef]
- Elliger, C.A.; Richmond, T.A.; Lebaric, Z.N.; Pierce, N.T.; Sweedler, J.V.; Gilly, W.F. Diversity of conotoxin types from Conus californicus reflects a diversity of prey types and a novel evolutionary history. Toxicon 2011, 57, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Duda, T.F., Jr.; Chang, D.; Lewis, B.D.; Lee, T. Geographic variation in venom allelic composition and diets of the widespread predatory marine gastropod Conus ebraeus. PLoS ONE 2009, 4, e6245. [Google Scholar] [CrossRef]
- Nentwig, W.; Kuhn-Nentwig, L. Spider Venoms Potentially Lethal to Humans. In Spider Ecophysiology; Nentwig, W., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 253–264. [Google Scholar] [CrossRef]
- Nentwig, W.; Kuhn-Nentwig, L. Main Components of Spider Venoms. In Spider Ecophysiology; Nentwig, W., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 191–202. [Google Scholar]
- Chippaux, J.P.; Goyffon, M. Epidemiology of scorpionism: A global appraisal. Acta Trop. 2008, 107, 71–79. [Google Scholar] [CrossRef]
- Chippaux, J.P. Snake-bites: Appraisal of the global situation. Bull. World Health Organ. 1998, 76, 515–524. [Google Scholar]
- Nentwig, W. (Ed.) Spider Ecophysiology, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 505–508. [Google Scholar]
- Herzig, V.; Wood, D.L.; Newell, F.; Chaumeil, P.A.; Kaas, Q.; Binford, G.J.; Nicholson, G.M.; Gorse, D.; King, G.F. ArachnoServer 2.0, an updated online resource for spider toxin sequences and structures. Nucleic Acids Res. 2011, 39, 653–657. [Google Scholar] [CrossRef]
- Pineda, S.S.; Chaumeil, P.A.; Kunert, A.; Kaas, Q.; Thang, M.W.C.; Li, L.; Nuhn, M.; Herzig, V.; Saez, N.J.; Cristofori-Armstrong, B.; et al. ArachnoServer 3.0: An online resource for automated discovery, analysis and annotation of spider toxins. Bioinformatics 2017, 34, 1074–1076. [Google Scholar] [CrossRef]
- World Spider Catalog Version 20.5. Available online: http://wsc.nmbe.ch (accessed on 27 August 2019).
- Savel-Niemann, A. Tarantula (Eurypelma californicum) venom, a multicomponent system. Biol. Chem. Hoppe Seyler 1989, 370, 485–498. [Google Scholar] [CrossRef]
- Kuhn-Nentwig, L.; Schaller, J.; Nentwig, W. Purification of toxic peptides and the amino acid sequence of CSTX-1 from the multicomponent venom of Cupiennius salei (Araneae: Ctenidae). Toxicon 1994, 32, 287–302. [Google Scholar] [CrossRef]
- Malli, H.; Kuhn-Nentwig, L.; Imboden, H.; Nentwig, W. Effects of size, motility and paralysation time of prey on the quantity of venom injected by the hunting spider Cupiennius salei. J. Exp. Biol. 1999, 202, 2083–2089. [Google Scholar]
- Kuhn-Nentwig, L.; Schaller, J.; Nentwig, W. Biochemistry, toxicology and ecology of the venom of the spider Cupiennius salei (Ctenidae). Toxicon 2004, 43, 543–553. [Google Scholar] [CrossRef]
- Wigger, E.; Kuhn-Nentwig, L.; Nentwig, W. The venom optimisation hypothesis: A spider injects large venom quantities only into difficult prey types. Toxicon 2002, 40, 749–752. [Google Scholar] [CrossRef]
- Malli, H.; Kuhn-Nentwig, L.; Imboden, H.; Moon, M.J.; Wyler, T. Immunocytochemical localization and secretion process of the toxin CSTX-1 in the venom gland of the wandering spider Cupiennius salei (Araneae: Ctenidae). Cell Tissue Res. 2000, 299, 417–426. [Google Scholar] [CrossRef]
- Silva, L.M.; Botelho, A.C.; Nacif-Pimenta, R.; Martins, G.F.; Alves, L.C.; Brayner, F.A.; Fortes-Dias, C.L.; Pimenta, P.F. Structural analysis of the venom glands of the armed spider Phoneutria nigriventer (Keyserling, 1891): Microanatomy, fine structure and confocal observations. Toxicon 2008, 51, 693–706. [Google Scholar] [CrossRef]
- Yigit, N.; Guven, T. Functional structure of Agelena labyrinthica’s (Araneae:Agelenidae) venom gland and electrophoresis of venom. Toxicon 2006, 47, 58–67. [Google Scholar] [CrossRef]
- Savel-Niemann, A.; Roth, D. Biochemical analysis of tarantula venom (Eurypelma californicum). Naturwissenschaften 1989, 76, 212–213. [Google Scholar] [CrossRef]
- Wullschleger, B.; Nentwig, W.; Kuhn-Nentwig, L. Spider venom: Enhancement of venom efficacy mediated by different synergistic strategies in Cupiennius salei. J. Exp. Biol. 2005, 208, 2115–2121. [Google Scholar] [CrossRef]
- Wullschleger, B.; Kuhn-Nentwig, L.; Tromp, J.; Kämpfer, U.; Schaller, J.; Schürch, S.; Nentwig, W. CSTX-13, a highly synergistically acting two-chain neurotoxic enhancer in the venom of the spider Cupiennius salei (Ctenidae). Proc. Natl. Acad. Sci. USA 2004, 101, 11251–11256. [Google Scholar] [CrossRef]
- Odell, G.V.; Fenton, A.W.; Ownby, C.L.; Doss, M.P.; Schmidt, J.O. The role of venom citrate. Toxicon 1999, 37, 407–409. [Google Scholar] [CrossRef]
- McCormick, J.; Li, Y.; McCormick, K.; Duynstee, H.I.; van Engen, A.K.; van der Marel, G.A.; Ganem, B.; van Boom, J.H.; Meinwald, J. Structure and Total Synthesis of HF-7, a Neuroactive Glyconucleoside Disulfate from the Funnel-Web Spider Hololena curta. J. Am. Chem. Soc. 1999, 121, 5661–5665. [Google Scholar] [CrossRef]
- Schroeder, F.C.; Taggi, A.E.; Gronquist, M.; Malik, R.U.; Grant, J.B.; Eisner, T.; Meinwald, J. NMR-spectroscopic screening of spider venom reveals sulfated nucleosides as major components for the brown recluse and related species. Proc. Natl. Acad. Sci. USA 2008, 105, 14283–14287. [Google Scholar] [CrossRef]
- Taggi, A.E.; Meinwald, J.; Schroeder, F.C. A new approach to natural products discovery exemplified by the identification of sulfated nucleosides in spider venom. J. Am. Chem. Soc. 2004, 126, 10364–10369. [Google Scholar] [CrossRef]
- Manzoli-Palma, M.d.F.; Gobbi, N.; Palma, M.S. The chelation of metal ions by the acylpolyamine toxins from the web-spider Nephilengys cruentata: Effects in the intoxication/detoxification of preys. Chemoecology 2006, 16, 203–208. [Google Scholar] [CrossRef]
- Bowie, D.; Mayer, M.L. Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron 1995, 15, 453–462. [Google Scholar] [CrossRef]
- Rash, L.D.; Hodgson, W.C. Pharmacology and biochemistry of spider venoms. Toxicon 2002, 40, 225–254. [Google Scholar] [CrossRef]
- Blenau, W.; Baumann, A. Molecular and pharmacological properties of insect biogenic amine receptors: Lessons from Drosophila melanogaster and Apis mellifera. Arch. Insect Biochem. Physiol. 2001, 48, 13–38. [Google Scholar] [CrossRef]
- Bontems, F.; Roumestand, C.; Gilquin, B.; Menez, A.; Toma, F. Refined structure of charybdotoxin: Common motifs in scorpion toxins and insect defensins. Science 1991, 254, 1521–1523. [Google Scholar] [CrossRef]
- Kuhn-Nentwig, L. Antimicrobial and cytolytic peptides of venomous arthropods. Cell. Mol. Life Sci. 2003, 60, 2651–2668. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef] [PubMed]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [PubMed]
- Kuhn-Nentwig, L.; Dathe, M.; Walz, A.; Schaller, J.; Nentwig, W. Cupiennin 1d *: The cytolytic activity depends on the hydrophobic N-terminus and is modulated by the polar C-terminus. FEBS Lett. 2002, 527, 193–198. [Google Scholar] [CrossRef]
- Corzo, G.; Villegas, E.; Gomez-Lagunas, F.; Possani, L.D.; Belokoneva, O.S.; Nakajima, T. Oxyopinins, large amphipathic peptides isolated from the venom of the wolf spider Oxyopes kitabensis with cytolytic properties and positive insecticidal cooperativity with spider neurotoxins. J. Biol. Chem. 2002, 277, 23627–23637. [Google Scholar] [CrossRef]
- Kozlov, S.A.; Vassilevski, A.A.; Feofanov, A.V.; Surovoy, A.Y.; Karpunin, D.V.; Grishin, E.V. Latarcins, antimicrobial and cytolytic peptides from the venom of the spider Lachesana tarabaevi (Zodariidae) that exemplify biomolecular diversity. J. Biol. Chem. 2006, 281, 20983–20992. [Google Scholar] [CrossRef]
- Kuhn-Nentwig, L.; Willems, J.; Seebeck, T.; Shalaby, T.; Kaiser, M.; Nentwig, W. Cupiennin 1a exhibits a remarkably broad, non-stereospecific cytolytic activity on bacteria, protozoan parasites, insects, and human cancer cells. Amino Acids 2011, 40, 69–76. [Google Scholar] [CrossRef][Green Version]
- Pukala, T.L.; Boland, M.P.; Gehman, J.D.; Kuhn-Nentwig, L.; Separovic, F.; Bowie, J.H. Solution structure and interaction of cupiennin 1a, a spider venom peptide, with phospholipid bilayers. Biochemistry 2007, 46, 3576–3585. [Google Scholar] [CrossRef]
- Belokoneva, O.S.; Satake, H.; Mal’tseva, E.L.; Pal’mina, N.P.; Villegas, E.; Nakajima, T.; Corzo, G. Pore formation of phospholipid membranes by the action of two hemolytic arachnid peptides of different size. Biochim. Biophys. Acta 2004, 1664, 182–188. [Google Scholar] [CrossRef][Green Version]
- Nomura, K.; Corzo, G. The effect of binding of spider-derived antimicrobial peptides, oxyopinins, on lipid membranes. Biochim. Biophys. Acta 2006, 1758, 1475–1482. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Volynsky, P.E.; Polyansky, A.A.; Karpunin, D.V.; Chupin, V.V.; Efremov, R.G.; Arseniev, A.S. Three-dimensional structure/hydrophobicity of latarcins specifies their mode of membrane activity. Biochemistry 2008, 47, 3525–3533. [Google Scholar] [CrossRef]
- Pukala, T.L.; Doyle, J.R.; Llewellyn, L.E.; Kuhn-Nentwig, L.; Apponyi, M.A.; Separovic, F.; Bowie, J.H. Cupiennin 1a, an antimicrobial peptide from the venom of the neotropical wandering spider Cupiennius salei, also inhibits the formation of nitric oxide by neuronal nitric oxide synthase. FEBS J. 2007, 274, 1778–1784. [Google Scholar] [CrossRef]
- Vassilevski, A.A.; Kozlov, S.A.; Grishin, E.V. Molecular diversity of spider venom. Biochemistry Biokhimiia 2009, 74, 1505–1534. [Google Scholar] [CrossRef]
- Pallaghy, P.K.; Nielsen, K.J.; Craik, D.J.; Norton, R.S. A common structural motif incorporating a cystine knot and a triple-stranded beta-sheet in toxic and inhibitory polypeptides. Protein Sci. 1994, 3, 1833–1839. [Google Scholar] [CrossRef]
- Norton, R.S.; Pallaghy, P.K. The cystine knot structure of ion channel toxins and related polypeptides. Toxicon 1998, 36, 1573–1583. [Google Scholar] [CrossRef]
- Escoubas, P.; Rash, L. Tarantulas: Eight-legged pharmacists and combinatorial chemists. Toxicon 2004, 43, 555–574. [Google Scholar] [CrossRef]
- Schaller, J.; Kämpfer, U.; Schürch, S.; Kuhn-Nentwig, L.; Haeberli, S.; Nentwig, W. CSTX-9, a toxic peptide from the spider Cupiennius salei: Amino acid sequence, disulphide bridge pattern and comparison with other spider toxins containing the cystine knot structure. Cell. Mol. Life Sci. 2001, 58, 1538–1545. [Google Scholar] [CrossRef]
- Gao, B.; Harvey, P.J.; Craik, D.J.; Ronjat, M.; De Waard, M.; Zhu, S. Functional evolution of scorpion venom peptides with an inhibitor cystine knot fold. Biosci. Rep. 2013, 33. [Google Scholar] [CrossRef]
- Kwon, S.; Bosmans, F.; Kaas, Q.; Cheneval, O.; Conibear, A.C.; Rosengren, K.J.; Wang, C.K.; Schroeder, C.I.; Craik, D.J. Efficient enzymatic cyclization of an inhibitory cystine knot-containing peptide. Biotechnol. Bioeng. 2016, 113, 2202–2212. [Google Scholar] [CrossRef]
- Le Nguyen, D.; Heitz, A.; Chiche, L.; Castro, B.; Boigegrain, R.A.; Favel, A.; Coletti-Previero, M.A. Molecular recognition between serine proteases and new bioactive microproteins with a knotted structure. Biochimie 1990, 72, 431–435. [Google Scholar] [CrossRef]
- Fujitani, N.; Kawabata, S.; Osaki, T.; Kumaki, Y.; Demura, M.; Nitta, K.; Kawano, K. Structure of the antimicrobial peptide tachystatin A. J. Biol. Chem. 2002, 277, 23651–23657. [Google Scholar] [CrossRef]
- Vervoort, J.; van den Hooven, H.W.; Berg, A.; Vossen, P.; Vogelsang, R.; Joosten, M.H.; de Wit, P.J. The race-specific elicitor AVR9 of the tomato pathogen Cladosporium fulvum: A cystine knot protein. Sequence-specific 1H NMR assignments, secondary structure and global fold of the protein. FEBS Lett. 1997, 404, 153–158. [Google Scholar] [CrossRef]
- Wang, X.; Connor, M.; Smith, R.; Maciejewski, M.W.; Howden, M.E.; Nicholson, G.M.; Christie, M.J.; King, G.F. Discovery and characterization of a family of insecticidal neurotoxins with a rare vicinal disulfide bridge. Nat. Struct. Biol. 2000, 7, 505–513. [Google Scholar] [CrossRef]
- Sunagar, K.; Undheim, E.A.; Chan, A.H.; Koludarov, I.; Munoz-Gomez, S.A.; Antunes, A.; Fry, B.G. Evolution stings: The origin and diversification of scorpion toxin peptide scaffolds. Toxins 2013, 5, 2456–2487. [Google Scholar] [CrossRef]
- Harvey, A.L.; Karlsson, E. Dendrotoxin from the venom of the green mamba, Dendroaspis angusticeps. A neurotoxin that enhances acetylcholine release at neuromuscular junction. Naunyn-Schmiedeberg’s Arch. Pharmacol. 1980, 312, 1–6. [Google Scholar] [CrossRef]
- Bayrhuber, M.; Vijayan, V.; Ferber, M.; Graf, R.; Korukottu, J.; Imperial, J.; Garrett, J.E.; Olivera, B.M.; Terlau, H.; Zweckstetter, M.; et al. Conkunitzin-S1 is the first member of a new Kunitz-type neurotoxin family. Structural and functional characterization. J. Biol. Chem. 2005, 280, 23766–23770. [Google Scholar] [CrossRef]
- Schweitz, H.; Bruhn, T.; Guillemare, E.; Moinier, D.; Lancelin, J.M.; Beress, L.; Lazdunski, M. Kalicludines and kaliseptine. Two different classes of sea anemone toxins for voltage sensitive K+ channels. J. Biol. Chem. 1995, 270, 25121–25126. [Google Scholar] [CrossRef]
- Peng, K.; Lin, Y.; Liang, S.P. Nuclear magnetic resonance studies on huwentoxin-XI from the Chinese bird spider Ornithoctonus huwena: 15N labeling and sequence-specific 1H, 15N nuclear magnetic resonance assignments. Acta Biochim. Biophys. Sin. 2006, 38, 457–466. [Google Scholar] [CrossRef]
- Schweitz, H.; Heurteaux, C.; Bois, P.; Moinier, D.; Romey, G.; Lazdunski, M. Calcicludine, a venom peptide of the Kunitz-type protease inhibitor family, is a potent blocker of high-threshold Ca2+ channels with a high affinity for L-type channels in cerebellar granule neurons. Proc. Natl. Acad. Sci. USA 1994, 91, 878–882. [Google Scholar] [CrossRef]
- Harvey, A.L. Twenty years of dendrotoxins. Toxicon 2001, 39, 15–26. [Google Scholar] [CrossRef]
- Boisbouvier, J.; Albrand, J.P.; Blackledge, M.; Jaquinod, M.; Schweitz, H.; Lazdunski, M.; Marion, D. A structural homologue of colipase in black mamba venom revealed by NMR floating disulphide bridge analysis. J. Mol. Biol. 1998, 283, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Joubert, F.J.; Strydom, D.J. Snake venom. The amino acid sequence of protein A from Dendroaspis polylepis polylepis (black mamba) venom. Hoppe Seyler Z. Phisol. Chem. 1980, 361, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Mollay, C.; Wechselberger, C.; Mignogna, G.; Negri, L.; Melchiorri, P.; Barra, D.; Kreil, G. Bv8, a small protein from frog skin and its homologue from snake venom induce hyperalgesia in rats. Eur. J. Pharmacol. (Mol. Pharmacol. Sect.) 1999, 374, 189–196. [Google Scholar] [CrossRef]
- Wechselberger, C.; Puglisi, R.; Engel, E.; Lepperdinger, G.; Boitani, C.; Kreil, G. The mammalian homologues of frog Bv8 are mainly expressed in spermatocytes. FEBS Lett. 1999, 462, 177–181. [Google Scholar] [CrossRef]
- Negri, L.; Lattanzi, R.; Giannini, E.; Melchiorri, P. Bv8/Prokineticin proteins and their receptors. Life Sci. 2007, 81, 1103–1116. [Google Scholar] [CrossRef]
- Kaser, A.; Winklmayr, M.; Lepperdinger, G.; Kreil, G. The AVIT protein family. Secreted cysteine-rich vertebrate proteins with diverse functions. EMBO Rep. 2003, 4, 469–473. [Google Scholar] [CrossRef]
- Szeto, T.H.; Wang, X.H.; Smith, R.; Connor, M.; Christie, M.J.; Nicholson, G.M.; King, G.F. Isolation of a funnel-web spider polypeptide with homology to mamba intestinal toxin 1 and the embryonic head inducer Dickkopf-1. Toxicon 2000, 38, 429–442. [Google Scholar] [CrossRef]
- Wen, S.; Wilson, D.T.; Kuruppu, S.; Korsinczky, M.L.; Hedrick, J.; Pang, L.; Szeto, T.; Hodgson, W.C.; Alewood, P.F.; Nicholson, G.M. Discovery of an MIT-like atracotoxin family: Spider venom peptides that share sequence homology but not pharmacological properties with AVIT family proteins. Peptides 2005, 26, 2412–2426. [Google Scholar] [CrossRef]
- Kuhn-Nentwig, L.; Langenegger, N.; Heller, M.; Koua, D.; Nentwig, W. The Dual Prey-Inactivation Strategy of Spiders-In-Depth Venomic Analysis of Cupiennius salei. Toxins 2019, 11, 167. [Google Scholar] [CrossRef]
- Undheim, E.A.; Grimm, L.L.; Low, C.F.; Morgenstern, D.; Herzig, V.; Zobel-Thropp, P.; Pineda, S.S.; Habib, R.; Dziemborowicz, S.; Fry, B.G.; et al. Weaponization of a Hormone: Convergent Recruitment of Hyperglycemic Hormone into the Venom of Arthropod Predators. Structure 2015, 23, 1283–1292. [Google Scholar] [CrossRef]
- McCowan, C.; Garb, J.E. Recruitment and diversification of an ecdysozoan family of neuropeptide hormones for black widow spider venom expression. Gene 2014, 536, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, S.; Kiyatkin, N.; Drevet, P.; Boulain, J.C.; Tacnet, F.; Ripoche, P.; Forest, E.; Grishin, E.; Menez, A. The low molecular weight protein which co-purifies with alpha-latrotoxin is structurally related to crustacean hyperglycemic hormones. J. Biol. Chem. 1994, 269, 19803–19809. [Google Scholar] [PubMed]
- Grishin, E.V.; Himmelreich, N.H.; Pluzhnikov, K.A.; Pozdnyakova, N.G.; Storchak, L.G.; Volkova, T.M.; Woll, P.G. Modulation of functional activities of the neurotoxin from black widow spider venom. FEBS Lett. 1993, 336, 205–207. [Google Scholar] [CrossRef]
- Kiyatkin, N.; Dulubova, I.; Chekhovskaya, I.; Lipkin, A.; Grishin, E. Structure of the low molecular weight protein copurified with alpha-latrotoxin. Toxicon 1992, 30, 771–774. [Google Scholar] [CrossRef]
- Volkova, T.M.; Pluzhnikov, K.A.; Woll, P.G.; Grishin, E.V. Low molecular weight components from black widow spider venom. Toxicon 1995, 33, 483–489. [Google Scholar] [CrossRef]
- Johnson, J.H.; Bloomquist, J.R.; Krapcho, K.J.; Kral, R.M., Jr.; Trovato, R.; Eppler, K.G.; Morgan, T.K.; DelMar, E.G. Novel insecticidal peptides from Tegenaria agrestis spider venom may have a direct effect on the insect central nervous system. Arch. Insect Biochem. Physiol. 1998, 38, 19–31. [Google Scholar] [CrossRef]
- Santos, A.D.; Imperial, J.S.; Chaudhary, T.; Beavis, R.C.; Chait, B.T.; Hunsperger, J.P.; Olivera, B.M.; Adams, M.E.; Hillyard, D.R. Heterodimeric structure of the spider toxin omega-agatoxin IA revealed by precursor analysis and mass spectrometry. J. Biol. Chem. 1992, 267, 20701–20705. [Google Scholar]
- Alewood, D.; Birinyi-Strachan, L.C.; Pallaghy, P.K.; Norton, R.S.; Nicholson, G.M.; Alewood, P.F. Synthesis and characterization of delta-atracotoxin-Ar1a, the lethal neurotoxin from venom of the Sydney funnel-web spider (Atrax robustus). Biochemistry 2003, 42, 12933–12940. [Google Scholar] [CrossRef]
- Bohlen, C.J.; Priel, A.; Zhou, S.; King, D.; Siemens, J.; Julius, D. A bivalent tarantula toxin activates the capsaicin receptor, TRPV1, by targeting the outer pore domain. Cell 2010, 141, 834–845. [Google Scholar] [CrossRef]
- Siemens, J.; Zhou, S.; Piskorowski, R.; Nikai, T.; Lumpkin, E.A.; Basbaum, A.I.; King, D.; Julius, D. Spider toxins activate the capsaicin receptor to produce inflammatory pain. Nature 2006, 444, 208–212. [Google Scholar] [CrossRef]
- King, G.F.; Gentz, M.C.; Escoubas, P.; Nicholson, G.M. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon 2008, 52, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.K.; Ligutti, J.; Liu, D.; Zou, A.; Poppe, L.; Li, H.; Andrews, K.L.; Moyer, B.D.; McDonough, S.I.; Favreau, P.; et al. Engineering potent and selective analogues of GpTx-1, a tarantula venom peptide antagonist of the Na(V)1.7 sodium channel. J. Med. Chem. 2015, 58, 2299–2314. [Google Scholar] [CrossRef] [PubMed]
- Revell, J.D.; Lund, P.E.; Linley, J.E.; Metcalfe, J.; Burmeister, N.; Sridharan, S.; Jones, C.; Jermutus, L.; Bednarek, M.A. Potency optimization of Huwentoxin-IV on hNav1.7: A neurotoxin TTX-S sodium-channel antagonist from the venom of the Chinese bird-eating spider Selenocosmia huwena. Peptides 2013, 44, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Cummins, T.R.; Alphy, S.; Blumenthal, K.M. Molecular interactions of the gating modifier toxin ProTx-II with NaV 1.5: Implied existence of a novel toxin binding site coupled to activation. J. Biol. Chem. 2007, 282, 12687–12697. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Luo, X.; Jiang, L.; Chen, H.; Wang, J.; He, H.; Liang, S. Synthesis and biological characterization of synthetic analogs of Huwentoxin-IV (Mu-theraphotoxin-Hh2a), a neuronal tetrodotoxin-sensitive sodium channel inhibitor. Toxicon 2013, 71, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Li, Z.; Jiang, Y.; Pan, X.; Wu, J.; Cristofori-Armstrong, B.; Smith, J.J.; Chin, Y.K.Y.; Lei, J.; Zhou, Q.; et al. Structural basis for the modulation of voltage-gated sodium channels by animal toxins. Science 2018, 362, eaau2596. [Google Scholar] [CrossRef]
- Xu, H.; Li, T.; Rohou, A.; Arthur, C.P.; Tzakoniati, F.; Wong, E.; Estevez, A.; Kugel, C.; Franke, Y.; Chen, J.; et al. Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin. Cell 2019, 176, 1238–1239. [Google Scholar] [CrossRef]
- Baconguis, I.; Gouaux, E. Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature 2012, 489, 400–405. [Google Scholar] [CrossRef]
- Deplazes, E.; Henriques, S.T.; Smith, J.J.; King, G.F.; Craik, D.J.; Mark, A.E.; Schroeder, C.I. Membrane-binding properties of gating modifier and pore-blocking toxins: Membrane interaction is not a prerequisite for modification of channel gating. Biochim. Biophys. Acta 2016, 1858, 872–882. [Google Scholar] [CrossRef]
- Agwa, A.J.; Peigneur, S.; Chow, C.Y.; Lawrence, N.; Craik, D.J.; Tytgat, J.; King, G.F.; Henriques, S.T.; Schroeder, C.I. Gating modifier toxins isolated from spider venom: Modulation of voltage-gated sodium channels and the role of lipid membranes. J. Biol. Chem. 2018, 293, 9041–9052. [Google Scholar] [CrossRef]
- Lee, S.Y.; MacKinnon, R. A membrane-access mechanism of ion channel inhibition by voltage sensor toxins from spider venom. Nature 2004, 430, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.H.; Jung, H.J.; Konishi, S.; Kim, H.H.; Park, Z.Y.; Kim, J.I. Structure-activity relationships of omega-Agatoxin IVA in lipid membranes. Biochem. Biophys. Res. Commun. 2017, 482, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Mihailescu, M.; Krepkiy, D.; Milescu, M.; Gawrisch, K.; Swartz, K.J.; White, S. Structural interactions of a voltage sensor toxin with lipid membranes. Proc. Natl. Acad. Sci. USA 2014, 111, E5463–E5470. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Lee, J.Y.; Kim, S.H.; Eu, Y.J.; Shin, S.Y.; Milescu, M.; Swartz, K.J.; Kim, J.I. Solution structure and lipid membrane partitioning of VSTx1, an inhibitor of the KvAP potassium channel. Biochemistry 2005, 44, 6015–6023. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Kim, J.I.; Min, H.J.; Sato, K.; Swartz, K.J.; Shimada, I. Solution structure of hanatoxin1, a gating modifier of voltage-dependent K(+) channels: Common surface features of gating modifier toxins. J. Mol. Biol. 2000, 297, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Henriques, S.T.; Deplazes, E.; Lawrence, N.; Cheneval, O.; Chaousis, S.; Inserra, M.; Thongyoo, P.; King, G.F.; Mark, A.E.; Vetter, I.; et al. Interaction of Tarantula Venom Peptide ProTx-II with Lipid Membranes Is a Prerequisite for Its Inhibition of Human Voltage-gated Sodium Channel NaV1.7. J. Biol. Chem. 2016, 291, 17049–17065. [Google Scholar] [CrossRef]
- Lau, C.H.Y.; King, G.F.; Mobli, M. Molecular basis of the interaction between gating modifier spider toxins and the voltage sensor of voltage-gated ion channels. Sci. Rep. 2016, 6, 34333. [Google Scholar] [CrossRef]
- Darnell, J.E.; Lodish, H.; Berk, A.; Zipursky, L.; Matsudaira, P.; Baltimore, D. Molecular Cell Biology, 4th ed.; W. H. Freeman: New York, NY, USA, 2000. [Google Scholar]
- Cardoso, F.C.; Lewis, R.J. Structure-Function and Therapeutic Potential of Spider Venom-Derived Cysteine Knot Peptides Targeting Sodium Channels. Front. Pharmacol. 2019, 10, 366. [Google Scholar] [CrossRef]
- Milescu, M.; Lee, H.C.; Bae, C.H.; Kim, J.I.; Swartz, K.J. Opening the shaker K+ channel with hanatoxin. J. Gen. Physiol. 2013, 141, 203–216. [Google Scholar] [CrossRef]
- Twomey, E.C.; Yelshanskaya, M.V.; Vassilevski, A.A.; Sobolevsky, A.I. Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors. Neuron 2018, 99, 956–968. [Google Scholar] [CrossRef]
- Dawson, R.J.; Benz, J.; Stohler, P.; Tetaz, T.; Joseph, C.; Huber, S.; Schmid, G.; Hugin, D.; Pflimlin, P.; Trube, G.; et al. Structure of the acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1. Nat. Commun. 2012, 3, 936. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Zamanian, M.; Bae, C.; Milescu, M.; Krepkiy, D.; Tilley, D.C.; Sack, J.T.; Yarov-Yarovoy, V.; Kim, J.I.; Swartz, K.J. Tarantula toxins use common surfaces for interacting with Kv and ASIC ion channels. Elife 2015, 4, e06774. [Google Scholar] [CrossRef] [PubMed]
- Vassilevski, A.A.; Sachkova, M.Y.; Ignatova, A.A.; Kozlov, S.A.; Feofanov, A.V.; Grishin, E.V. Spider toxins comprising disulfide-rich and linear amphipathic domains: A new class of molecules identified in the lynx spider Oxyopes takobius. FEBS J. 2013, 280, 6247–6261. [Google Scholar] [CrossRef] [PubMed]
- Oparin, P.B.; Nadezhdin, K.D.; Berkut, A.A.; Arseniev, A.S.; Grishin, E.V.; Vassilevski, A.A. Structure of purotoxin-2 from wolf spider: Modular design and membrane-assisted mode of action in arachnid toxins. Biochem. J. 2016, 473, 3113–3126. [Google Scholar] [CrossRef] [PubMed]
- Sachkova, M.Y.; Slavokhotova, A.A.; Grishin, E.V.; Vassilevski, A.A. Genes and evolution of two-domain toxins from lynx spider venom. FEBS Lett. 2014, 588, 740–745. [Google Scholar] [CrossRef]
- Kuhn-Nentwig, L.; Nentwig, W. The Cytotoxic Mode of Action of the Venom of Cupiennius salei (Ctenidae). In Spider Ecophysiology; Nentwig, W., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 217–228. [Google Scholar]
- Diego-Garcia, E.; Abdel-Mottaleb, Y.; Schwartz, E.F.; de la Vega, R.C.; Tytgat, J.; Possani, L.D. Cytolytic and K+ channel blocking activities of beta-KTx and scorpine-like peptides purified from scorpion venoms. Cell. Mol. Life Sci. 2008, 65, 187–200. [Google Scholar] [CrossRef]
- Chassagnon, I.R.; McCarthy, C.A.; Chin, Y.K.; Pineda, S.S.; Keramidas, A.; Mobli, M.; Pham, V.; De Silva, T.M.; Lynch, J.W.; Widdop, R.E.; et al. Potent neuroprotection after stroke afforded by a double-knot spider-venom peptide that inhibits acid-sensing ion channel 1a. Proc. Natl. Acad. Sci. USA 2017, 114, 3750–3755. [Google Scholar] [CrossRef]
- Vassilevski, A.A.; Fedorova, I.M.; Maleeva, E.E.; Korolkova, Y.V.; Efimova, S.S.; Samsonova, O.V.; Schagina, L.V.; Feofanov, A.V.; Magazanik, L.G.; Grishin, E.V. Novel class of spider toxin: Active principle from the yellow sac spider Cheiracanthium punctorium venom is a unique two-domain polypeptide. J. Biol. Chem. 2010, 285, 32293–32302. [Google Scholar] [CrossRef]
- Sachkova, M.Y.; Slavokhotova, A.A.; Grishin, E.V.; Vassilevski, A.A. Structure of the yellow sac spider Cheiracanthium punctorium genes provides clues to evolution of insecticidal two-domain knottin toxins. Insect Mol. Biol. 2014, 23, 527–538. [Google Scholar] [CrossRef]
- Vassilevski, A.A.; Kozlov, S.A.; Samsonova, O.V.; Egorova, N.S.; Karpunin, D.V.; Pluzhnikov, K.A.; Feofanov, A.V.; Grishin, E.V. Cyto-insectotoxins, a novel class of cytolytic and insecticidal peptides from spider venom. Biochem. J. 2008, 411, 687–696. [Google Scholar] [CrossRef]
- Binford, G.J.; Bodner, M.R.; Cordes, M.H.; Baldwin, K.L.; Rynerson, M.R.; Burns, S.N.; Zobel-Thropp, P.A. Molecular evolution, functional variation, and proposed nomenclature of the gene family that includes sphingomyelinase D in sicariid spider venoms. Mol. Biol. Evol. 2009, 26, 547–566. [Google Scholar] [CrossRef] [PubMed]
- Zobel-Thropp, P.A.; Kerins, A.E.; Binford, G.J. Sphingomyelinase D in sicariid spider venom is a potent insecticidal toxin. Toxicon 2012, 60, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Shikata, Y.; Watanabe, T.; Teramoto, T.; Inoue, A.; Kawakami, Y.; Nishizawa, Y.; Katayama, K.; Kuwada, M. Isolation and characterization of a peptide isomerase from funnel web spider venom. J. Biol. Chem. 1995, 270, 16719–16723. [Google Scholar] [CrossRef] [PubMed]
- Diniz, M.R.V.; Paiva, A.L.B.; Guerra-Duarte, C.; Nishiyama, M.Y., Jr.; Mudadu, M.A.; Oliveira, U.; Borges, M.H.; Yates, J.R.; Junqueira-de-Azevedo, I.L. An overview of Phoneutria nigriventer spider venom using combined transcriptomic and proteomic approaches. PLoS ONE 2018, 13, e0200628. [Google Scholar] [CrossRef]
- Akhunov, A.A.; Makevnina, L.G.; Golubenko, Z.; Paskhina, T.S. Kininase of the Latrodectus tredecimguttatus venom: A study of its enzyme substrate specificity. Immunopharmacology 1996, 32, 160–162. [Google Scholar] [CrossRef]
- Cajado-Carvalho, D.; Kuniyoshi, A.K.; Duzzi, B.; Iwai, L.K.; Oliveira, U.C.; Junqueira de Azevedo, I.L.; Kodama, R.T.; Portaro, F.V. Insights into the Hypertensive Effects of Tityus serrulatus Scorpion Venom: Purification of an Angiotensin-Converting Enzyme-Like Peptidase. Toxins 2016, 8, 348. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Hyodo, F.; Morita, T. Wide distribution of cysteine-rich secretory proteins in snake venoms: Isolation and cloning of novel snake venom cysteine-rich secretory proteins. Arch. Biochem. Biophys. 2003, 412, 133–141. [Google Scholar] [CrossRef]
- Gibbs, G.M.; O’Bryan, M.K. Cysteine rich secretory proteins in reproduction and venom. Soc. Reprod. Fertil. Suppl. 2007, 65, 261–267. [Google Scholar]
- Da Ros, V.; Busso, D.; Cohen, D.J.; Maldera, J.; Goldweic, N.; Cuasnicu, P.S. Molecular mechanisms involved in gamete interaction: Evidence for the participation of cysteine-rich secretory proteins (CRISP) in sperm-egg fusion. Soc. Reprod. Fertil. Suppl. 2007, 65, 353–356. [Google Scholar]
- Kjeldsen, L.; Cowland, J.B.; Johnsen, A.H.; Borregaard, N. SGP28, a novel matrix glycoprotein in specific granules of human neutrophils with similarity to a human testis-specific gene product and a rodent sperm-coating glycoprotein. FEBS Lett. 1996, 380, 246–250. [Google Scholar] [CrossRef]
- Guo, M.; Teng, M.; Niu, L.; Liu, Q.; Huang, Q.; Hao, Q. Crystal structure of the cysteine-rich secretory protein stecrisp reveals that the cysteine-rich domain has a K+ channel inhibitor-like fold. J. Biol. Chem. 2005, 280, 12405–12412. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.L.; Haley, T.L.; West, K.A.; Crabb, J.W. Pseudechetoxin: A peptide blocker of cyclic nucleotide-gated ion channels. Proc. Natl. Acad. Sci. USA 1999, 96, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, H.; Liu, M.N.; Song, H.; Han, H.M.; Wang, Q.L.; Yin, C.C.; Zhou, Y.C.; Qi, Z.; Shu, Y.Y.; et al. Structural and functional analysis of natrin, a venom protein that targets various ion channels. Biochem. Biophys. Res. Commun. 2006, 351, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Assumpcao, T.C.; Ma, D.; Schwarz, A.; Reiter, K.; Santana, J.M.; Andersen, J.F.; Ribeiro, J.M.; Nardone, G.; Yu, L.L.; Francischetti, I.M. Salivary antigen-5/CAP family members are Cu2+-dependent antioxidant enzymes that scavenge O(2)(-). and inhibit collagen-induced platelet aggregation and neutrophil oxidative burst. J. Biol. Chem. 2013, 288, 14341–14361. [Google Scholar] [CrossRef] [PubMed]
- Grishin, E.V. Black widow spider toxins: The present and the future. Toxicon 1998, 36, 1693–1701. [Google Scholar] [CrossRef]
- Garb, J.E.; Hayashi, C.Y. Molecular evolution of alpha-latrotoxin, the exceptionally potent vertebrate neurotoxin in black widow spider venom. Mol. Biol. Evol. 2013, 30, 999–1014. [Google Scholar] [CrossRef]
- Gendreau, K.L.; Haney, R.A.; Schwager, E.E.; Wierschin, T.; Stanke, M.; Richards, S.; Garb, J.E. House spider genome uncovers evolutionary shifts in the diversity and expression of black widow venom proteins associated with extreme toxicity. BMC Genom. 2017, 18, 178. [Google Scholar] [CrossRef]
- Mironov, S.L.; Sokolov Yu, V.; Chanturiya, A.N.; Lishko, V.K. Channels produced by spider venoms in bilayer lipid membrane: Mechanisms of ion transport and toxic action. Biochim. Biophys. Acta 1986, 862, 185–198. [Google Scholar] [CrossRef]
- Shatursky, O.; Pashkov, V.N.; Bulgacov, O.V.; Grishin, E.V. Interaction of alpha-latroinsectotoxin from Latrodectus mactans venom with bilayer lipid membranes. Biochim. Biophys. Acta 1995, 1233, 14–20. [Google Scholar] [CrossRef]
- Ushkaryov, Y.A.; Volynski, K.E.; Ashton, A.C. The multiple actions of black widow spider toxins and their selective use in neurosecretion studies. Toxicon 2004, 43, 527–542. [Google Scholar] [CrossRef]
- Yan, S.; Wang, X. Recent Advances in Research on Widow Spider Venoms and Toxins. Toxins 2015, 7, 5055–5067. [Google Scholar] [CrossRef] [PubMed]
- Mosavi, L.K.; Cammett, T.J.; Desrosiers, D.C.; Peng, Z.Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004, 13, 1435–1448. [Google Scholar] [CrossRef] [PubMed]
- Dulubova, I.E.; Krasnoperov, V.G.; Khvotchev, M.V.; Pluzhnikov, K.A.; Volkova, T.M.; Grishin, E.V.; Vais, H.; Bell, D.R.; Usherwood, P.N. Cloning and structure of delta-latroinsectotoxin, a novel insect-specific member of the latrotoxin family: Functional expression requires C-terminal truncation. J. Biol. Chem. 1996, 271, 7535–7543. [Google Scholar] [CrossRef] [PubMed]
- Vassilevski, A.A.; Kozlov, S.A.; Grishin, E.V. Antimicrobial peptide precursor structures suggest effective production strategies. Recent Pat. Inflamm. Allergy Drug Discov. 2008, 2, 58–63. [Google Scholar] [CrossRef]
- Kuhn-Nentwig, L. Cytolytic and antimicrobial peptides in the venom of scorpions and spiders. In Animal Toxins: State of the Art—Perspectives in Health and Biotechnology; Lima, M., Pimenta, A., France Martin-Eauclaire, M., Zingali, R., Rochat, H., Eds.; Universidade Federal de Minas Gerais: Belo Horizonte, Brasil, 2009; Volume 15, pp. 249–256. [Google Scholar]
- Dubovskii, P.V.; Vassilevski, A.A.; Kozlov, S.A.; Feofanov, A.V.; Grishin, E.V.; Efremov, R.G. Latarcins: Versatile spider venom peptides. Cell. Mol. Life Sci. 2015, 72, 4501–4522. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S.A.; Grishin, E.V. The universal algorithm of maturation for secretory and excretory protein precursors. Toxicon 2007, 49, 721–726. [Google Scholar] [CrossRef]
- Langenegger, N.; Koua, D.; Schürch, S.; Heller, M.; Nentwig, W.; Kuhn-Nentwig, L. Identification of a precursor processing protease from the spider Cupiennius salei essential for venom neurotoxin maturation. J. Biol. Chem. 2018, 293, 2079–2090. [Google Scholar] [CrossRef]
- Rholam, M.; Fahy, C. Processing of peptide and hormone precursors at the dibasic cleavage sites. Cell. Mol. Life Sci. 2009, 66, 2075–2091. [Google Scholar] [CrossRef]
- Duckert, P.; Brunak, S.; Blom, N. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 2004, 17, 107–112. [Google Scholar] [CrossRef]
- Rouille, Y.; Duguay, S.J.; Lund, K.; Furuta, M.; Gong, Q.; Lipkind, G.; Oliva, A.A., Jr.; Chan, S.J.; Steiner, D.F. Proteolytic processing mechanisms in the biosynthesis of neuroendocrine peptides: The subtilisin-like proprotein convertases. Front. Neuroendocrinol. 1995, 16, 322–361. [Google Scholar] [CrossRef]
- Zhu, S.; Peigneur, S.; Gao, B.; Luo, L.; Jin, D.; Zhao, Y.; Tytgat, J. Molecular diversity and functional evolution of scorpion potassium channel toxins. Mol. Cell. Proteom. 2011, 10, M110.002832. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Zeng, X.C.; Hahin, R.; Cao, Z.J.; Liu, H.; Li, W.X. Genomic organization of four novel nondisulfide-bridged peptides from scorpion Mesobuthus martensii Karsch: Gaining insight into evolutionary mechanism. Peptides 2005, 26, 2427–2433. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Sherman, P.; Luo, L.; Bowie, J.; Zhu, S. Structural and functional characterization of two genetically related meucin peptides highlights evolutionary divergence and convergence in antimicrobial peptides. FASEB J. 2009, 23, 1230–1245. [Google Scholar] [CrossRef] [PubMed]
- Lorenzini, D.M.; Fukuzawa, A.H.; da Silva, P.I., Jr.; Machado-Santelli, G.; Bijovsky, A.T.; Daffre, S. Molecular cloning, expression analysis and cellular localization of gomesin, an anti-microbial peptide from hemocytes of the spider Acanthoscurria gomesiana. Insect Biochem. Mol. Biol. 2003, 33, 1011–1016. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, J.; Shen, J.H.; Jin, Y.; Lee, W.H.; Zhang, Y. Maximins S, a novel group of antimicrobial peptides from toad Bombina maxima. Biochem. Biophys. Res. Commun. 2005, 327, 945–951. [Google Scholar] [CrossRef]
- Osmakov, D.I.; Kozlov, S.A.; Andreev, Y.A.; Koshelev, S.G.; Sanamyan, N.P.; Sanamyan, K.E.; Dyachenko, I.A.; Bondarenko, D.A.; Murashev, A.N.; Mineev, K.S.; et al. Sea anemone peptide with uncommon beta-hairpin structure inhibits acid-sensing ion channel 3 (ASIC3) and reveals analgesic activity. J. Biol. Chem. 2013, 288, 23116–23127. [Google Scholar] [CrossRef]
- Undheim, E.A.; Sunagar, K.; Hamilton, B.R.; Jones, A.; Venter, D.J.; Fry, B.G.; King, G.F. Multifunctional warheads: Diversification of the toxin arsenal of centipedes via novel multidomain transcripts. J. Proteom. 2014, 102, 1–10. [Google Scholar] [CrossRef]
- Casteels-Josson, K.; Capaci, T.; Casteels, P.; Tempst, P. Apidaecin multipeptide precursor structure: A putative mechanism for amplification of the insect antibacterial response. EMBO J. 1993, 12, 1569–1578. [Google Scholar] [CrossRef]
- Marcus, J.P.; Green, J.L.; Goulter, K.C.; Manners, J.M. A family of antimicrobial peptides is produced by processing of a 7S globulin protein in Macadamia integrifolia kernels. Plant J. 1999, 19, 699–710. [Google Scholar] [CrossRef]
- Adams, M.E.; Bindokas, V.P.; Hasegawa, L.; Venema, V.J. Omega-agatoxins: Novel calcium channel antagonists of two subtypes from funnel web spider (Agelenopsis aperta) venom. J. Biol. Chem. 1990, 265, 861–867. [Google Scholar]
- Branton, W.D.; Rudnick, M.S.; Zhou, Y.; Eccleston, E.D.; Fields, G.B.; Bowers, L.D. Fatty acylated toxin structure. Nature 1993, 365, 496–497. [Google Scholar] [CrossRef] [PubMed]
- Kuwada, M.; Teramoto, T.; Kumagaye, K.Y.; Nakajima, K.; Watanabe, T.; Kawai, T.; Kawakami, Y.; Niidome, T.; Sawada, K.; Nishizawa, Y.; et al. Omega-agatoxin-TK containing D-serine at position 46, but not synthetic omega-[L-Ser46] agatoxin-TK, exerts blockade of P-type calcium channels in cerebellar Purkinje neurons. Mol. Pharmacol. 1994, 46, 587–593. [Google Scholar] [PubMed]
- Pimenta, A.M.; Rates, B.; Bloch, C., Jr.; Gomes, P.C.; Santoro, M.M.; de Lima, M.E.; Richardson, M.; Cordeiro Mdo, N. Electrospray ionization quadrupole time-of-flight and matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometric analyses to solve micro-heterogeneity in post-translationally modified peptides from Phoneutria nigriventer (Aranea, Ctenidae) venom. Rapid Commun. Mass Spectrom. 2005, 19, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.A.; Alves, W.E.; Lucas, M.S.; Habermehl, G.G. Isolation and characterization of a bradykinin potentiating peptide (BPP-S) isolated from Scaptocosa raptoria venom. Toxicon 1996, 34, 599–603. [Google Scholar] [CrossRef]
- Ferreira, L.A.; Lucas, S.M.; Alves, E.W.; Hermann, V.V.; Reichl, A.P.; Habermehl, G.; Zingali, R.B. Isolation, characterization and biological properties of two kinin-like peptides (peptide-S and peptide-r) from Scaptocosa raptoria venom. Toxicon 1998, 36, 31–39. [Google Scholar] [CrossRef]
- Akchunov, A.A.; Golubenko, Z.; Sosnina, N. Isolation and characterization of biological properties of inhibitors angiotensin-1-converting enzyme from the spider venom Latrodectus tredecimguttatus. Agents Actions Suppl. 1992, 38 Pt 1, 469–474. [Google Scholar]
- King, G.F.; Hardy, M.C. Spider-venom peptides: Structure, pharmacology, and potential for control of insect pests. Annu. Rev. Entomol. 2013, 58, 475–496. [Google Scholar] [CrossRef]
- Milescu, M.; Bosmans, F.; Lee, S.; Alabi, A.A.; Kim, J.I.; Swartz, K.J. Interactions between lipids and voltage sensor paddles detected with tarantula toxins. Nat. Struct. Mol. Biol. 2009, 16, 1080–1085. [Google Scholar] [CrossRef]
- Ramu, Y.; Xu, Y.; Lu, Z. Enzymatic activation of voltage-gated potassium channels. Nature 2006, 442, 696–699. [Google Scholar] [CrossRef]
- McCrone, J.D. Spider venoms: Biochemical aspects. Am. Zool. 1969, 9, 153–156. [Google Scholar] [CrossRef]
- Lucas, S.M. The history of venomous spider identification, venom extraction methods and antivenom production: A long journey at the Butantan Institute, Sao Paulo, Brazil. J. Venom. Anim. Tox. Incl. Trop. Dis. 2015, 21, 21. [Google Scholar] [CrossRef] [PubMed]
- Barrio, A.; Brazil, O.V. Ein neues Verfahren der Giftentnahme bei Spinnen. Experientia 1950, 6, 112–113. [Google Scholar] [CrossRef]
- Bachmann, M. Isolation and partial characterization of a toxin from the venom of the East African orthognath spider Pterinochilus spec. Toxicon 1982, 20, 547–552. [Google Scholar] [CrossRef]
- Entwistle, I.D.; Johnstone, R.A.; Medzihradszky, D.; May, T.E. Isolation of a pure toxic polypeptide from the venom of the spider Phoneutria nigriventer and its neurophysiological activity on an insect femur preparation. Toxicon 1982, 20, 1059–1067. [Google Scholar] [CrossRef]
- Skinner, W.S.; Adams, M.E.; Quistad, G.B.; Kataoka, H.; Cesarin, B.J.; Enderlin, F.E.; Schooley, D.A. Purification and characterization of two classes of neurotoxins from the funnel web spider, Agelenopsis aperta. J. Biol. Chem. 1989, 264, 2150–2155. [Google Scholar]
- Lange, C.; Paris, C.; Celerier, M.L. The components of the venom of a spider Scodra griseipes. 2. Structural information on biogenic amines using tandem mass spectrometry. Rapid Commun. Mass Spectrom. 1992, 6, 517–519. [Google Scholar] [CrossRef]
- Saez, N.J.; Senff, S.; Jensen, J.E.; Er, S.Y.; Herzig, V.; Rash, L.D.; King, G.F. Spider-venom peptides as therapeutics. Toxins 2010, 2, 2851–2871. [Google Scholar] [CrossRef]
- Windley, M.J.; Herzig, V.; Dziemborowicz, S.A.; Hardy, M.C.; King, G.F.; Nicholson, G.M. Spider-venom peptides as bioinsecticides. Toxins 2012, 4, 191–227. [Google Scholar] [CrossRef]
- Pineda, S.S.; Undheim, E.A.; Rupasinghe, D.B.; Ikonomopoulou, M.P.; King, G.F. Spider venomics: Implications for drug discovery. Future Med. Chem. 2014, 6, 1699–1714. [Google Scholar] [CrossRef]
- Ostrow, K.L.; Mammoser, A.; Suchyna, T.; Sachs, F.; Oswald, R.; Kubo, S.; Chino, N.; Gottlieb, P.A. cDNA sequence and in vitro folding of GsMTx4, a specific peptide inhibitor of mechanosensitive channels. Toxicon 2003, 42, 263–274. [Google Scholar] [CrossRef]
- Kiyatkin, N.I.; Dulubova, I.E.; Chekhovskaya, I.A.; Grishin, E.V. Cloning and structure of cDNA encoding alpha-latrotoxin from black widow spider venom. FEBS Lett. 1990, 270, 127–131. [Google Scholar] [CrossRef]
- Wilson, D.; Daly, N.L. Venomics: A Mini-Review. High-Throughput 2018, 7, 19. [Google Scholar] [CrossRef]
- Sanggaard, K.W.; Bechsgaard, J.S.; Fang, X.; Duan, J.; Dyrlund, T.F.; Gupta, V.; Jiang, X.; Cheng, L.; Fan, D.; Feng, Y.; et al. Spider genomes provide insight into composition and evolution of venom and silk. Nat. Commun. 2014, 5, 3765. [Google Scholar] [CrossRef] [PubMed]
- Babb, P.L.; Lahens, N.F.; Correa-Garhwal, S.M.; Nicholson, D.N.; Kim, E.J.; Hogenesch, J.B.; Kuntner, M.; Higgins, L.; Hayashi, C.Y.; Agnarsson, I.; et al. The Nephila clavipes genome highlights the diversity of spider silk genes and their complex expression. Nat. Genet. 2017, 49, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Schwager, E.E.; Sharma, P.P.; Clarke, T.; Leite, D.J.; Wierschin, T.; Pechmann, M.; Akiyama-Oda, Y.; Esposito, L.; Bechsgaard, J.; Bilde, T.; et al. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 2017, 15, 62. [Google Scholar] [CrossRef] [PubMed]
- Garb, J.E.; Sharma, P.P.; Ayoub, N.A. Recent progress and prospects for advancing arachnid genomics. Curr. Opin. Insect Sci. 2018, 25, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Connor, M.; Wilson, D.; Wilson, H.I.; Nicholson, G.M.; Smith, R.; Shaw, D.; Mackay, J.P.; Alewood, P.F.; Christie, M.J.; et al. Discovery and structure of a potent and highly specific blocker of insect calcium channels. J. Biol. Chem. 2001, 276, 40306–40312. [Google Scholar] [CrossRef]
- Diao, J.; Lin, Y.; Tang, J.; Liang, S. cDNA sequence analysis of seven peptide toxins from the spider Selenocosmia huwena. Toxicon 2003, 42, 715–723. [Google Scholar] [CrossRef]
- Pescatori, M.; Bradbury, A.; Bouet, F.; Gargano, N.; Mastrogiacomo, A.; Grasso, A. The cloning of a cDNA encoding a protein (latrodectin) which co-purifies with the alpha-latrotoxin from the black widow spider Latrodectus tredecimguttatus (Theridiidae). Eur. J. Biochem. 1995, 230, 322–328. [Google Scholar] [CrossRef]
- Frohman, M.A.; Dush, M.K.; Martin, G.R. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 1988, 85, 8998–9002. [Google Scholar] [CrossRef]
- Paiva, A.L.B.; Mudadu, M.A.; Pereira, E.H.T.; Marri, C.A.; Guerra-Duarte, C.; Diniz, M.R.V. Transcriptome analysis of the spider Phoneutria pertyi venom glands reveals novel venom components for the genus Phoneutria. Toxicon 2019, 163, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Chen, B.; Xiao, Z.; Zhou, X.; Liu, Z. Transcriptomic Analysis of the Spider Venom Gland Reveals Venom Diversity and Species Consanguinity. Toxins 2019, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wang, Z.; Yu, N.; Li, J.; Liu, Z. Toxin diversity revealed by the venom gland transcriptome of Pardosa pseudoannulata, a natural enemy of several insect pests. Comp. Biochem. Physiol. D Gen. Prot. 2018, 28, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Oldrati, V.; Koua, D.; Allard, P.M.; Hulo, N.; Arrell, M.; Nentwig, W.; Lisacek, F.; Wolfender, J.L.; Kuhn-Nentwig, L.; Stocklin, R. Peptidomic and transcriptomic profiling of four distinct spider venoms. PLoS ONE 2017, 12, e0172966. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.C.; Long, R.W.; Wu, Y.Q.; Guo, Y.B.; Liu, D.L.; Peng, L.; Li, D.Q.; Yang, D.W.; Xu, X.; Liu, F.X.; et al. Identification and characterization of toxins in the venom gland of the Chinese bird spider, Haplopelma hainanum, by transcriptomic analysis. Insect Sci. 2016, 23, 487–499. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, C.; Tan, H.; Wang, H.; Jiang, Y.; Liang, S.; Zhang, F.; Liu, Z. A survey of the venom of the spider Lycosa vittata by biochemical, pharmacological and transcriptomic analyses. Toxicon 2015, 107, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, Y.; He, Q.Z.; Luo, J.; Zhu, L.; Lu, S.S.; Liu, J.Y.; Huang, P.F.; Zeng, X.Z.; Liang, S.P. Structural and Functional Diversity of Peptide Toxins from Tarantula Haplopelma hainanum (Ornithoctonus hainana) Venom Revealed by Transcriptomic, Peptidomic, and Patch Clamp Approaches. J. Biol. Chem. 2015, 290, 14192–14207. [Google Scholar] [CrossRef]
- Xu, X.; Wang, H.; Zhang, F.; Hu, Z.; Liang, S.; Liu, Z. A Comparative Analysis of the Venom Gland Transcriptomes of the Fishing Spiders Dolomedes mizhoanus and Dolomedes sulfurous. PLoS ONE 2015, 10, e0139908. [Google Scholar] [CrossRef]
- Zobel-Thropp, P.A.; Thomas, E.Z.; David, C.L.; Breci, L.A.; Binford, G.J. Plectreurys tristis venome: A proteomic and transcriptomic analysis. J. Venom Res. 2014, 5, 33–47. [Google Scholar]
- Zhang, Y.; Huang, Y.; He, Q.; Liu, J.; Luo, J.; Zhu, L.; Lu, S.; Huang, P.; Chen, X.; Zeng, X.; et al. Toxin diversity revealed by a transcriptomic study of Ornithoctonus huwena. PLoS ONE 2014, 9, e100682. [Google Scholar] [CrossRef]
- Clarke, T.H.; Garb, J.E.; Hayashi, C.Y.; Haney, R.A.; Lancaster, A.K.; Corbett, S.; Ayoub, N.A. Multi-tissue transcriptomics of the black widow spider reveals expansions, co-options, and functional processes of the silk gland gene toolkit. BMC Genom. 2014, 15, 365. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S.A.; Lazarev, V.N.; Kostryukova, E.S.; Selezneva, O.V.; Ospanova, E.A.; Alexeev, D.G.; Govorun, V.M.; Grishin, E.V. Comprehensive analysis of the venom gland transcriptome of the spider Dolomedes fimbriatus. Sci. Data 2014, 1, 140023. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.S.; Hardy, M.C.; Wood, D.; Bailey, T.; King, G.F. SVM-based prediction of propeptide cleavage sites in spider toxins identifies toxin innovation in an Australian tarantula. PLoS ONE 2013, 8, e66279. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Liu, C.; Duan, Z.; Deng, M.; Tang, X.; Liang, S. Transcriptome analysis of venom glands from a single fishing spider Dolomedes mizhoanus. Toxicon 2013, 73, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Cao, R.; Jiang, L.; Liang, S. A combined de novo protein sequencing and cDNA library approach to the venomic analysis of Chinese spider Araneus ventricosus. J. Proteom. 2013, 78, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Ono, S.; Kubo, T. Molecular Cloning and Sequence Analysis of the cDNAs Encoding Toxin-Like Peptides from the Venom Glands of Tarantula Grammostola rosea. Int. J. Pept. 2012, 2012, 731293. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, J.; Tang, X.; Wang, F.; Jiang, L.; Xiong, X.; Wang, M.; Rong, M.; Liu, Z.; Liang, S. Transcriptome analysis of the venom glands of the Chinese wolf spider Lycosa Singoriensis. Zoology 2010, 113, 10–18. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, Y.; Hu, W.; Xu, D.; Tao, H.; Yang, X.; Li, Y.; Jiang, L.; Liang, S. Molecular diversification of peptide toxins from the tarantula Haplopelma hainanum (Ornithoctonus hainana) venom based on transcriptomic, peptidomic, and genomic analyses. J. Proteome Res. 2010, 9, 2550–2564. [Google Scholar] [CrossRef]
- Gremski, L.H.; da Silveira, R.B.; Chaim, O.M.; Probst, C.M.; Ferrer, V.P.; Nowatzki, J.; Weinschutz, H.C.; Madeira, H.M.; Gremski, W.; Nader, H.B.; et al. A novel expression profile of the Loxosceles intermedia spider venomous gland revealed by transcriptome analysis. Mol. Biosyst. 2010, 6, 2403–2416. [Google Scholar] [CrossRef]
- Diego-Garcia, E.; Peigneur, S.; Waelkens, E.; Debaveye, S.; Tytgat, J. Venom components from Citharischius crawshayi spider (Family Theraphosidae): Exploring transcriptome, venomics, and function. Cell. Mol. Life Sci. 2010, 67, 2799–2813. [Google Scholar] [CrossRef]
- Jiang, L.; Peng, L.; Chen, J.; Zhang, Y.; Xiong, X.; Liang, S. Molecular diversification based on analysis of expressed sequence tags from the venom glands of the Chinese bird spider Ornithoctonus huwena. Toxicon 2008, 51, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. 2012, 2012, 251364. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Undheim, E.A.B. True Lies: Using Proteomics to Assess the Accuracy of Transcriptome-Based Venomics in Centipedes Uncovers False Positives and Reveals Startling Intraspecific Variation in Scolopendra Subspinipes. Toxins 2018, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez de la Vega, R.C.; Giraud, T. Intragenome Diversity of Gene Families Encoding Toxin-like Proteins in Venomous Animals. Integr. Comp. Biol. 2016, 56, 938–949. [Google Scholar] [CrossRef] [PubMed]
- Von Reumont, B.M.; Undheim, E.A.B.; Jauss, R.T.; Jenner, R.A. Venomics of Remipede Crustaceans Reveals Novel Peptide Diversity and Illuminates the Venom’s Biological Role. Toxins 2017, 9, 234. [Google Scholar] [CrossRef] [PubMed]
- Barghi, N.; Concepcion, G.P.; Olivera, B.M.; Lluisma, A.O. High conopeptide diversity in Conus tribblei revealed through analysis of venom duct transcriptome using two high-throughput sequencing platforms. Mar. Biotechnol. 2015, 17, 81–98. [Google Scholar] [CrossRef]
- Giordano, F.; Aigrain, L.; Quail, M.A.; Coupland, P.; Bonfield, J.K.; Davies, R.M.; Tischler, G.; Jackson, D.K.; Keane, T.M.; Li, J.; et al. De novo yeast genome assemblies from MinION, PacBio and MiSeq platforms. Sci. Rep. 2017, 7, 3935. [Google Scholar] [CrossRef]
- Bayega, A.; Wang, Y.C.; Oikonomopoulos, S.; Djambazian, H.; Fahiminiya, S.; Ragoussis, J. Transcript Profiling Using Long-Read Sequencing Technologies. Methods Mol. Biol. 2018, 1783, 121–147. [Google Scholar] [CrossRef]
- Goodwin, S.; Gurtowski, J.; Ethe-Sayers, S.; Deshpande, P.; Schatz, M.C.; McCombie, W.R. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 2015, 25, 1750–1756. [Google Scholar] [CrossRef]
- Au, K.F.; Underwood, J.G.; Lee, L.; Wong, W.H. Improving PacBio long read accuracy by short read alignment. PLoS ONE 2012, 7, e46679. [Google Scholar] [CrossRef]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.-C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Highly-accurate long-read sequencing improves variant detection and assembly of a human genome. BioRxiv 2019, 519025. [Google Scholar] [CrossRef]
- Pacific Biosciences of California, Inc. Pacific Biosciences Announces a New Paradigm in DNA Sequencing—Highly Accurate Single-Molecule Long Reads; Pacific Biosciences of California, Inc.: Menlo Park, CA, USA, 2018. [Google Scholar]
- Kircher, M.; Sawyer, S.; Meyer, M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012, 40, e3. [Google Scholar] [CrossRef] [PubMed]
- Illumina, Inc. Effects of Index Misassignment on Multiplexing and Downstream Analysis; Illumina, Inc.: San Diego, CA, USA, 2018. [Google Scholar]
- Costello, M.; Fleharty, M.; Abreu, J.; Farjoun, Y.; Ferriera, S.; Holmes, L.; Granger, B.; Green, L.; Howd, T.; Mason, T.; et al. Characterization and remediation of sample index swaps by non-redundant dual indexing on massively parallel sequencing platforms. BMC Genom. 2018, 19, 332. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.; Stanley, G.; Gulati, G.S.; Ezran, C.; Travaglini, K.J.; Wei, E.; Chan, C.K.F.; Nabhan, A.N.; Su, T.; Morganti, R.M.; et al. Index switching causes “spreading-of-signal” among multiplexed samples in Illumina HiSeq 4000 DNA sequencing. BioRxiv 2017, 125724. [Google Scholar] [CrossRef]
- Griffiths, J.A.; Richard, A.C.; Bach, K.; Lun, A.T.L.; Marioni, J.C. Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun. 2018, 9, 2667. [Google Scholar] [CrossRef]
- Owens, G.L.; Todesco, M.; Drummond, E.B.M.; Yeaman, S.; Rieseberg, L.H. A novel post hoc method for detecting index switching finds no evidence for increased switching on the Illumina HiSeq X. Mol. Ecol. Resour. 2018, 18, 169–175. [Google Scholar] [CrossRef]
- Lomonte, B.; Calvete, J.J. Strategies in ‘snake venomics’ aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J. Venom. Anim. Tox. Incl. Trop. Dis. 2017, 23, 26. [Google Scholar] [CrossRef]
- Petras, D.; Sanz, L.; Segura, A.; Herrera, M.; Villalta, M.; Solano, D.; Vargas, M.; Leon, G.; Warrell, D.A.; Theakston, R.D.; et al. Snake venomics of African spitting cobras: Toxin composition and assessment of congeneric cross-reactivity of the pan-African EchiTAb-Plus-ICP antivenom by antivenomics and neutralization approaches. J. Proteome Res. 2011, 10, 1266–1280. [Google Scholar] [CrossRef]
- Correa-Netto, C.; Teixeira-Araujo, R.; Aguiar, A.S.; Melgarejo, A.R.; De-Simone, S.G.; Soares, M.R.; Foguel, D.; Zingali, R.B. Immunome and venome of Bothrops jararacussu: A proteomic approach to study the molecular immunology of snake toxins. Toxicon 2010, 55, 1222–1235. [Google Scholar] [CrossRef]
- Calvete, J.J.; Petras, D.; Calderon-Celis, F.; Lomonte, B.; Encinar, J.R.; Sanz-Medel, A. Protein-species quantitative venomics: Looking through a crystal ball. J. Venom. Anim. Tox. Incl. Trop. Dis. 2017, 23, 27. [Google Scholar] [CrossRef]
- Liao, Z.; Cao, J.; Li, S.; Yan, X.; Hu, W.; He, Q.; Chen, J.; Tang, J.; Xie, J.; Liang, S. Proteomic and peptidomic analysis of the venom from Chinese tarantula Chilobrachys jingzhao. Proteomics 2007, 7, 1892–1907. [Google Scholar] [CrossRef] [PubMed]
- Santana, R.C.; Perez, D.; Dobson, J.; Panagides, N.; Raven, R.J.; Nouwens, A.; Jones, A.; King, G.F.; Fry, B.G. Venom Profiling of a Population of the Theraphosid Spider Phlogius crassipes Reveals Continuous Ontogenetic Changes from Juveniles through Adulthood. Toxins 2017, 9, 116. [Google Scholar] [CrossRef] [PubMed]
- Trevisan-Silva, D.; Bednaski, A.V.; Fischer, J.S.G.; Veiga, S.S.; Bandeira, N.; Guthals, A.; Marchini, F.K.; Leprevost, F.V.; Barbosa, V.C.; Senff-Ribeiro, A.; et al. A multi-protease, multi-dissociation, bottom-up-to-top-down proteomic view of the Loxosceles intermedia venom. Sci. Data 2017, 4, 170090. [Google Scholar] [CrossRef] [PubMed]
- Melani, R.D.; Nogueira, F.C.S.; Domont, G.B. It is time for top-down venomics. J. Venom. Anim. Tox. Incl. Trop. Dis. 2017, 23, 44. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I.; Aebersold, R. Interpretation of shotgun proteomic data: The protein inference problem. Mol. Cell. Proteom. 2005, 4, 1419–1440. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Gomez, S.; Vargas-Munoz, L.J.; Saldarriaga-Cordoba, M.; Cifuentes, Y.; Perafan, C. Identifying different transcribed proteins in the newly described Theraphosidae Pamphobeteus verdolaga. Toxicon 2017, 129, 81–88. [Google Scholar] [CrossRef]
- Duan, Z.; Yan, X.; Cao, R.; Liu, Z.; Wang, X.; Liang, S. Proteomic analysis of Latrodectus tredecimguttatus venom for uncovering potential latrodectism-related proteins. J. Biochem. Mol. Toxicol. 2008, 22, 328–336. [Google Scholar] [CrossRef]
- Ghezellou, P.; Garikapati, V.; Kazemi, S.M.; Strupat, K.; Ghassempour, A.; Spengler, B. A perspective view of top-down proteomics in snake venom research. Rapid Commun. Mass Spectrom. 2018. [Google Scholar] [CrossRef]
- Pla, D.; Petras, D.; Saviola, A.J.; Modahl, C.M.; Sanz, L.; Perez, A.; Juarez, E.; Frietze, S.; Dorrestein, P.C.; Mackessy, S.P.; et al. Transcriptomics-guided bottom-up and top-down venomics of neonate and adult specimens of the arboreal rear-fanged Brown Treesnake, Boiga irregularis, from Guam. J. Proteom. 2018, 174, 71–84. [Google Scholar] [CrossRef]
- Petras, D.; Heiss, P.; Harrison, R.A.; Sussmuth, R.D.; Calvete, J.J. Top-down venomics of the East African green mamba, Dendroaspis angusticeps, and the black mamba, Dendroaspis polylepis, highlight the complexity of their toxin arsenals. J. Proteom. 2016, 146, 148–164. [Google Scholar] [CrossRef]
- Petras, D.; Heiss, P.; Sussmuth, R.D.; Calvete, J.J. Venom Proteomics of Indonesian King Cobra, Ophiophagus hannah: Integrating Top-Down and Bottom-Up Approaches. J. Proteome Res. 2015, 14, 2539–2556. [Google Scholar] [CrossRef] [PubMed]
- Melani, R.D.; Skinner, O.S.; Fornelli, L.; Domont, G.B.; Compton, P.D.; Kelleher, N.L. Mapping Proteoforms and Protein Complexes from King Cobra Venom Using Both Denaturing and Native Top-down Proteomics. Mol. Cell. Proteom. 2016, 15, 2423–2434. [Google Scholar] [CrossRef] [PubMed]
- Gocmen, B.; Heiss, P.; Petras, D.; Nalbantsoy, A.; Sussmuth, R.D. Mass spectrometry guided venom profiling and bioactivity screening of the Anatolian Meadow Viper, Vipera anatolica. Toxicon 2015, 107, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Calderon-Celis, F.; Cid-Barrio, L.; Encinar, J.R.; Sanz-Medel, A.; Calvete, J.J. Absolute venomics: Absolute quantification of intact venom proteins through elemental mass spectrometry. J. Proteom. 2017, 164, 33–42. [Google Scholar] [CrossRef]
- Calderon-Celis, F.; Diez-Fernandez, S.; Costa-Fernandez, J.M.; Encinar, J.R.; Calvete, J.J.; Sanz-Medel, A. Elemental Mass Spectrometry for Absolute Intact Protein Quantification without Protein-Specific Standards: Application to Snake Venomics. Anal. Chem. (Wash.) 2016, 88, 9699–9706. [Google Scholar] [CrossRef]
- Jungo, F.; Bougueleret, L.; Xenarios, I.; Poux, S. The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon 2012, 60, 551–557. [Google Scholar] [CrossRef]
- Bateman, A.; Haft, D.H. HMM-based databases in InterPro. Brief. Bioinform. 2002, 3, 236–245. [Google Scholar] [CrossRef][Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef]
- Koua, D.; Kuhn-Nentwig, L. Spider Neurotoxins, Short Linear Cationic Peptides and Venom Protein Classification Improved by an Automated Competition between Exhaustive Profile HMM Classifiers. Toxins 2017, 9, 245. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef] [PubMed]

| Prefix | Target | Action | Example |
|---|---|---|---|
| ω (omega) | CaV channels | Inhibits CaV channels | omega-agatoxin-1a, P15969 |
| κ (kappa) | KV channels | Inhibits KV channels | kappa-hexatoxin-Hv1c, P82228 |
| β (beta) | NaV channels | Shifts voltage dependence of NaV channel activation | beta-hexatoxin-Mg1a, P83561 |
| δ (delta) | NaV channels | Delays inactivation of NaV channels | delta-miturgitoxin-Cp1b, C0HKG8 |
| µ (mu) | NaV channels | Inhibits NaV channels | mu-diguetoxin-Dc1a, P49126 |
| M | membrane | Membranolytic activity | M-ctenitoxin-Cs1a, P83619 |
| U | unknown | Unknown activity | U2-ctenitoxin-Cs1a, P83919 |
| Spider Species | Sex/Number of Specimens/Time Since Last Milking | Sequencing Method | Total EST/Contigs | Isolation of Sequences Based on | Identified Toxins/Transcripts | Ref. |
|---|---|---|---|---|---|---|
| Parasteatoda tepidariorum */** (Theridiidae) |  /adult/3 d since last meal, no milking /adult/3 d since last meal, no milking | Illumina HiSeq 4000 (2 × 100 bp) 2 replicates | NA, aligned to genome assembly | differential expression analysis | 1318 upregulated transcripts | [23] |
| Phoneutria pertyi (Ctenidae) |  /10 ad./48 h /10 ad./48 h | Sanger (1305 clones) | 106 contigs, 189 singletons | Basic Local Alignment Search Tool (BLAST) (UniProtKB) non-hits manual | 63 transcripts of cys-rich peptides | [222] |
| Cupiennius salei * (Trechaleidae) |  /20 ad./24, 48, 62 h; 8, 14 d /20 ad./24, 48, 62 h; 8, 14 d | 454 GS-FLX | 34,107 contigs | BLAST (UniProtKB), signal peptide, HMM, cys-pattern | 81 transcripts of cys-rich peptides, 56 mature peptides | [107] |
| Selenocosmia jiafu (Theraphosidae) | NA/6/4 d | Sanger (1299 clones) | 752 ESTs, 61 contigs, 196 singletons | BLAST (UniProtKB, nrNCBI), signal peptide | 257 transcripts, 99 mature peptides | [223] |
| Tetragnatha versicolor (Tetragnathidae) |  /10/2, 3 d /10/2, 3 d /2/2, 3 d /2/2, 3 danalysed by sex | Illumina (2 × 50 bp) HiSeq 2500 |  16,799 contigs, 16,799 contigs,  24,351 contigs 24,351 contigs |  / / Reads remapped against combined assembly, TRINOTATE pipeline Reads remapped against combined assembly, TRINOTATE pipeline | NA, 9177 dimorphic, 1404 non-dimorphic | [35] |
| Pardosa pseudoannulata ** (Lycosidae) |  /15 ad./NA /15 ad./NA  /15 ad./NA /15 ad./NA  /45 ad./NA (tissue expression profiling) /45 ad./NA (tissue expression profiling) | Illumina HiSeq | 75,980 contigs | BLAST (nrNCBI, ntNCBI, SwissProt), domain prediction, GO, cys pattern | 48 potential peptide toxins | [224] |
| Phoneutria nigriventer * (Ctenidae) |  /20 ad./48 h /20 ad./48 h /10 ad./48 h /10 ad./48 h | Illumina (2 × 151 bp) HiSeq 1500 Sanger (1476 electrograms) | 49,992 contigs 1224 ESTs, 132 contigs, 162 singletons | BLAST (UniProtKB, TSA), domain prediction, GO, expression level BLAST (UniProtKB), ORF, signal peptide, propeptide | 99 or 98 cys-rich peptide toxins | [154] |
| Poecilotheria Formosa * (Theraphosidae) Viridasius fasciatus * (Viridasiidae) Latrodectus mactans * (Theridiidae) Heteropoda davidbowie * (Sparassidae) | NA/9 ad./ 0, 2, 3, 4 d NA/10 ad./ 0, 2, 3, 4 d NA/39 ad./ 0, 2, 3, 4 d NA/9 ad./ 0, 3, 7 d | Ion Torrent | 94,148 contigs 330,060 contigs 301,423 contigs 239,749 contigs | private HMMs based on ArachnoServer sequences, read count threshold | 37 toxins 41 toxins 10 toxins29 toxins | [225] |
| Cyriopagopus hainanus (sub Haplopelma hainana) (Theraphosidae) |  /1 ad./NA /1 ad./NA | Illumina (2 × 101 bp) HiSeq 2000 | 57,181 contigs | BLAST (Toxprot, UniProtKB), cys count ≥ 5, domain prediction | 201 potential toxins | [226] |
| Lycosa vittata (Lycosidae) |  /6 ad./NA /6 ad./NA | Sanger (500 clones) | NA | NA | 51 toxin-like peptides | [227] |
| Cyriopagopus hainanus (sub Haplopelma hainana) (Theraphosidae) | NA/NA/2 d | 454 GS-FLX | 65,432 contigs | BLAST (EST NCBI, nrNCBI), ORF, cys count ≥ 4, length ≥ 45 amino acids | 1136 potential precursors | [228] |
| Dolomedes sulfureus (Pisauridae) |  /10 ad./4 d /10 ad./4 d | Sanger (500 clones) | 267 ESTs, 25 contigs, 58 singletons | BLAST (nrNCBI, UniProtKB) | 127 putative toxin precursors, 90 mature peptides | [229] |
| Plectreurys tristis * (Plectreuridae) |  /5/3, 4 d /5/3, 4 d | Sanger (1717 clones) | 307 ESTs, 37 contigs, 105 singletons | BLAST (ArachnoServer, NCBI, private dbs) | 19 putative peptide transcripts | [230] |
| Cyriopagopus schmidti (sub Ornithoctonus huwena) (Theraphosidae) | NA/3/2 d | 454 GS FLX Titanium | 4224 contigs | BLAST (UniProtKB, ToxRelDB, Repbase), cys pattern | 626 toxin precursors, 90 mature peptides | [231] |
| Latrodectus Hesperus */** (Theridiidae) |  /7/NA /7/NA | Illumina (2 × 100 bp) | 85,193 contigs | BLAST (UniProtKB), ORF prediction, differential expression analysis | 695 venom gland specific transcripts | [22] [232] |
| Dolomedes fimbriatus (Pisauridae) | NA/several/7 d | Sanger (5952 clones) | 451 contigs | cys pattern search, signal peptide, propeptide | 451 transcripts, 163 mature peptides | [233] |
| Selenotypus plumipes (Theraphosidae) | NA/2/NA | 454 GS-FLX | 136,469 six-frame translated sequences | ORF prediction, BLAST (ArachnoServer) signal peptide, propeptide, cys count | 970 mature (likely to be an overestimate) | [234] |
| Trittame lok i * (Barychelidae) |  /9/4 d /9/4 d | 454 GS FLX Titanium | 4711 contigs | BLAST UniProtKB | 46 full-length toxin precursors | [21] |
| Dolomedes mizhoanus (Pisauridae) |  /1/4 d /1/4 d | Sanger | 356 ESTs, 19 contigs, 26 singletons | BLAST (nrNCBI, UniProtKB) signal peptide, SpiderP | 53 or 55 cys-knot toxin precursors, 48 mature peptides | [235] |
| Latrodectus tredecimguttatus (Theridiidae) | NA/3 ad./NA  /15/4 d /15/4 d | Illumina (2 × 90 bp) HiSeq 2000 Sanger | 34,334 contigs 1015 unique ESTs | ORF prediction, BLAST (UniProtKB), Cys-pattern, domain prediction (SMART/Pfam) | 146 toxin-like proteins | [24] |
| Araneus ventricosus * (Araneidae) |  /20/3 d /20/3 d | Sanger | 886 ESTs | ≥4 cys, signal peptide | 200 toxin-like precursors | [236] |
| Grammostola rosea (Theraphosidae) | NA/30/NA | Sanger (1500 clones) | 869 ESTs | BLAST | 48 peptides | [237] |
| Lycosa singoriensis (Lycosidae) | NA/20/4 d | Sanger | 833 ESTs | BLAST (nrNCBI, UniProtKB) | 223 toxin-like transcripts | [238] |
| Cyriopagopus hainanus * (sub Ornithoctonus hainana) (Theraphosidae) |  /20 ad./NA /20 ad./NA | Sanger (1049 clones) | NA | BLAST, pairing with proteomic data from N-term sequencing (Edman) | 88 peptide toxins | [239] |
| Loxosceles intermedia (Sicariidae) |   /350/5 d /350/5 d | Sanger (2400 clones) | 1843 ESTs, 257 contigs, 281 singletons | BLAST(GenBank) | 88 contigs, 80 singletons (toxin sequences) | [240] |
| Pelinobius muticus * (sub Citharischius crawshayi) (Theraphosidae) | NA/1/2 d | Sanger (282 clones) | 236 ESTs, 14 contigs, 30 singletons | BLAST (GenBank, ArachnoServer) | 11 toxin-like, 3 putative toxin transcripts | [241] |
| Cyriopagopus schmidti (sub Ornithoctonus huwena) (Theraphosidae) | NA/20/4 d | Sanger | 468 ESTs, 24 contigs, 65 singletons | BLAST (nrNCBI, UniProtKB) | 31 mature peptides | [242] |
| Loxosceles laeta (Sicariidae) |  /100/5 d /100/5 d | Sanger | 3008 ESTs, 326 contigs, 1031 singletons | BLAST, domain prediction (SMART/Pfam), signal peptide | 93 clusters of known toxins, 117 clusters of possible toxins | [18] |
| Agelena orientalis (Agelenidae) | NA/NA/NA | Sanger (2166 clones) | 37 contigs, 332 singletons | BLAST (GenBank, peptide sequence databases) | 48 toxin-like structures | [17] |
| Macrothele gigas * (Dipluridae) | NA/2 glands/NA | Sanger (300 clones) | NA | NA | 10 multi-cys peptides | [16] |
| Spider Species | Top-Down | Bottom-Up | Venom Pre-Fractionation | Digest | Post-Fractionation | Data Analysis Type | Database | Identified Toxins | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Parasteatoda tepidariorum * (Theridiidae) | X | SDS-PAGE (8 fractions) | in-gel trypsin | online RP-HPLC | db-dependend | predicted transcripts from genome assembly | ≥99 distinct proteins | [23] | |
| Cupiennius salei * (Trechaleidae) | X | SEC, desalting | trypsin, chymotrypsin | online RP-HPLC | db-dependend | UniProtKB Araneae sequences + identified transcripts | 49 peptides, ≥23 proteins | [107] | |
| X | SEC, desalting, online RP-HPLC | - | - | db-dependend | |||||
| Tetragnatha versicolor * (Tetragnathidae) | X | SDS-PAGE (3 fractions) | in-gel trypsin | NA, (probably online RP-HPLC) | db-dependend | translated transcriptome sequences + NCBI chelicerate sequences | 62 distinct proteins in 31 clusters | [35] | |
| Phoneutria nigriventer * (Ctenidae) | X | - | trypsin | MudPIT | db-dependend | predicted proteins from transcriptome | 29 cys-rich peptide toxins | [154] | |
| Selenocosmia crassipes (sub Phlogius crassipes) (Theraphosidae) | X | - | trypsin | online RP-HPLC | db-dependend | UniProtKB Arthropoda sequences | Ontogenetic analysis, depending on group: 12, 11, 8, or 8 | [265] | |
| X | SDS-PAGE | in-gel trypsin | online RP-HPLC | ||||||
| Pamphobeteus verdolaga * (Theraphosidae) | X | RP-HPLC | Lys-C/trypsin | online RP-HPLC | db-dependend | some with sequences from UniProt, NCBI or the ArachnoServer | 16 peptides | [269] | |
| Loxosceles intermedia (Sicariidae) | X | 10 kDa cutoff spin-column | trypsin, pepsin, chymotrypsin | online RP-HPLC/ MudPIT | de-novo, spectral-network algorithmes | de-novo sequences against Loxosceles UniProt sequences + L. intermedia vg transcriptome | 190 proteins | [266] | |
| Poecilotheria Formosa * (Theraphosidae) Viridasius fasciatus * (Viridasiidae) Latrodectus mactans * (Theridiidae) Heteropoda davidbowie * (Sparassidae) | X | solid phase extraction | NA | online RP-HPLC | db-dependend | assembled contigs from transcriptome sequencing | 103 9 4 68 | [225] | |
| Plectreurys tristis * (Plectreuridae) | X | SDS-PAGE | in-gel trypsin | - | db-dependend | plectreurid transcriptome, private haplogyne vg cDNA libraries, chelicerate seq. in NCBI | 7 astacins-like groups, 7 peptide groups, 1 unknown | [230] | |
| Latrodectus Hesperus * (Theridiidae) | X | NA | trypsin | MudPIT | db-dependend | Latrodectus hesperus seq. on NCBI, ESTs (vg specific) + assembled transcriptome | 61 | [22] | |
| Trittame loki * (Barychelidae) | X | NA | trypsin | online RP-HPLC | db-dependend | translated cDNA library | 45 | [21] | |
| X | online RP-HPLC | - | - | ||||||
| Araneus ventricosus * (Araneidae) | X | 2D gel electrophoresis (106 spots) | in-gel trypsin | online RP-HPLC | db-dependend, de-novo | NCBInr with animal species restriction, de-novo sequences matched against transcriptome | db-dependend (functional analysis): 65 de-novo: 130 | [236] | |
| X | - | trypsin | online RP-HPLC | ||||||
| Latrodectus tredecimguttatus (Theridiidae) | X | SDS-PAGE | in-gel trypsin | online RP-HPLC | db-dependend, de-novo | NCBInr with animal species restriction | 75 venom proteins | [270] | |
| X | - | trypsin | online RP-HPLC | ||||||
| Chilobrachys guangxiensis (sub Chilobrachys jingzhao) (Theraphosidae) | X | SEC, > 10 kDa: 2D gel electrophoreses, | in-gel trypsin | online RP-HPLC, - | db-dependend manually de-novo | raw genome data of the arthropod; de-novo: MS-BLAST | 47 from in-gel digestion | [264] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Langenegger, N.; Nentwig, W.; Kuhn-Nentwig, L. Spider Venom: Components, Modes of Action, and Novel Strategies in Transcriptomic and Proteomic Analyses. Toxins 2019, 11, 611. https://doi.org/10.3390/toxins11100611
Langenegger N, Nentwig W, Kuhn-Nentwig L. Spider Venom: Components, Modes of Action, and Novel Strategies in Transcriptomic and Proteomic Analyses. Toxins. 2019; 11(10):611. https://doi.org/10.3390/toxins11100611
Chicago/Turabian StyleLangenegger, Nicolas, Wolfgang Nentwig, and Lucia Kuhn-Nentwig. 2019. "Spider Venom: Components, Modes of Action, and Novel Strategies in Transcriptomic and Proteomic Analyses" Toxins 11, no. 10: 611. https://doi.org/10.3390/toxins11100611
APA StyleLangenegger, N., Nentwig, W., & Kuhn-Nentwig, L. (2019). Spider Venom: Components, Modes of Action, and Novel Strategies in Transcriptomic and Proteomic Analyses. Toxins, 11(10), 611. https://doi.org/10.3390/toxins11100611




