Purification and Characterization of a Novel Insecticidal Toxin, μ-sparatoxin-Hv2, from the Venom of the Spider Heteropoda venatoria
Abstract
1. Introduction
2. Results
2.1. Characterization of μ-SPRTX-Hv2 as a Cockroach NaVs Toxin
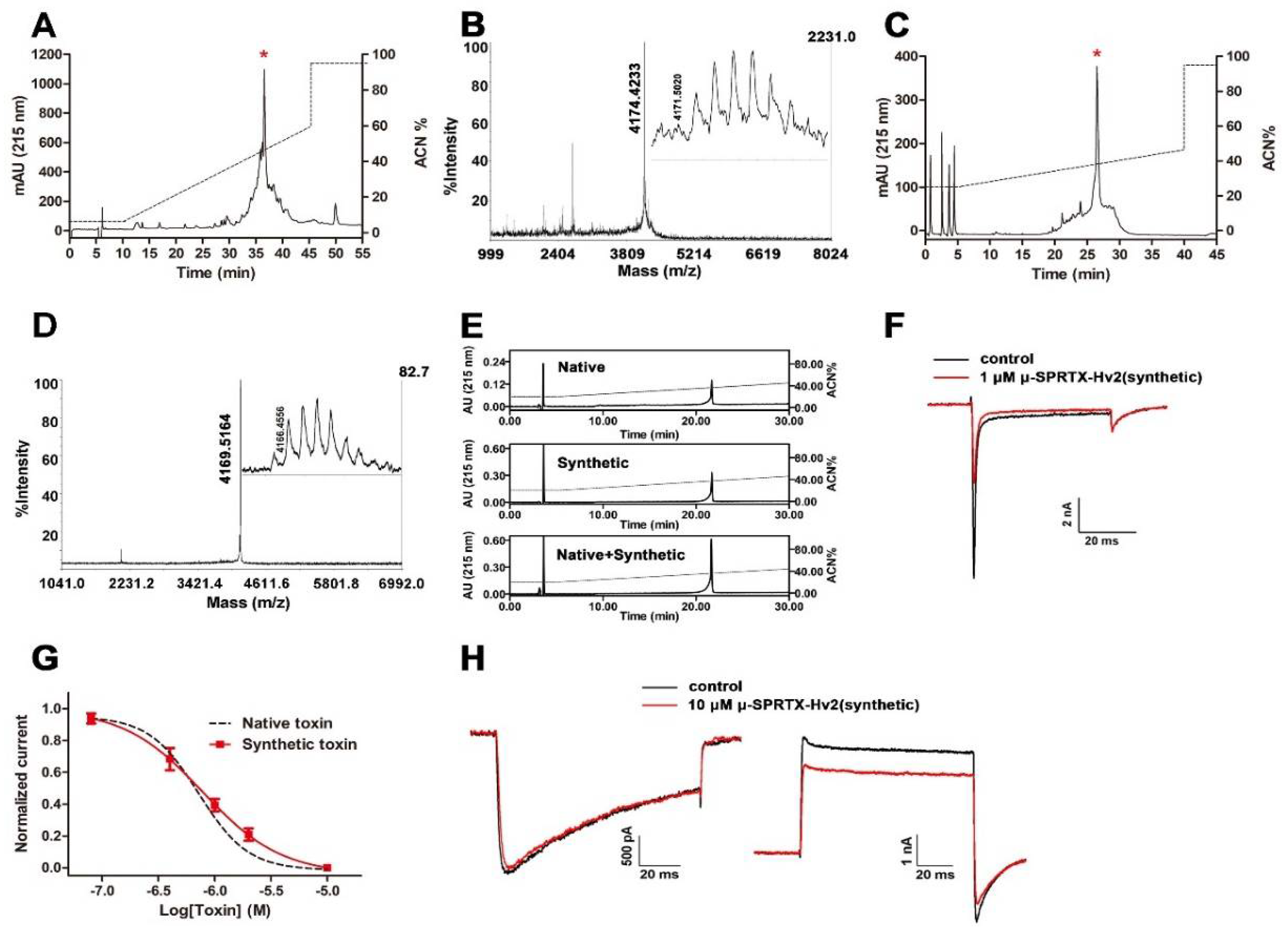
2.2. μ-SPRTX-Hv2 Synthesis and Activity Assay
2.3. μ-SPRTX-Hv2 did not Affect Gating Kinetics of DUM NaVs
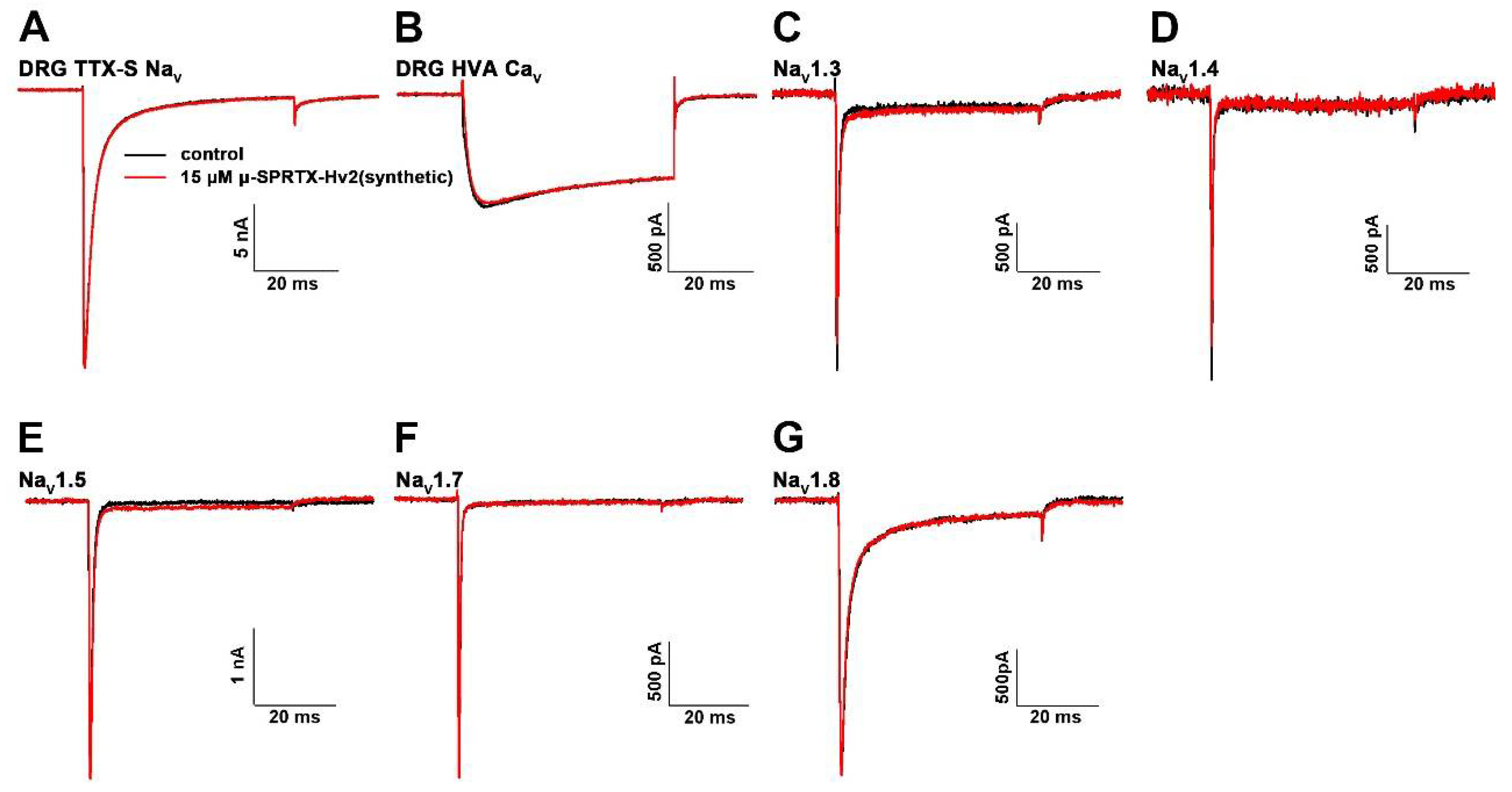
2.4. μ-SPRTX-Hv2 did not Act on Mammalian NaVs and CaVs
3. Discussion
4. Materials and Methods
4.1. Venom and Toxin Purification
4.2. Toxin Sequence Determination
4.3. Solid-Phase Peptide Synthesis
4.4. Bioactivity Assays
4.5. Acute Dissociation and Culture of Rat DRG and Insect DUM Neurons
4.6. Whole-Cell Currents Recording
4.7. Data Analysis
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Vassilevski, A.A.; Kozlov, S.A.; Grishin, E.V. Molecular diversity of spider venom. Biochemistry (Mosc.) 2009, 74, 1505–1534. [Google Scholar] [CrossRef] [PubMed]
- Escoubas, P.; Sollod, B.; King, G.F. Venom landscapes: Mining the complexity of spider venoms via a combined cdna and mass spectrometric approach. Toxicon 2006, 47, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Pineda, S.S.; Chaumeil, P.A.; Kunert, A.; Kaas, Q.; Thang, M.W.C.; Le, L.; Nuhn, M.; Herzig, V.; Saez, N.J.; Cristofori-Armstrong, B.; et al. Arachnoserver 3.0: An online resource for automated discovery, analysis and annotation of spider toxins. Bioinformatics 2018, 34, 1074–1076. [Google Scholar] [CrossRef] [PubMed]
- Casida, J.E. Pest toxicology: The primary mechanisms of pesticide action. Chem. Res. Toxicol. 2009, 22, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Sattelle, D.B.; Cordova, D.; Cheek, T.R. Insect ryanodine receptors: Molecular targets for novel pest control chemicals. Invertebr. Neurosci. 2008, 8, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, S.G.; Garcia, M.E.; Valentim, A.C.; Cordeiro, M.N.; Diniz, C.R.; Richardson, M. Purification and amino acid sequence of the insecticidal neurotoxin T×4(6-1) from the venom of the ‘armed’ spider Phoneutria nigriventer (keys). Toxicon 1995, 33, 83–93. [Google Scholar] [CrossRef]
- Corzo, G.; Gilles, N.; Satake, H.; Villegas, E.; Dai, L.; Nakajima, T.; Haupt, J. Distinct primary structures of the major peptide toxins from the venom of the spider macrothele gigas that bind to sites 3 and 4 in the sodium channel. FEBS Lett. 2003, 547, 43–50. [Google Scholar] [CrossRef]
- Corzo, G.; Escoubas, P.; Stankiewicz, M.; Pelhate, M.; Kristensen, C.P.; Nakajima, T. Isolation, synthesis and pharmacological characterization of delta-palutoxins IT, novel insecticidal toxins from the spider Paracoelotes luctuosus (Amaurobiidae). Eur. J. Biochem. 2000, 267, 5783–5795. [Google Scholar] [CrossRef] [PubMed]
- Bende, N.S.; Dziemborowicz, S.; Herzig, V.; Ramanujam, V.; Brown, G.W.; Bosmans, F.; Nicholson, G.M.; King, G.F.; Mobli, M. The insecticidal spider toxin sfi1 is a knottin peptide that blocks the pore of insect voltage-gated sodium channels via a large beta-hairpin loop. FEBS J. 2015, 282, 904–920. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.; Peigneur, S.; Tytgat, J. Neurotoxins and their binding areas on voltage-gated sodium channels. Front. Pharmacol. 2011, 2, 71. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Fitches, E.; Pyati, P.; Gatehouse, J.A. Effect of insecticidal fusion proteins containing spider toxins targeting sodium and calcium ion channels on pyrethroid-resistant strains of peach-potato aphid (Myzus persicae). Pest Manag. Sci. 2015, 71, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Bende, N.S.; Dziemborowicz, S.; Mobli, M.; Herzig, V.; Gilchrist, J.; Wagner, J.; Nicholson, G.M.; King, G.F.; Bosmans, F. A distinct sodium channel voltage-sensor locus determines insect selectivity of the spider toxin dc1a. Nat. Commun. 2014, 5, 4350. [Google Scholar] [CrossRef] [PubMed]
- Herzig, V.; Ikonomopoulou, M.; Smith, J.J.; Dziemborowicz, S.; Gilchrist, J.; Kuhn-Nentwig, L.; Rezende, F.O.; Moreira, L.A.; Nicholson, G.M.; Bosmans, F.; et al. Molecular basis of the remarkable species selectivity of an insecticidal sodium channel toxin from the african spider Augacephalus ezendami. Sci. Rep. 2016, 6, 29538. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Smith, R.; Fletcher, J.I.; Wilson, H.; Wood, C.J.; Howden, M.E.; King, G.F. Structure-function studies of omega-atracotoxin, a potent antagonist of insect voltage-gated calcium channels. Eur. J. Biochem. 1999, 264, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Connor, M.; Wilson, D.; Wilson, H.I.; Nicholson, G.M.; Smith, R.; Shaw, D.; Mackay, J.P.; Alewood, P.F.; Christie, M.J.; et al. Discovery and structure of a potent and highly specific blocker of insect calcium channels. J. Biol. Chem. 2001, 276, 40306–40312. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Luo, X.; Meng, E.; Xiao, Y.; Liang, S. Inhibition of insect calcium channels by huwentoxin-v, a neurotoxin from Chinese tarantula Ornithoctonus huwena venom. Eur. J. Pharmacol. 2008, 582, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Dhadialla, T.S.; Gill, S.S. Insect midgut and insecticidal proteins. In Advances in Insect Physiology; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Jin, L.; Fang, M.; Chen, M.; Zhou, C.; Ombati, R.; Hakim, M.A.; Mo, G.; Lai, R.; Yan, X.; Wang, Y.; et al. An insecticidal toxin from nephila clavata spider venom. Amino Acids 2017, 49, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Herzig, V.; Ikonomopoulou, M.P.; Dziemborowicz, S.; Bosmans, F.; Nicholson, G.M.; King, G.F. Insect-active toxins with promiscuous pharmacology from the African theraphosid spider Monocentropus balfouri. Toxins (Basel) 2017, 9, 155. [Google Scholar] [CrossRef] [PubMed]
- Ikonomopoulou, M.P.; Smith, J.J.; Herzig, V.; Pineda, S.S.; Dziemborowicz, S.; Er, S.Y.; Durek, T.; Gilchrist, J.; Alewood, P.F.; Nicholson, G.M.; et al. Isolation of two insecticidal toxins from venom of the Australian theraphosid spider Coremiocnemis tropix. Toxicon 2016, 123, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Song, B.; Mo, G.; Yuan, M.; Li, H.; Wang, P.; Yuan, M.; Lu, Q. A novel neurotoxin from venom of the spider, Brachypelma albopilosum. PLoS ONE 2014, 9, e110221. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wu, X.; Peng, Z.; Duan, Z.; Xi, Z.; Chen, M.; Farooq, A.; Liang, S.; Liu, Z. Peptide-rich venom from the spider Heteropoda venatoria potently inhibits insect voltage-gated sodium channels. Toxicon 2016, 125, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.C.; Johnson, J.H.; Hammerland, L.G.; Kelbaugh, P.R.; Volkmann, R.A.; Saccomano, N.A.; Mueller, A.L. Heteropodatoxins: Peptides isolated from spider venom that block kv4.2 potassium channels. Mol. Pharmacol. 1997, 51, 491–498. [Google Scholar] [PubMed]
- King, G.F.; Gentz, M.C.; Escoubas, P.; Nicholson, G.M. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon 2008, 52, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Peigneur, S.; Beress, L.; Moller, C.; Mari, F.; Forssmann, W.G.; Tytgat, J. A natural point mutation changes both target selectivity and mechanism of action of sea anemone toxins. FASEB J. 2012, 26, 5141–5151. [Google Scholar] [CrossRef] [PubMed]
- Loughney, K.; Kreber, R.; Ganetzky, B. Molecular analysis of the para locus, a sodium channel gene in drosophila. Cell 1989, 58, 1143–1154. [Google Scholar] [CrossRef]
- Dong, K.; Du, Y.; Rinkevich, F.; Nomura, Y.; Xu, P.; Wang, L.; Silver, K.; Zhorov, B.S. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem. Mol. Biol. 2014, 50, 1–17. [Google Scholar] [CrossRef] [PubMed]
- King, G.F.; Escoubas, P.; Nicholson, G.M. Peptide toxins that selectively target insect NaV and CaV channels. Channels 2008, 2, 100–116. [Google Scholar] [CrossRef] [PubMed]
- Dong, K. A single amino acid change in the para sodium channel protein is associated with knockdown-resistance (kdr) to pyrethroid insecticides in German cockroach. Insect Biochem. Mol. Biol. 1997, 27, 93–100. [Google Scholar] [CrossRef]
- Tan, J.; Liu, Z.; Nomura, Y.; Goldin, A.L.; Dong, K. Alternative splicing of an insect sodium channel gene generates pharmacologically distinct sodium channels. J. Neurosci. 2002, 22, 5300–5309. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Liu, Z.; Tan, J.; Nomura, Y.; Dong, K. RNA editing generates tissue-specific sodium channels with distinct gating properties. J. Biol. Chem. 2004, 279, 32554–32561. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Bingham, J.P.; Zhu, W.; Moczydlowski, E.; Liang, S.; Cummins, T.R. Tarantula huwentoxin-IV inhibits neuronal sodium channels by binding to receptor site 4 and trapping the domain II voltage sensor in the closed configuration. J. Biol. Chem. 2008, 283, 27300–27313. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Xiao, Z.; Xu, Y.; Zeng, J.; Peng, D.; Liang, S.; Tang, C.; Liu, Z. The peptide toxin delta-hexatoxin-MrIX inhibits fast inactivation of NaVs in mouse cerebellar granule cells. Peptides 2018, 102, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Wigger, E.; Kuhn-Nentwig, L.; Nentwig, W. The venom optimisation hypothesis: A spider injects large venom quantities only into difficult prey types. Toxicon 2002, 40, 749–752. [Google Scholar] [CrossRef]
- Cooper, A.M.; Nelsen, D.R.; Hayes, W.K. The strategic use of venom by spiders. Evol. Venom. Anim. Their Toxins 2015, 1–18. [Google Scholar]
- Hardy, M.C.; Daly, N.L.; Mobli, M.; Morales, R.A.V.; King, G.F. Isolation of an orally active insecticidal toxin from the venom of an Australian tarantula. PLoS ONE 2013, 8, e73136. [Google Scholar] [CrossRef] [PubMed]
- Fitches, E.; Audsley, N.; Gatehouse, J.A.; Edwards, J.P. Fusion proteins containing neuropeptides as novel insect contol agents: Snowdrop lectin delivers fused allatostatin to insect haemolymph following oral ingestion. Insect Biochem. Mol. Biol. 2002, 32, 1653–1661. [Google Scholar] [CrossRef]
- Fitches, E.; Woodhouse, S.D.; Edwards, J.P.; Gatehouse, J.A. In vitro and in vivo binding of snowdrop (Galanthus nivalis agglutinin; GNA) and jackbean (Canavalia ensiformis; Con A) lectins within tomato moth (Lacanobia oleracea) larvae; mechanisms of insecticidal action. J. Insect Physiol. 2001, 47, 777–787. [Google Scholar] [CrossRef]
- Nakasu, E.Y.; Williamson, S.M.; Edwards, M.G.; Fitches, E.C.; Gatehouse, J.A.; Wright, G.A.; Gatehouse, A.M. Novel biopesticide based on a spider venom peptide shows no adverse effects on honeybees. Proc. Biol. Sci. 2014, 281, 20140619. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Leger, R.J.S. A scorpion neurotoxin increases the potency of a fungal insecticide. Nat. Biotechnol. 2007, 25, 1455. [Google Scholar] [CrossRef] [PubMed]
- Lovett, B.; St Leger, R.J. Genetically engineering better fungal biopesticides. Pest Manag. Sci. 2018, 74, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhou, X.; Chen, J.; Tang, C.; Xiao, Z.; Ying, D.; Liu, Z.; Liang, S. The venom of the spider selenocosmia jiafu contains various neurotoxins acting on voltage-gated ion channels in rat dorsal root ganglion neurons. Toxins 2014, 6, 988–1001. [Google Scholar] [CrossRef] [PubMed]
- Pi, J.; Quan, M.; Zeng, X. Solid-phase chemical synthesis and oxidative refolding of hainantoxin-III. Chin. J. Chromatogr. 2007, 25, 399–403. [Google Scholar] [CrossRef]
- Liang, S.; Qin, Y.; Zhang, D.; Pan, X.; Chen, X.; Xie, J. Biological characterization of spider (Selenocosmia huwena) crude venom. Zool. Res. 1993, 14, 60–65. [Google Scholar]
- Hu, H.Z.; Li, Z.W. Substance p potentiates ATP-activated currents in rat primary sensory neurons. Brain Res. 1996, 739, 163–168. [Google Scholar] [CrossRef]
- Forsyth, P.; Sevcik, C.; Martinez, R.; Castillo, C.; D’Suze, G. Bactridine’s effects on dum cricket neurons under voltage clamp conditions. J. Insect Physiol. 2012, 58, 1676–1685. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Z.; Zhang, Y.; Zeng, J.; Liang, S.; Tang, C.; Liu, Z. Purification and Characterization of a Novel Insecticidal Toxin, μ-sparatoxin-Hv2, from the Venom of the Spider Heteropoda venatoria. Toxins 2018, 10, 233. https://doi.org/10.3390/toxins10060233
Xiao Z, Zhang Y, Zeng J, Liang S, Tang C, Liu Z. Purification and Characterization of a Novel Insecticidal Toxin, μ-sparatoxin-Hv2, from the Venom of the Spider Heteropoda venatoria. Toxins. 2018; 10(6):233. https://doi.org/10.3390/toxins10060233
Chicago/Turabian StyleXiao, Zhen, Yunxiao Zhang, Jiao Zeng, Songping Liang, Cheng Tang, and Zhonghua Liu. 2018. "Purification and Characterization of a Novel Insecticidal Toxin, μ-sparatoxin-Hv2, from the Venom of the Spider Heteropoda venatoria" Toxins 10, no. 6: 233. https://doi.org/10.3390/toxins10060233
APA StyleXiao, Z., Zhang, Y., Zeng, J., Liang, S., Tang, C., & Liu, Z. (2018). Purification and Characterization of a Novel Insecticidal Toxin, μ-sparatoxin-Hv2, from the Venom of the Spider Heteropoda venatoria. Toxins, 10(6), 233. https://doi.org/10.3390/toxins10060233





