Estrogen Receptor α Is Crucial in Zearalenone-Induced Invasion and Migration of Prostate Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. ERα is Responsible for ZEA- Induced PC3 Cell Invasion
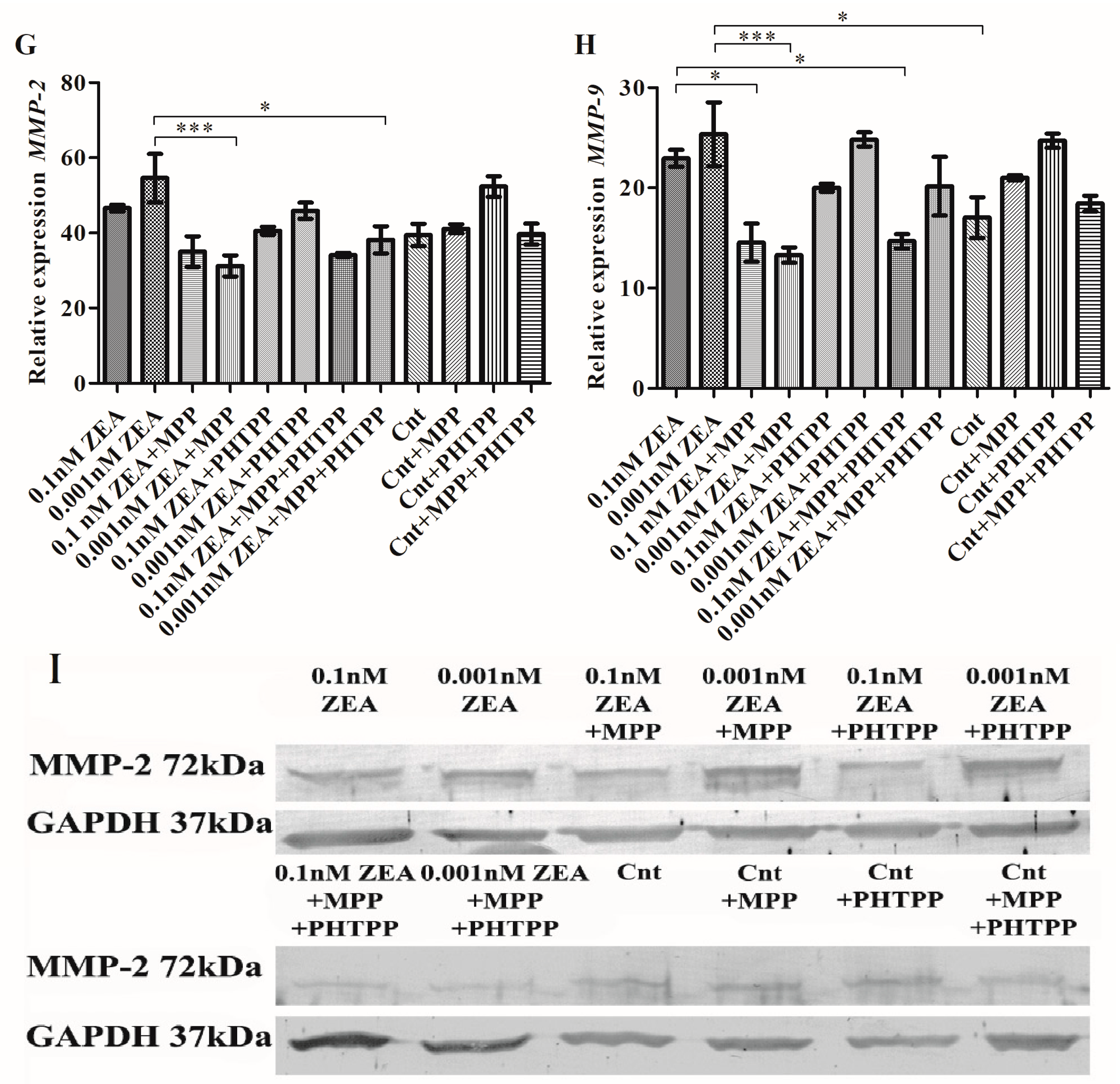
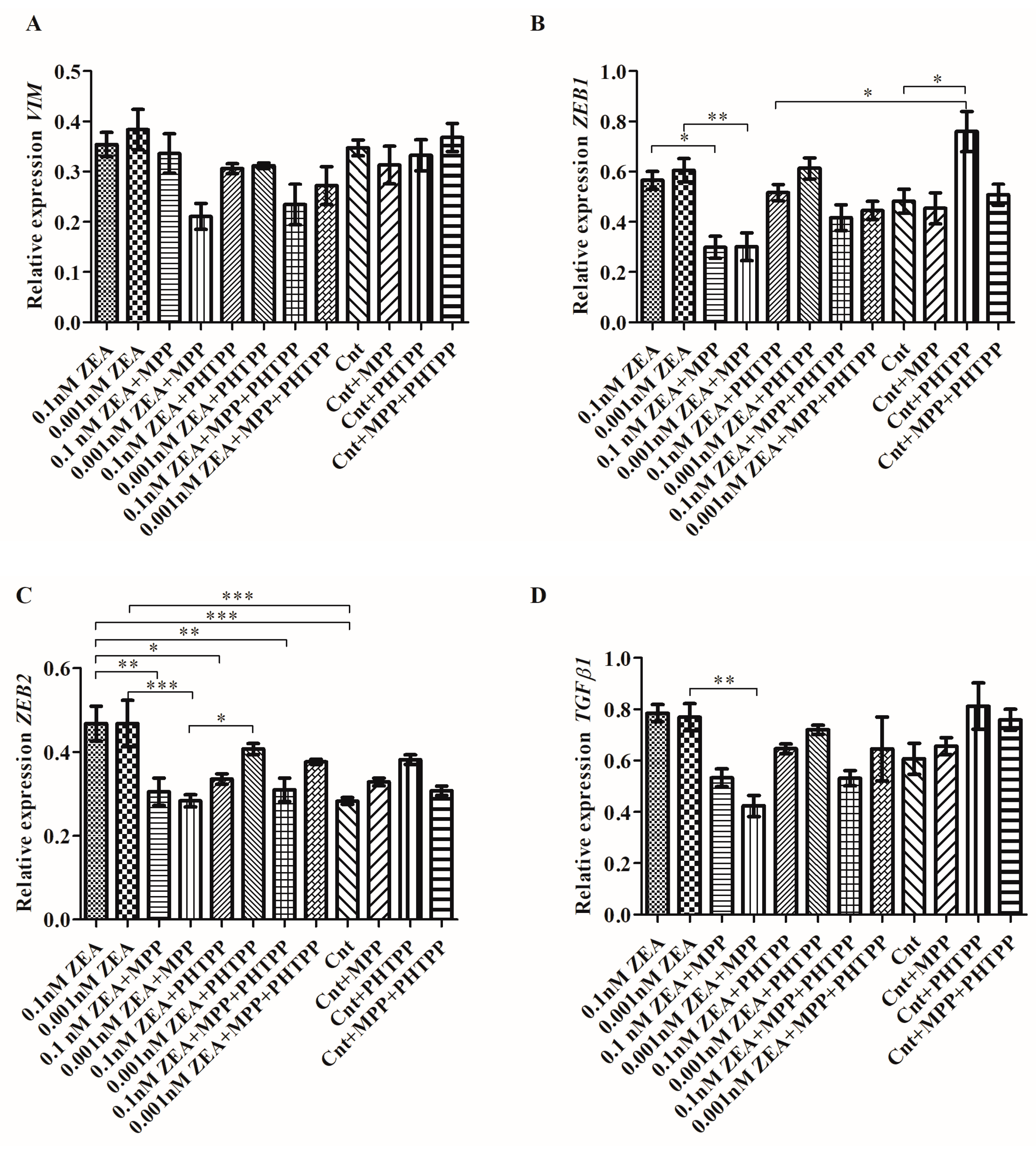
2.2. ZEA Modulates the Expression of EMT Genes Dependent on ERα
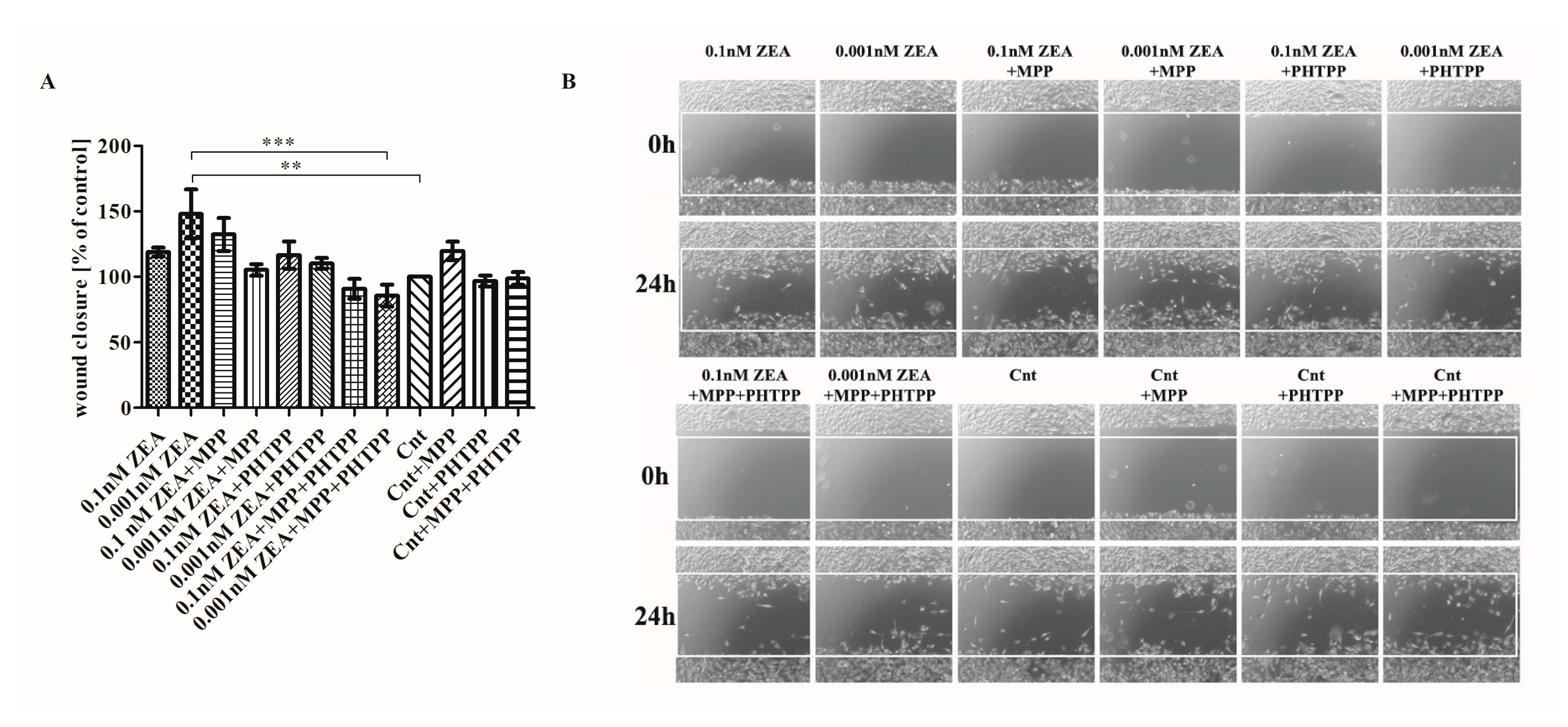
2.3. ERs Are Crucial in ZEA-Induced PC3 Cell Migration
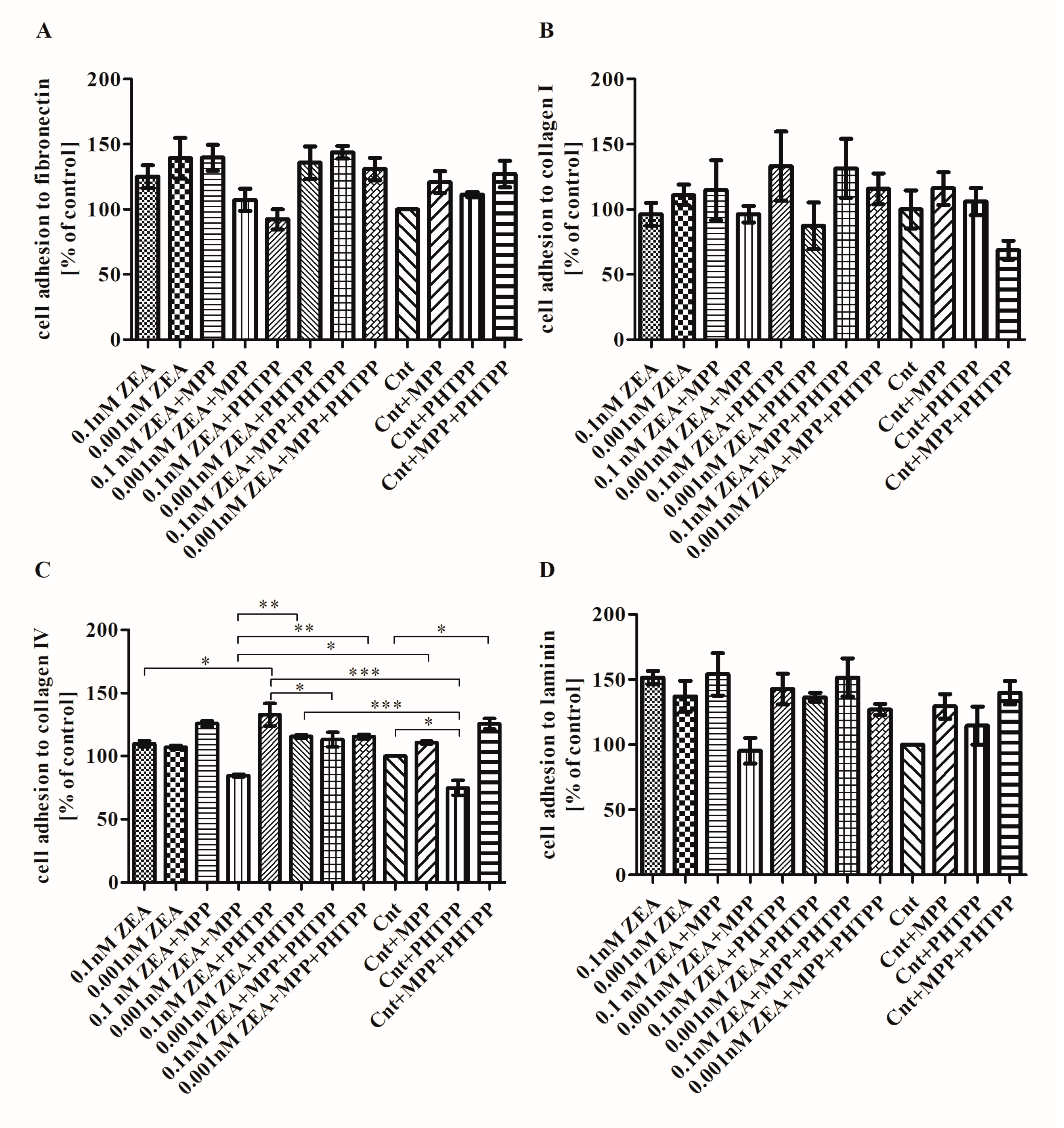
2.4. The Effect of ZEA on Cell Adhesion to ECM Proteins is Associated with ERα
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cell Culture
5.2. Immunocytochemistry (ICC)
5.3. Cell Adhesion Assay
5.4. Monolayer Wound Migration Assay
5.5. Cell Invasion Assay
5.6. Gelatin Zymography
5.7. Western Blot
5.8. Real Time qPCR (RTqPCR)
5.9. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Alshannaq, A.; Yu, J.H. Occurrence, Toxicity, and Analysis of Major Mycotoxins in Food. Int. J. Environ. Res. Public Health 2017, 14, E632. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.J.; Shen, H.H.; Zhang, X.F.; Yang, X.L.; Qiu, F.; Ou-Yang, Z.; Yang, M.H. Analysis of zearalenone and alpha-zearalenol in 100 foods and medicinal plants determined by HPLC-FLD and positive confirmation by LC-MS-MS. J. Sci. Food Agric. 2013, 93, 1584–1590. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, K.; Habrowska-Gorczynska, D.E.; Piastowska-Ciesielska, A.W. Zearalenone as an endocrine disruptor in humans. Environ. Toxicol. Pharmacol. 2016, 48, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Yli-Mattila, T. Ecology and evolution of toxigenic Fusarium species in cereals in Northern Europe and Asia. J. Plant Pathol. 2010, 92, 7–18. [Google Scholar] [CrossRef]
- Kriszt, R. A new zearalenone biodegradation strategy using non-pathogenic Rhodococcuspyridinivorans K408 strain. PLoS ONE 2012, 7, e43608. [Google Scholar] [CrossRef] [PubMed]
- Brera, C.; Debegnach, F.; De, S.B.; Di, I.S.; Gregori, E.; Neuhold, S.; Valitutti, F. Exposure assessment to mycotoxins in gluten-free diet for celiac patients. Food Chem. Toxicol. 2014, 69, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Frizzell, C. Endocrine disrupting effects of zearalenone, alpha- and beta-zearalenol at the level of nuclear receptor binding and steroidogenesis. Toxicol. Lett. 2011, 206, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, Y.; Gu, J.; Yuan, Y.; Liu, X.; Zheng, W.; Huang, Q.; Liu, Z.; Bian, J. Zearalenone inhibits testosterone biosynthesis in mouse Leydig cells via the crosstalk of estrogen receptor signaling and orphan nuclear receptor Nur77 expression. Toxicol. In Vitro 2014, 28, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Ayed-Boussema, I.; Bouaziz, C.; Rjiba, K.; Valenti, K.; Laporte, F.; Bacha, H.; Hassen, W. The mycotoxin Zearalenone induces apoptosis in human hepatocytes (HepG2) via p53-dependent mitochondrial signaling pathway. Toxicol. In Vitro 2008, 22, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Ahamed, S.; Foster, J.S.; Bukovsky, A.; Wimalasena, J. Signal transduction through the Ras/Erk pathway is essential for the mycoestrogen zearalenone-induced cell-cycle progression in MCF-7 cells. Mol. Carcinog. 2001, 30, 88–98. [Google Scholar] [CrossRef]
- Pitt, J.I. Toxigenic fungi and mycotoxins. Br. Med. Bull. 2000, 56, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, K.; Habrowska-Gorczynska, D.E.; Dominska, K.; Piastowska-Ciesielska, A.W. The dose-dependent effect of zearalenone on mitochondrial metabolism, plasma membrane permeabilization and cell cycle in human prostate cancer cell lines. Chemosphere 2017, 180, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Abassi, H.; Ayed-Boussema, I.; Shirley, S.; Abid, S.; Bacha, H.; Micheau, O. The mycotoxin zearalenone enhances cell proliferation, colony formation and promotes cell migration in the human colon carcinoma cell line HCT116. Toxicol. Lett. 2016, 254, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Palm, D.; Lang, K.; Brandt, B.; Zaenker, K.S.; Entschladen, F. In vitro and in vivo imaging of cell migration: Two interdepending methods to unravel metastasis formation. Semin. Cancer Biol. 2005, 15, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.A.; Kraut, N.; Beug, H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 2005, 17, 548–558. [Google Scholar] [CrossRef] [PubMed]
- Pluciennik, E. The role of WWOX tumor suppressor gene in the regulation of EMT process via regulation of CDH1-ZEB1-VIM expression in endometrial cancer. Int. J. Oncol. 2015, 46, 2639–2648. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, K.; Piastowska-Ciesielska, A.W. Oestrogens and oestrogen receptors in prostate cancer. SpringerPlus 2016, 5, 522. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowska, U.; Didkowska, J. Zachorowania i zgony na nowotwory zlosliwe w Polsce. Krajowy Rejestr Nowotworów, Centrum Onkologii—Instytut Marii Sklodowskiej—Curie. 2015. Available online: http://onkologia.org.pl/raporty/08/06/2015 (accessed on 30 June 2017).
- Mahmoud, A.M.; Al-Alem, U.; Ali, M.M.; Bosland, M.C. Genistein increases estrogen receptor beta expression in prostate cancer via reducing its promoter methylation. J. Steroid Biochem. Mol. Biol. 2015, 152, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Cooke, P.S.; Nanjappa, M.K.; Ko, C.; Prins, G.S.; Hess, R.A. Estrogens in Male Physiology. Physiol. Rev. 2017, 97, 995–1043. [Google Scholar] [CrossRef] [PubMed]
- Christoforou, P.; Christopoulos, P.F.; Koutsilieris, M. The role of estrogen receptor beta in prostate cancer. Mol. Med. 2014, 20, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.W.; Tilley, W.D.; Neal, D.E.; Carroll, J.S. Estrogen receptor beta in prostate cancer: Friend or foe? Endocr. Relat. Cancer 2014, 21, T219–T234. [Google Scholar] [CrossRef] [PubMed]
- Khosrokhavar, R.; Rahimifard, N.; Shoeibi, S.; Hamedani, M.P.; Hosseini, M.J. Effects of zearalenone and alpha-Zearalenol in comparison with Raloxifene on T47D cells. Toxicol. Mech. Methods 2009, 19, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Parveen, M.; Zhu, Y.; Kiyama, R. Expression profiling of the genes responding to zearalenone and its analogues using estrogen-responsive genes. FEBS Lett. 2009, 583, 2377–2384. [Google Scholar] [CrossRef] [PubMed]
- Yip, K.Y.; Wan, M.L.Y.; Wong, A.S.T.; Korach, K.S.; El-Nezami, H. Combined low-dose zearalenone and aflatoxin B1 on cell growth and cell-cycle progression in breast cancer MCF-7 cells. Toxicol. Lett. 2017, 281, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Ferraiuolo, R.M.; Tubman, J.; Sinha, I.; Hamm, C.; Porter, L.A. The cyclin-like protein, SPY1, regulates the ERalpha and ERK1/2 pathways promoting tamoxifen resistance. Oncotarget 2017, 8, 23337–23352. [Google Scholar] [CrossRef] [PubMed]
- Tatay, E.; Espin, S.; Garcia-Fernandez, A.J.; Ruiz, M.J. Estrogenic activity of zearalenone, alpha-zearalenol and beta-zearalenol assessed using the E-Screen assay in MCF-7 cells. Toxicol. Mech. Methods 2017, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, S.; Lelong, M.; Bourgine, G.; Efstathiou, T.; Saligaut, C.; Pakdel, F. Assessment of the potential activity of major dietary compounds as selective estrogen receptor modulators in two distinct cell models for proliferation and differentiation. Toxicol. Appl. Pharmacol. 2017, 325, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, C.Y.; Shi, J.; Miao, L.; Du, X.; Mayer, D.; Zhang, J. Estrogens promote invasion of prostate cancer cells in a paracrine manner through up-regulation of matrix metalloproteinase 2 in prostatic stromal cells. Endocrinology 2011, 152, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Slavin, S.; Yeh, C.R.; Da, J.; Yu, S.; Miyamoto, H.; Messing, E.M.; Guancial, E.; Yeh, S. Estrogen receptor alpha in cancer-associated fibroblasts suppresses prostate cancer invasion via modulation of thrombospondin 2 and matrix metalloproteinase 3. Carcinogenesis 2014, 35, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Dey, P.; Velazquez-Villegas, L.A.; Faria, M.; Turner, A.; Jonsson, P.; Webb, P.; Williams, C.; Gustafsson, J.A.; Strom, A.M. Estrogen Receptor beta2 Induces Hypoxia Signature of Gene Expression by Stabilizing HIF-1alpha in Prostate Cancer. PLoS ONE 2015, 10, e0128239. [Google Scholar] [CrossRef]
- Slusarz, A. Aggressive prostate cancer is prevented in ERalphaKO mice and stimulated in ERbetaKO TRAMP mice. Endocrinology 2012, 153, 4160–4170. [Google Scholar] [CrossRef] [PubMed]
- Takizawa, I. Estrogen receptor alpha drives proliferation in PTEN-deficient prostate carcinoma by stimulating survival signaling, MYC expression and altering glucose sensitivity. Oncotarget 2015, 6, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Piastowska-Ciesielska, A.W.; Domińska, K.; Nowakowska, M.; Gajewska, M.; Gajos-Michniewicz, A.; Ochędalski, T. Angiotensin modulates human mammary epithelial cell motility. J Renin Angiotensin Aldosterone Syst. 2014, 15, 419–429. [Google Scholar] [CrossRef] [PubMed]
- ImageJ software. Available online: http://rsb. info.nih.gov/ij/ (accessed on 31 January 2015).
- Primer3 software. Available online: http://frodo.wi.mit.edu/ (accessed on 31 January 2013).

| Gene | Primer Sequence (5’-3’) | Product Size (bp) |
|---|---|---|
| H3F3A | AGGACTTTAAAAGATCTGCGCTTCCAGAG ACCAGATAGGCCTCACTTGCCTCCTGC | 74 |
| MMP-2 | ACCAGCTGGCCTAGTGATGATGTT TGTCCTTCAGCACAAACAGGTTGC | 184 |
| MMP-9 | CTGGCAGGGTTTCCCATCAG GCAGTACCACGGCCAACTAC | 101 |
| RPLP0 | ACGGATTACACCTTCCCACTTGCTAAAAGGTC AGCCACAAAGGCAGATGGATCAGCCAAG | 69 |
| RPS17 | AAGCGCGTGTGCGAGGAGATCG TCGCTTCATCAGATGCGTGACATAACCTG | 87 |
| TGFβ1 | CAATTCCTGGCGATACCTCAG GCACAACTCCGGTGACATCAA | 86 |
| ZEB1 | GGAAATCAGGATGAAAGACA CACACAAATCACAAGCATAC | 136 |
| ZEB2 | CTAACCCAAGGAGCAGGTAATC GTGAATTCGCAGGTGTTCTTTC | 96 |
| VIM | AGCCGAAAACACCCTGCAAT CGTTCAAGGTCAAGACGTC | 72 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kowalska, K.; Habrowska-Górczyńska, D.E.; Urbanek, K.A.; Domińska, K.; Piastowska-Ciesielska, A.W. Estrogen Receptor α Is Crucial in Zearalenone-Induced Invasion and Migration of Prostate Cancer Cells. Toxins 2018, 10, 98. https://doi.org/10.3390/toxins10030098
Kowalska K, Habrowska-Górczyńska DE, Urbanek KA, Domińska K, Piastowska-Ciesielska AW. Estrogen Receptor α Is Crucial in Zearalenone-Induced Invasion and Migration of Prostate Cancer Cells. Toxins. 2018; 10(3):98. https://doi.org/10.3390/toxins10030098
Chicago/Turabian StyleKowalska, Karolina, Dominika Ewa Habrowska-Górczyńska, Kinga Anna Urbanek, Kamila Domińska, and Agnieszka Wanda Piastowska-Ciesielska. 2018. "Estrogen Receptor α Is Crucial in Zearalenone-Induced Invasion and Migration of Prostate Cancer Cells" Toxins 10, no. 3: 98. https://doi.org/10.3390/toxins10030098
APA StyleKowalska, K., Habrowska-Górczyńska, D. E., Urbanek, K. A., Domińska, K., & Piastowska-Ciesielska, A. W. (2018). Estrogen Receptor α Is Crucial in Zearalenone-Induced Invasion and Migration of Prostate Cancer Cells. Toxins, 10(3), 98. https://doi.org/10.3390/toxins10030098






