True Lies: Using Proteomics to Assess the Accuracy of Transcriptome-Based Venomics in Centipedes Uncovers False Positives and Reveals Startling Intraspecific Variation in Scolopendra subspinipes
Abstract
1. Introduction
2. Results
2.1. Transcriptome Generation
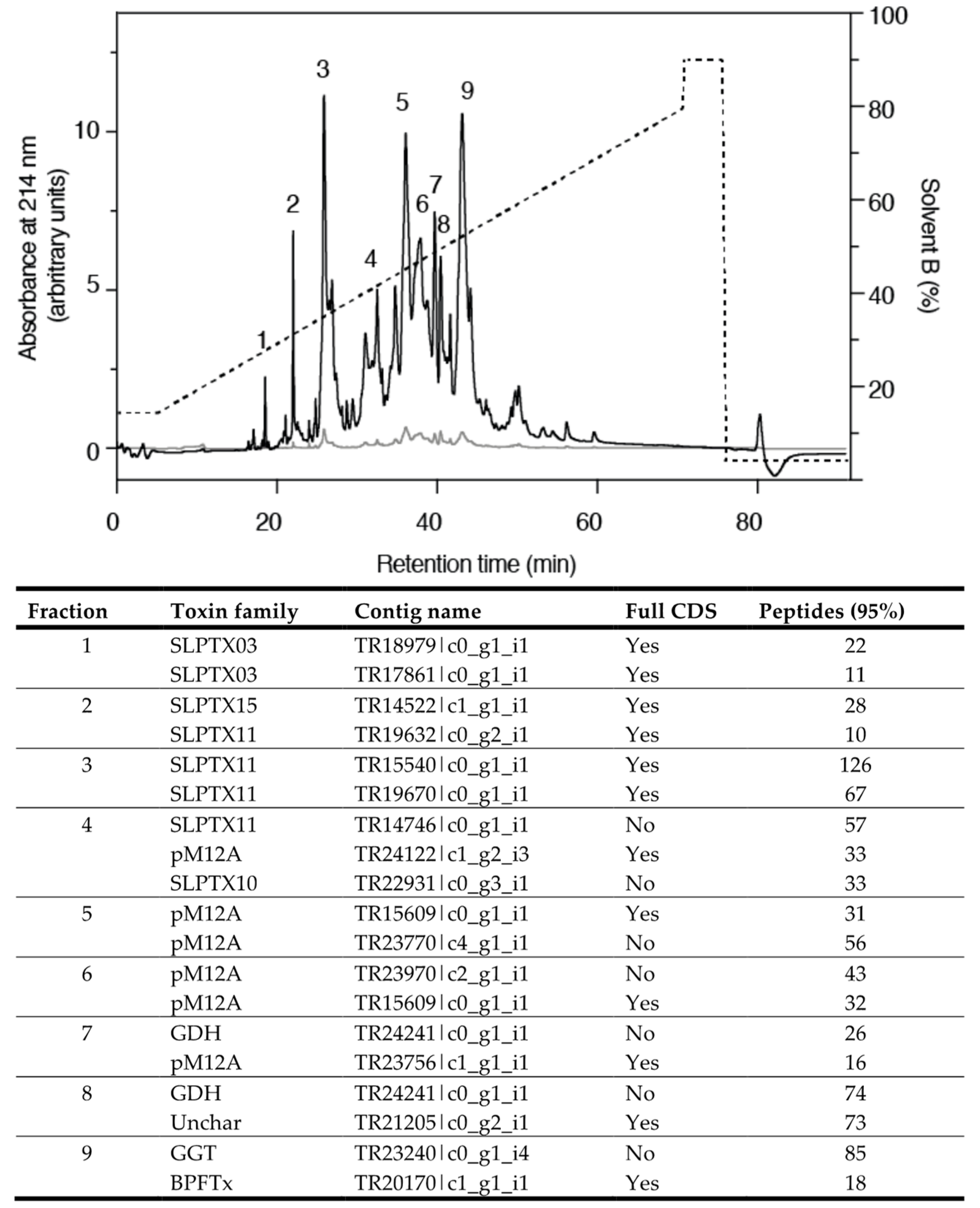
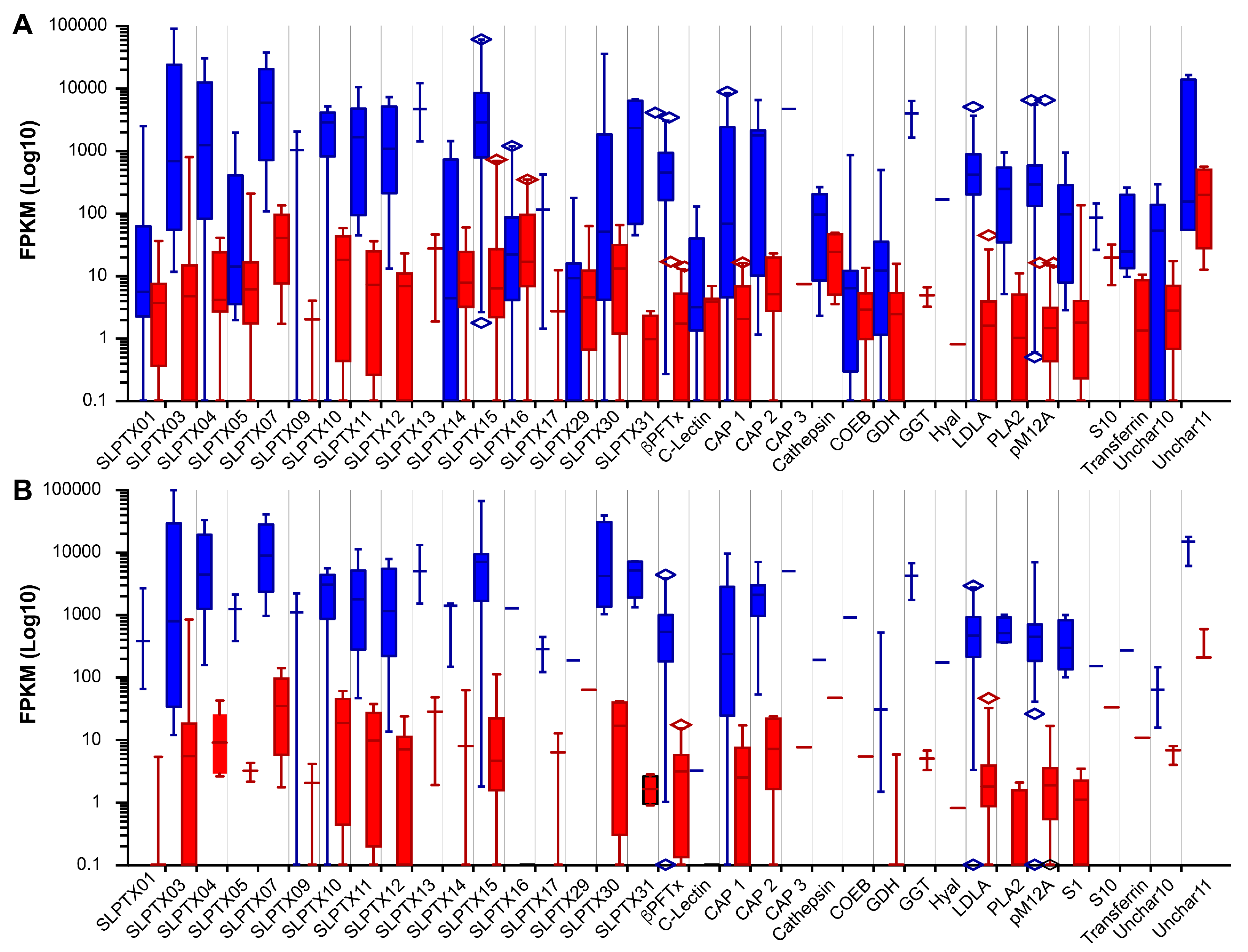
2.2. Venom Composition of S. subspinipes
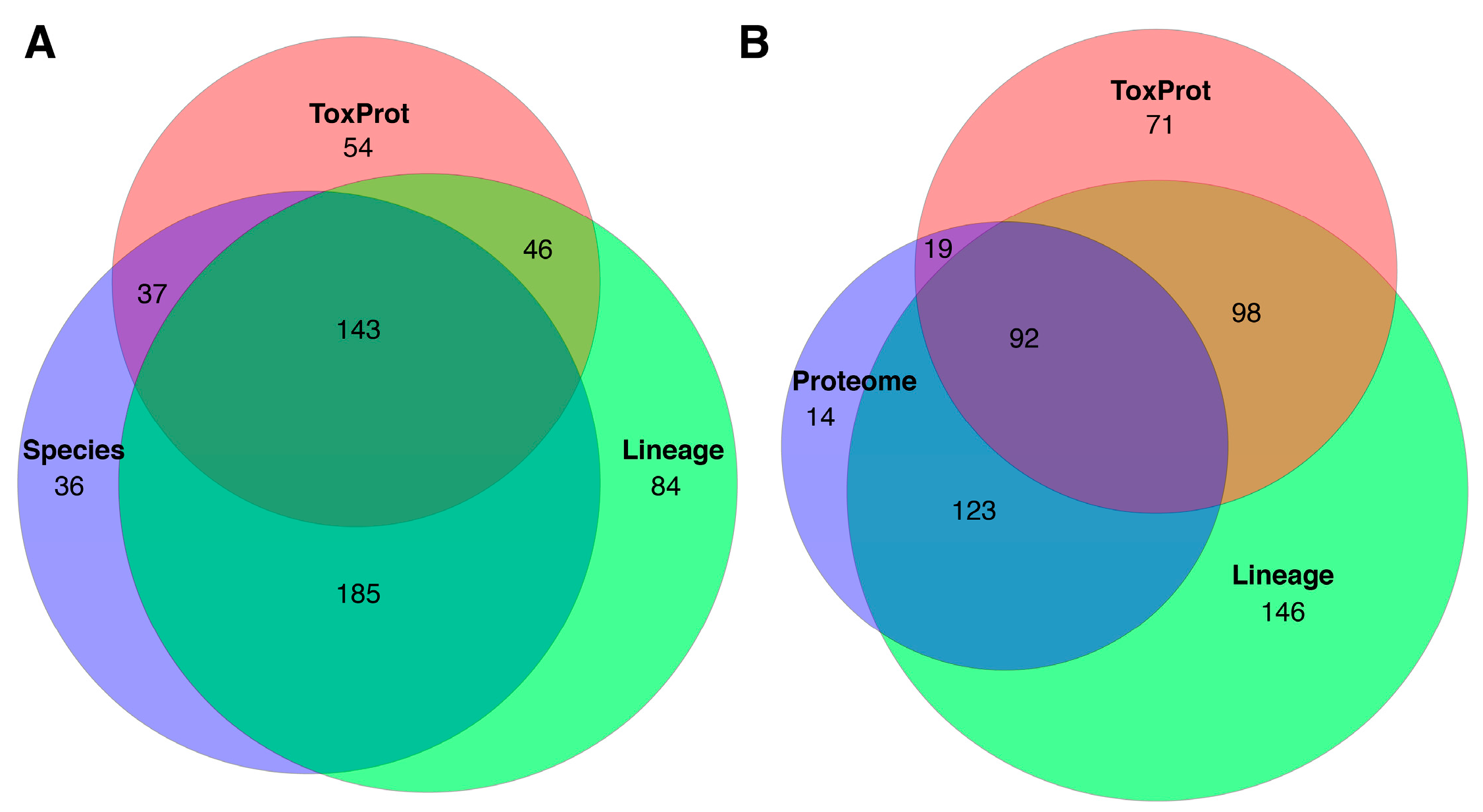
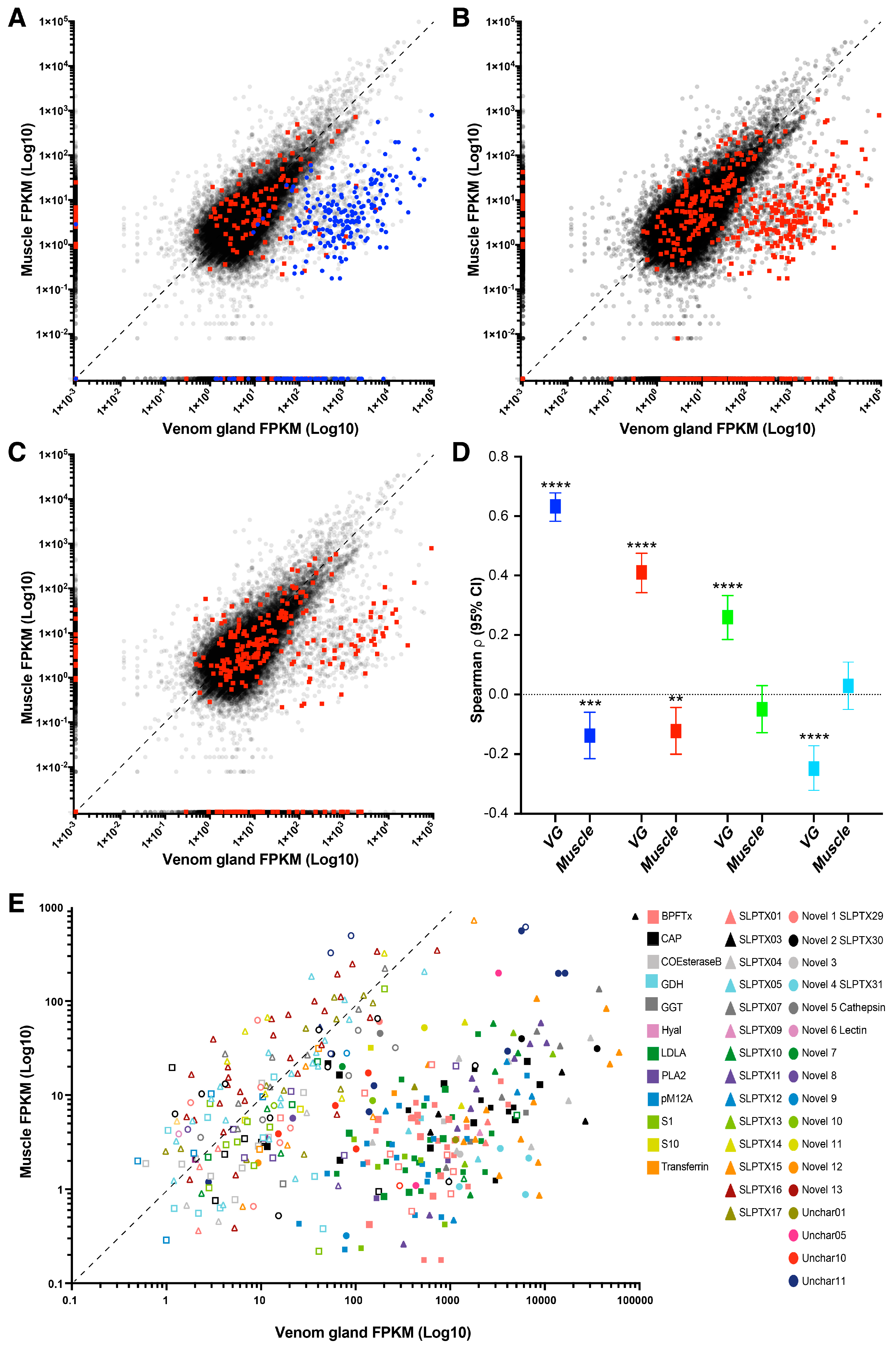
2.3. Identifying Toxins by BLAST Annotation
2.4. Identifying Putative Toxins by Comparative Transcriptomics
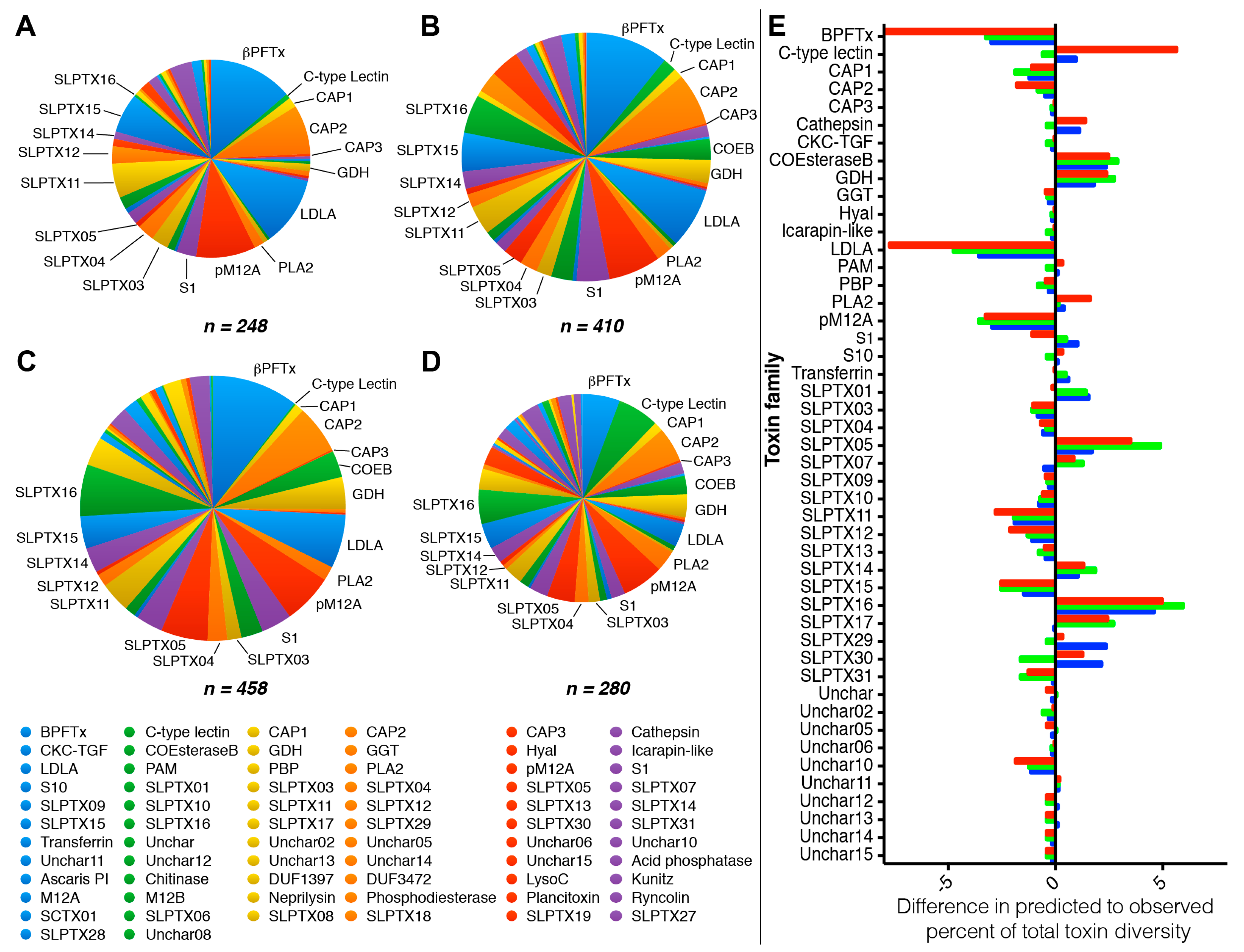
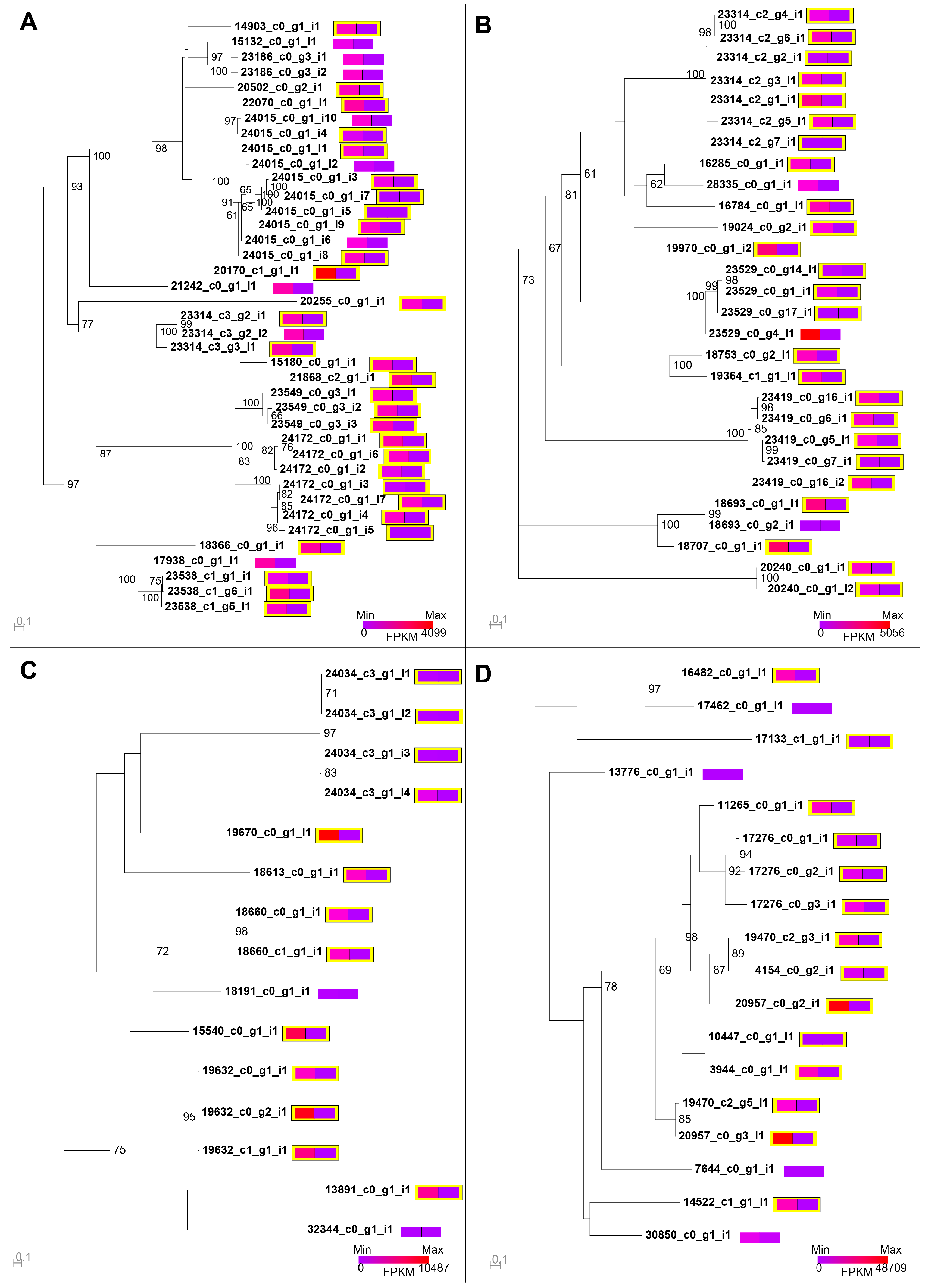
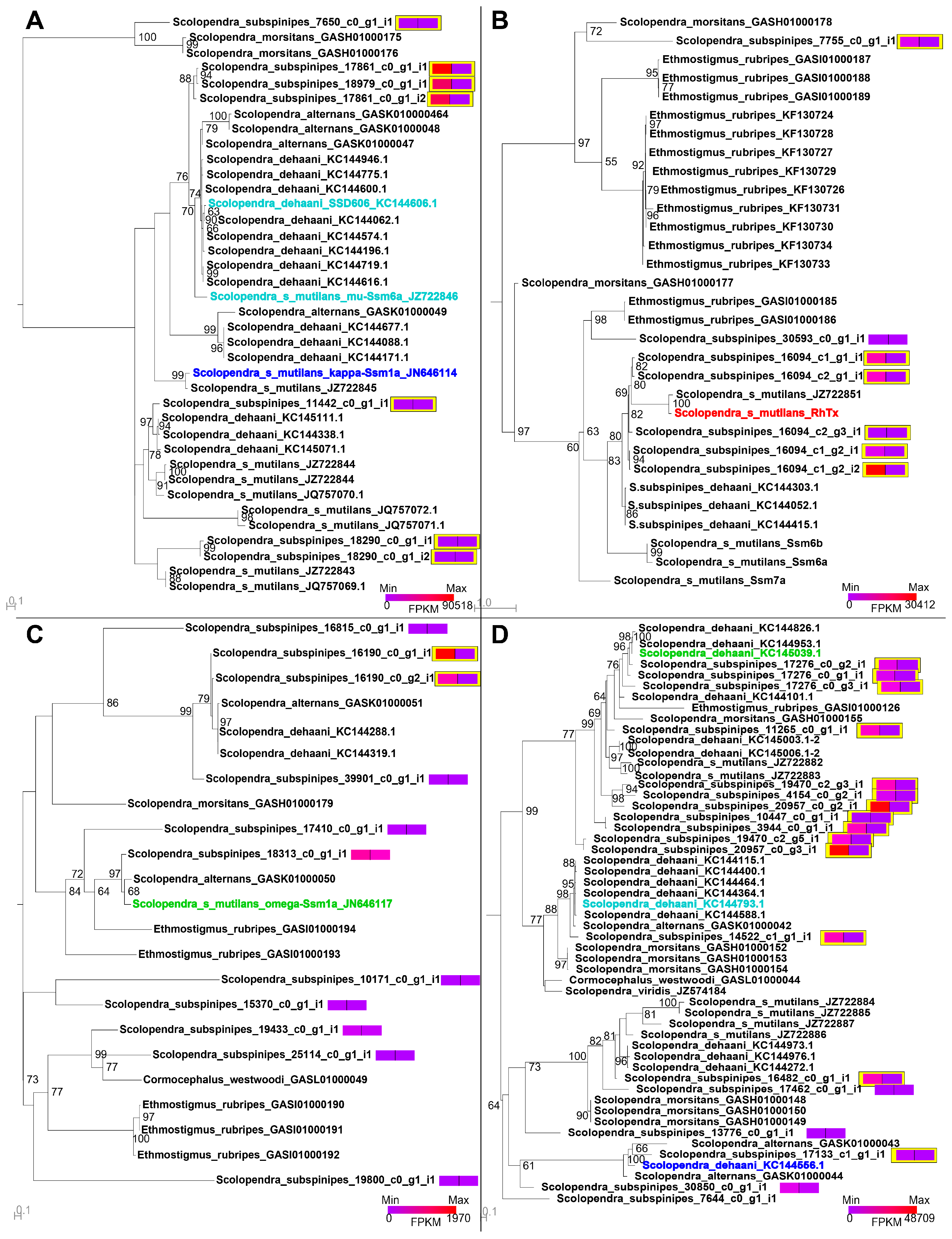
2.5. Intraspecific Variation in the Venom of S. subspinipes
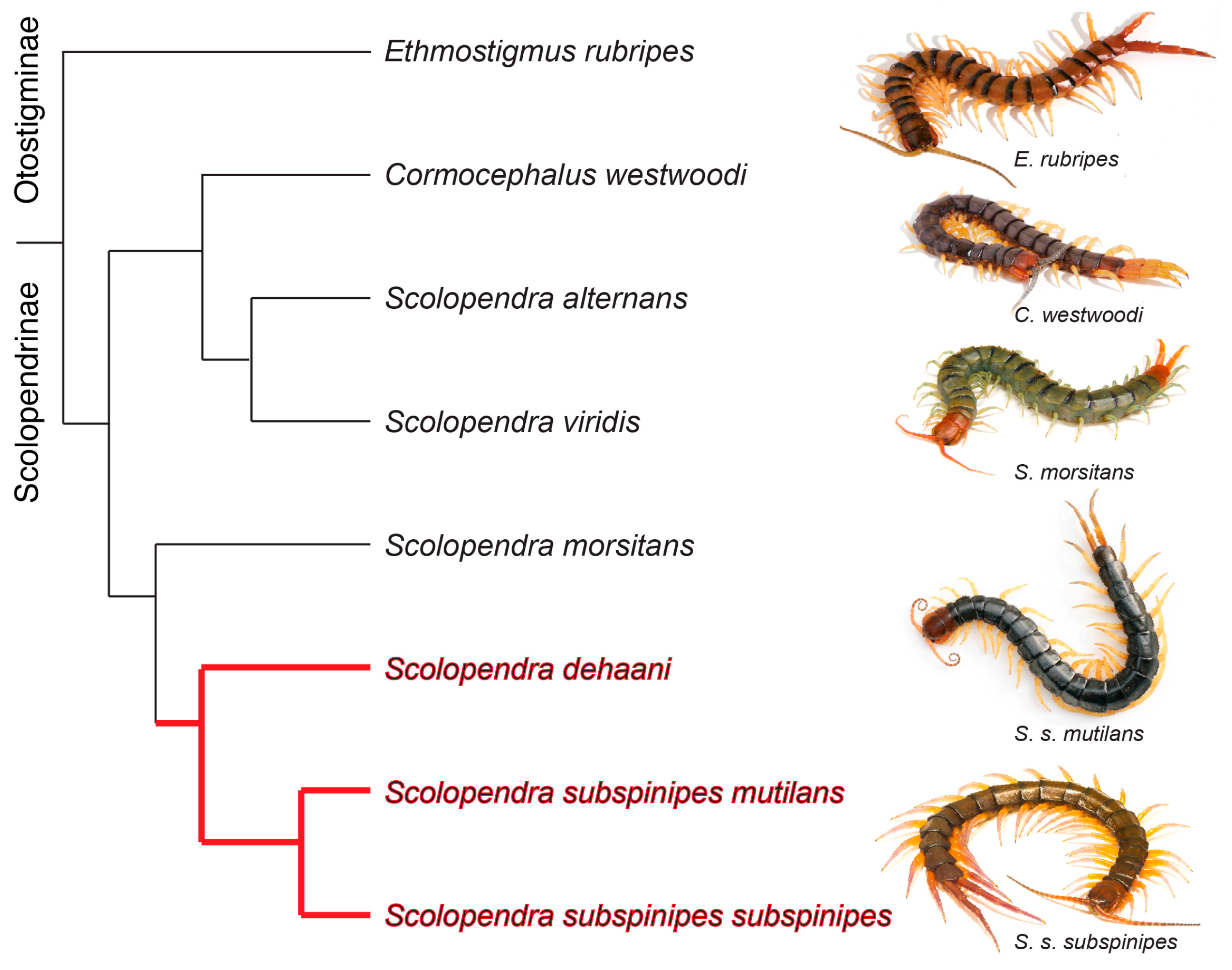
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Venom Collection and Transcriptome Sequencing
5.2. Transcriptome Assembly and Analyses
5.3. Venom Proteome Analyses
5.4. Toxin Annotation
5.5. Phylogenetic Analyses
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kalia, J.; Milescu, M.; Salvatierra, J.; Wagner, J.; Klint, J.K.; King, G.F.; Olivera, B.M.; Bosmans, F. From foe to friend: Using animal toxins to investigate ion channel function. J. Mol. Biol. 2015, 427, 158–175. [Google Scholar] [CrossRef] [PubMed]
- King, G.F. Venoms to drugs: Translating venom peptides into therapeutics. Aust. Biochem. 2013, 44, 13–15. [Google Scholar]
- Smith, J.J.; Herzig, V.; King, G.F.; Alewood, P.F. The insecticidal potential of venom peptides. Cell. Mol. Life Sci. 2013, 70, 3665–3693. [Google Scholar] [CrossRef] [PubMed]
- Pineda, S.S.; Undheim, E.A.; Rupasinghe, D.B.; Ikonomopoulou, M.P.; King, G.F. Spider venomics: Implications for drug discovery. Futur. Med. Chem. 2014, 6, 1699–1714. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.A.B.; King, G.F. On the venom system of centipedes (Chilopoda), a neglected group of venomous animals. Toxicon 2011, 57, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.A.B.; Grimm, L.L.; Low, C.-F.; Morgenstern, D.; Herzig, V.; Zobel-Thropp, P.; Pineda, S.S.; Habib, R.; Dziemborowicz, S.; Fry, B.G.; et al. Weaponization of a hormone: Convergent recruitment of hyperglycemic hormone into the venom of arthropod predators. Structure 2015, 23, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.A.B.; Jenner, R.A.; King, G.F. Centipede venoms as a source of drug leads. Expert Opin. Drug Discov. 2016, 11, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, Z.; Xiao, Y.; Li, Y.; Rong, M.; Liang, S.; Zhang, Z.; Yu, H.; King, G.F.; Lai, R. Chemical punch packed in venoms makes centipedes excellent predators. Mol. Cell. Proteom. 2012, 11, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-C.; Zhang, R.; Zhao, F.; Chen, Z.-M.; Liu, H.-W.; Wang, Y.-J.; Jiang, P.; Zhang, Y.; Wu, Y.; Ding, J.-P.; et al. Venomic and transcriptomic analysis of centipede Scolopendra subspinipes dehaani. J. Proteome Res. 2012, 11, 6197–6212. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Wu, F.; Wen, M.; Yang, X.; Wang, C.; Li, Y.; He, S.; Zhang, L.; Zhang, Y.; Tian, C. A distinct three-helix centipede toxin SSD609 inhibits Iks channels by interacting with the KCNE1 auxiliary subunit. Sci. Rep. 2015, 5, 13399. [Google Scholar] [CrossRef] [PubMed]
- Ikonomopoulou, M.P.; Smith, J.J.; Herzig, V.; Pineda, S.S.; Dziemborowicz, S.; Er, S.Y.; Durek, T.; Gilchrist, J.; Alewood, P.F.; Nicholson, G.M.; et al. Isolation of two insecticidal toxins from venom of the Australian theraphosid spider Coremiocnemis tropix. Toxicon 2016, 123, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Chassagnon, I.R.; McCarthy, C.A.; Chin, Y.K.; Pineda, S.S.; Keramidas, A.; Mobli, M.; Pham, V.; De Silva, T.M.; Lynch, J.W.; Widdop, R.E.; et al. Potent neuroprotection after stroke afforded by a double-knot spider-venom peptide that inhibits acid-sensing ion channel 1a. Proc. Natl. Acad. Sci. USA 2017, 114, 3750–3755. [Google Scholar] [CrossRef] [PubMed]
- Madio, B.; Undheim, E.A.B.; King, G.F. Revisiting venom of the sea anemone Stichodactyla haddoni: Omics techniques reveal the complete toxin arsenal of a well-studied sea anemone genus. J. Proteom. 2017, 166, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Von Reumont, B.M.; Undheim, E.A.B.; Jauss, R.T.; Jenner, R.A. Venomics of remipede crustaceans reveals novel peptide diversity and illuminates the venom’s biological role. Toxins 2017, 9, 234. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez de la Vega, R.C.; Giraud, T. Intragenome diversity of gene families encoding toxin-like proteins in venomous animals. Integr. Comp. Biol. 2016, 56, 938–949. [Google Scholar] [CrossRef] [PubMed]
- Siriwut, W.; Edgecombe, G.D.; Sutcharit, C.; Tongkerd, P.; Panha, S. A taxonomic review of the centipede genus Scolopendra Linnaeus, 1758 (Scolopendromorpha, Scolopendridae) in mainland Southeast Asia, with description of a new species from Laos. ZooKeys 2016, 1–124. [Google Scholar] [CrossRef]
- Siriwut, W.; Edgecombe, G.D.; Sutcharit, C.; Panha, S. The centipede genus Scolopendra in mainland Southeast Asia: Molecular phylogenetics, geometric morphometrics and external morphology as tools for species delimitation. PLoS ONE 2015, 10, e0135355. [Google Scholar] [CrossRef] [PubMed]
- Vahtera, V.; Edgecombe, G.; Giribet, G. Phylogenetics of scolopendromorph centipedes: Can denser taxon sampling improve an artificial classification? Invertebr. Syst. 2013, 27, 578–602. [Google Scholar] [CrossRef]
- Kang, S.; Liu, Y.; Zeng, X.; Deng, H.; Luo, Y.; Chen, K.; Chen, S. Taxonomy and identification of the genus Scolopendra in China using integrated methods of external morphology and molecular phylogenetics. Sci. Rep. 2017, 7, 16032. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Moran, Y. The rise and fall of an evolutionary innovation: Contrasting strategies of venom evolution in ancient and young animals. PLoS Genet. 2015, 11, e1005596. [Google Scholar] [CrossRef] [PubMed]
- Rong, M.; Yang, S.; Wen, B.; Mo, G.; Kang, D.; Liu, J.; Lin, Z.; Jiang, W.; Li, B.; Du, C.; et al. Peptidomics combined with cDNA library unravel the diversity of centipede venom. J. Proteom. 2014, 114, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.A.B.; Jones, A.; Clauser, K.R.; Holland, J.H.; Pineda, S.S.; King, G.F.; Fry, B.G. Clawing through evolution: Toxin diversification and convergence in the ancient lineage Chilopoda (Centipedes). Mol. Biol. Evol. 2014, 31, 2124–2148. [Google Scholar] [CrossRef] [PubMed]
- Malta, M.B.; Lira, M.S.; Soares, S.L.; Rocha, G.C.; Knysak, I.; Martins, R.; Guizze, S.P.G.; Santoro, M.L.; Barbaro, K.C. Toxic activities of Brazilian centipede venoms. Toxicon 2008, 52, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.A.B.; Fry, B.; King, G.F. Centipede venom: Recent discoveries and current state of knowledge. Toxins 2015, 7, 679–704. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Ferreira, M.; Magalhães, G.S.; Fernandez, J.H.; Junqueira-de-Azevedo, I.d.L.M.; Le Ho, P.; Lima, C.; Valente, R.H.; Moura-da-Silva, A.M. Structural and biological characterization of Nattectin, a new C-type lectin from the venomous fish Thalassophryne nattereri. Biochimie 2011, 93, 971–980. [Google Scholar] [CrossRef] [PubMed]
- Morita, T. Structures and functions of snake venom CLPs (C-type lectin-like proteins) with anticoagulant-, procoagulant-, and platelet-modulating activities. Toxicon 2005, 45, 1099–1114. [Google Scholar] [CrossRef] [PubMed]
- Dowd, A.J.; McGonigle, S.; Dalton, J.P. Fasciola hepatica cathepsin L proteinase cleaves fibrinogen and produces a novel type of fibrin clot. Eur. J. Biochem. 1995, 232, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Peiren, N.; de Graaf, D.C.; Brunain, M.; Bridts, C.H.; Ebo, D.G.; Stevens, W.J.; Jacobs, F.J. Molecular cloning and expression of icarapin, a novel IgE-binding bee venom protein. FEBS Lett. 2006, 580, 4895–4899. [Google Scholar] [CrossRef] [PubMed]
- Bouzid, W.; Verdenaud, M.; Klopp, C.; Ducancel, F.; Noirot, C.; Vetillard, A. De novo sequencing and transcriptome analysis for Tetramorium bicarinatum: A comprehensive venom gland transcriptome analysis from an ant species. BMC Genom. 2014, 15, 987. [Google Scholar] [CrossRef] [PubMed]
- Haney, R.A.; Clarke, T.H.; Gadgil, R.; Fitzpatrick, R.; Hayashi, C.Y.; Ayoub, N.A.; Garb, J.E. Effects of gene duplication, positive selection, and shifts in gene expression on the evolution of the venom gland transcriptome in widow spiders. Genome Biol. Evol. 2016, 8, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.D.; Undheim, E.A.B.; Ueberheide, B.; King, G.F. Venom peptides as therapeutics: Advances, challenges and the future of venom-peptide discovery. Expert Rev. Proteom. 2017, 14, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Von Reumont, B.M.; Campbell, L.I.; Richter, S.; Hering, L.; Sykes, D.; Hetmank, J.; Jenner, R.A.; Bleidorn, C. A Polychaete’s powerful punch: Venom gland transcriptomics of Glycera reveals a complex cocktail of toxin homologs. Genome Biol. Evol. 2014, 6, 2406–2423. [Google Scholar] [CrossRef] [PubMed]
- Whelan, N.V.; Kocot, K.M.; Santos, S.R.; Halanych, K.M. Nemertean toxin genes revealed through transcriptome sequencing. Genome Biol. Evol. 2014, 6, 3314–3325. [Google Scholar] [CrossRef] [PubMed]
- Junqueira-de-Azevedo, I.L.; Bastos, C.M.; Ho, P.L.; Luna, M.S.; Yamanouye, N.; Casewell, N.R. Venom-related transcripts from Bothrops jararaca tissues provide novel molecular insights into the production and evolution of snake venom. Mol. Biol. Evol. 2015, 32, 754–766. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.A.; Gruning, B.A.; Paszkiewicz, K.; Pritchard, L. Galaxy tools and workflows for sequence analysis with applications in molecular plant pathology. PeerJ 2013, 1, e167. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar]
- Jungo, F.; Bougueleret, L.; Xenarios, I.; Poux, S. The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon 2012, 60, 551–557. [Google Scholar] [CrossRef] [PubMed]
- González-Morales, L.; Pedraza-Escalona, M.; Diego-Garcia, E.; Restano-Cassulini, R.; Batista, C.V.F.; Gutiérrez, M.C.; Possani, L.D. Proteomic characterization of the venom and transcriptomic analysis of the venomous gland from the Mexican centipede Scolopendra viridis. J. Proteom. 2014, 111, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.; Engelbrecht, J.; Brunak, S.; von Heijne, G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997, 10, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. CD-HIT: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Sloggett, C.; Goonasekera, N.; Makunin, I.; Benson, D.; Crowe, M.; Gladman, S.; Kowsar, Y.; Pheasant, M.; Horst, R.; et al. Genomics Virtual Laboratory: A practical bioinformatics workbench for the cloud. PLoS ONE 2015, 10, e0140829. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Han, M.V.; Zmasek, C.M. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinform. 2009, 10, 356. [Google Scholar] [CrossRef] [PubMed]

| Group | Description | Number Cysteines | CDS Length | Contigs in Transcriptome | Contigs in Venom |
|---|---|---|---|---|---|
| Novel 1 | SLPTX29—Lepidopteran putative fungal protease inhibitor-like peptide. Structurally related to insulin-like growth factor domain containing protein (sIGFBP) | 12–14 | 97–128 | 11 | 1 |
| Novel 2 | SLPTX30—Pheromone binding protein-like peptide (IPR006170) | 3–6 | 88–180 | 16 | 4 |
| Novel 3 | Cystine-knot cytokine domain containing TGF-beta-like peptide | 10–12 | 127 | 1 | 1 |
| Novel 4 | SLPTX31—No BLAST or InterPro hits | 0 | 45–70 | 6 | 4 |
| Novel 5 | Cathepsin L-type cysteine-protease | 7 | 334–335 | 2 | 1 |
| Novel 6 | C-type lectin-like | 8 | 15–236 | 7 | 2 |
| Novel 7 | Phosphatidylethanolamine-binding protein (IPR008914) | 4 | 183–213 | 2 | 2 |
| Novel 8 | No BLAST or InterPro hits (Uncharacterized family 12) | 10–12 | 190–204 | 2 | 1 |
| Novel 9 | No InterPro hits, similar to uncharacterised protein from Strigamia maritima (T1J4X8) (Uncharacterised family 13) | 6 | 300–313 | 2 | 1 |
| Novel 10 | Peptidylglycine alpha-hydroxylating monooxygenase | 8–13 | 344–919 | 2 | 1 |
| Novel 11 | Icarapin-like (Venom carbohydrate-rich protein) | 0 | 227 | 1 | 1 |
| Novel 12 | No InterPro hits, similar to uncharacterised protein from Strigamia maritima (T1J2G0) (Uncharacterized family 14) | 5 | 222 | 1 | 1 |
| Novel 13 | No BLAST or InterPro hits (Uncharacterized family 15) | 8 | 275 | 1 | 1 |
| Proteome | Species Specific | Lineage Specific | ToxProt | |
|---|---|---|---|---|
| Unique contigs | 550 | 956 | 1357 | 1214 |
| Unique CDS | 469 | 811 | 1115 | 1058 |
| Unique CDS with signal peptide | 248 | 401 | 458 | 280 |
| Unique full-length CDS | 156 | 269 | 291 | 184 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, J.J.; Undheim, E.A.B. True Lies: Using Proteomics to Assess the Accuracy of Transcriptome-Based Venomics in Centipedes Uncovers False Positives and Reveals Startling Intraspecific Variation in Scolopendra subspinipes. Toxins 2018, 10, 96. https://doi.org/10.3390/toxins10030096
Smith JJ, Undheim EAB. True Lies: Using Proteomics to Assess the Accuracy of Transcriptome-Based Venomics in Centipedes Uncovers False Positives and Reveals Startling Intraspecific Variation in Scolopendra subspinipes. Toxins. 2018; 10(3):96. https://doi.org/10.3390/toxins10030096
Chicago/Turabian StyleSmith, Jennifer J., and Eivind A. B. Undheim. 2018. "True Lies: Using Proteomics to Assess the Accuracy of Transcriptome-Based Venomics in Centipedes Uncovers False Positives and Reveals Startling Intraspecific Variation in Scolopendra subspinipes" Toxins 10, no. 3: 96. https://doi.org/10.3390/toxins10030096
APA StyleSmith, J. J., & Undheim, E. A. B. (2018). True Lies: Using Proteomics to Assess the Accuracy of Transcriptome-Based Venomics in Centipedes Uncovers False Positives and Reveals Startling Intraspecific Variation in Scolopendra subspinipes. Toxins, 10(3), 96. https://doi.org/10.3390/toxins10030096




