Impact of Dinophysis acuminata Feeding Mesodinium rubrum on Nutrient Dynamics and Bacterial Composition in a Microcosm
Abstract
1. Introduction
2. Results
2.1. Predator-Prey Population Dynamics and Environmental Changes
2.2. Composition and Structure of the Microbial Community throughout the Growth Curve
3. Discussion
4. Materials and Methods
4.1. Cultures
4.2. Batch Culture Setup
4.3. Nutrient Sample Collection and Preparation
4.4. Toxin Analysis
4.5. DNA Extraction and Illumine Sequencing
4.6. Bioinformatics Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Hallegraeff, G.M. A review of harmful algal blooms and their apparent global increase. Phycologia 1993, 32, 79–99. [Google Scholar] [CrossRef]
- Reguera, B.; Riobo, P.; Rodriguez, F.; Diaz, P.A.; Pizarro, G.; Paz, B.; Franco, J.M.; Blanco, J. Dinophysis toxins: Causative organisms, distribution and fate in shellfish. Mar. Drugs 2014, 12, 394–461. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, H.; Suganuma, M. Tumor promotion by inhibitors of protein phosphatase 1 and 2A: The okadaic acid class of compounds. Adv Cancer Res. 1993, 61, 143–194. [Google Scholar] [PubMed]
- Marasigan, A.N.; Sato, S.; Fukuyo, Y.; Kodama, M. Accumulation of a high level of diarrhetic shellfish toxins in the green mussel Perna viridis during a bloom of Dinophysis caudata and Dinophysis miles in Sapian bay, Panay island, the Philippines. Fish. Sci. 2001, 67, 994–996. [Google Scholar] [CrossRef]
- Bodero, M.; Hoogenboom, R.; Bovee, T.F.H.; Portier, L.; de Haan, L.; Peijnenburg, A.; Hendriksen, P.J.M. Whole genome mRNA transcriptomics analysis reveals different modes of action of the diarrheic shellfish poisons okadaic acid and dinophysistoxin-1 versus azaspiracid-1 in Caco-2 cells. Toxicol. In Vitro 2017, 46, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Miles, C.O.; Wilkins, A.L.; Munday, R.; Dines, M.H.; Hawkes, A.D.; Briggs, L.R.; Sandvik, M.; Jensen, D.J.; Cooney, J.M.; Holland, P.T.; et al. Isolation of pectenotoxin-2 from Dinophysis acuta and its conversion to pectenotoxin-2 seco acid, and preliminary assessment of their acute toxicities. Toxicon 2004, 43, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Park, M.G.; Kim, S. First successful culture of the marine dinoflagellate Dinophysis acuminata. Aquat. Microb. Ecol. 2006, 45, 101–106. [Google Scholar] [CrossRef]
- Riisgaard, K.; Hansen, P.J. Role of food uptake for photosynthesis, growth and survival of the mixotrophic dinoflagellate Dinophysis acuminata. Mar. Ecol. Prog. Ser. 2009, 381, 51–62. [Google Scholar] [CrossRef]
- Stoecker, D.K.; Hansen, P.J.; Caron, D.A.; Mitra, A. Mixotrophy in the Marine Plankton. Ann. Rev. Mar. Sci. 2017, 9, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Campbell, L.; Olson, R.J.; Sosik, H.M.; Abraham, A.; Henrichs, D.W.; Hyatt, C.J.; Buskey, E.J. First Harmful Dinophysis (Dinophyceae, Dinophysiales) bloom in the U.S. Is Revealed by Automated Imaging Flow Cytometry1. J. Phycol. 2010, 46, 66–75. [Google Scholar] [CrossRef]
- Hattenrath-Lehmann, T.K.; Marcoval, M.A.; Berry, D.L.; Fire, S.; Wang, Z.; Morton, S.L.; Gobler, C.J. The emergence of Dinophysis acuminata blooms and DSP toxins in shellfish in New York waters. Harmful Algae 2013, 26, 33–44. [Google Scholar] [CrossRef]
- Moita, M.T.; Pazos, Y.; Rocha, C.; Nolasco, R.; Oliveira, P.B. Toward predicting Dinophysis blooms off NW Iberia: A decade of events. Harmful Algae 2016, 53, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; An, X.; Liu, L.; Zhang, K.; Zheng, D.; Tong, M. Characterization of Dinophysis acuminata from the Yellow Sea, China, and its response to different temperatures and Mesodinium prey. Oceanol. Hydrobiol. Stud. 2017, 46, 439–450. [Google Scholar] [CrossRef]
- Tong, M.; Smith, J.L.; Kulis, D.M.; Anderson, D.M. Role of dissolved nitrate and phosphate in isolates of Mesodinium rubrum and toxin-producing Dinophysis acuminata. Aquat. Microb. Ecol. 2015, 75, 169–185. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Tong, M.; Kulis, D.; Anderson, D.M. Effect of ciliate strain, size, and nutritional content on the growth and toxicity of mixotrophic Dinophysis acuminata. Harmful Algae 2018, 78, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Kulis, D.M.; Fux, E.; Smith, J.L.; Hess, P.; Zhou, Q.; Anderson, D.M. The effects of growth phase and light intensity on toxin production by Dinophysis acuminata from the northeastern United States. Harmful Algae 2011, 10, 254–264. [Google Scholar] [CrossRef]
- Lundgren, V.M.; Glibert, P.M.; Granéli, E.; Vidyarathna, N.K.; Fiori, E.; Ou, L.; Flynn, K.J.; Mitra, A.; Stoecker, D.K.; Hansen, P.J. Metabolic and physiological changes in Prymnesium parvum when grown under, and grazing on prey of, variable nitrogen: Phosphorus stoichiometry. Harmful Algae 2016, 55, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nagai, S.; Nitshitani, G.; Tomaru, Y.; Sakiyama, S.; Kamiyama, T. Predation by the toxic dinoflagellate Dinophysis fortii on the ciliate Myrionecta rubra and observation of sequestration of ciliate chloroplasts. J. Phycol. 2008, 44, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Ojamäe, K.; Hansen, P.J.; Lips, I. Mass entrapment and lysis of Mesodinium rubrum cells in mucus threads observed in cultures with Dinophysis. Harmful Algae 2016, 55, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Giménez Papiol, G.; Beuzenberg, V.; Selwood, A.I.; MacKenzie, L.; Packer, M.A. The use of a mucus trap by Dinophysis acuta for the capture of Mesodinium rubrum prey under culture conditions. Harmful Algae 2016, 58, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nagai, S.; Suzuki, T.; Nishikawa, T.; Kamiyama, T. Differences in the Production and Excretion Kinetics of Okadaic Acid, Dinophysistoxin-1, and Pectenotoxin-2 between Cultures of Dinophysis acuminata and Dinophysis fortii Isolated from Western Japan1. J. Phycol. 2011, 47, 1326–1337. [Google Scholar] [CrossRef] [PubMed]
- Mafra, L.L., Jr.; Nagai, S.; Uchida, H.; Tavares, C.P.S.; Escobar, B.P.; Suzuki, T. Harmful effects of Dinophysis to the ciliate Mesodinium rubrum: Implications for prey capture. Harmful Algae 2016, 59, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Hattenrath-Lehmann, T.; Gobler, C.J. The contribution of inorganic and organic nutrients to the growth of a North American isolate of the mixotrophic Dinoflagellate, Dinophysis acuminata. Limnol. Oceanogr. 2015, 60, 1588–1630. [Google Scholar] [CrossRef]
- Sarmento, H.; Morana, C.; Gasol, J.M. Bacterioplankton niche partitioning in the use of phytoplankton-derived dissolved organic carbon: Quantity is more important than quality. ISME J. 2016, 10, 2582. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Hmelo, L.R.; van Tol, H.M.; Durham, B.P.; Carlson, L.T.; Heal, K.R.; Morales, R.L.; Berthiaume, C.T.; Parker, M.S.; Djunaedi, B.; et al. Interaction and signaling between a cosmopolitan phytoplankton and associated bacteria. Nature 2015, 522, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Jauzein, C.; Evans, A.N.; Erdner, D.L. The impact of associated bacteria on morphology and physiology of the dinoflagellate Alexandrium tamarense. Harmful Algae 2015, 50, 65–75. [Google Scholar] [CrossRef]
- Park, B.S.; Kim, J.-H.; Kim, J.H.; Gobler, C.J.; Baek, S.H.; Han, M.-S. Dynamics of bacterial community structure during blooms of Cochlodinium polykrikoides (Gymnodiniales, Dinophyceae) in Korean coastal waters. Harmful Algae 2015, 48, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Joo, J.-H.; Baek, K.-D.; Han, M.-S. A mutualistic interaction between the bacterium Pseudomonas asplenii and the harmful algal species Chattonella marina (Raphidophyceae). Harmful Algae 2016, 56, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Buhmann, M.T.; Schulze, B.; Forderer, A.; Schleheck, D.; Kroth, P.G. Bacteria may induce the secretion of mucin-like proteins by the diatom Phaeodactylum tricornutum. J. Phycol. 2016, 52, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Myung, G.; Yih, W.; Kim, H.S.; Park, J.S.; Cho, B.C. Ingestion of bacterial cells by the marine photosynthetic ciliate Myrionecta rubra. Aquat. Microb. Ecol. 2006, 44, 175–180. [Google Scholar] [CrossRef]
- Hattenrath-Lehmann, T.K.; Gobler, C.J. Identification of unique microbiomes associated with harmful algal blooms caused by Alexandrium fundyense and Dinophysis acuminata. Harmful Algae 2017, 68, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Herfort, L.; Peterson, T.D.; Prahl, F.G.; McCue, L.A.; Needoba, J.A.; Crump, B.C.; Roegner, G.C.; Campbell, V.; Zuber, P. Red Waters of Myrionecta rubra are Biogeochemical Hotspots for the Columbia River Estuary with Impacts on Primary/Secondary Productions and Nutrient Cycles. Estuaries Coasts 2012, 35, 878–891. [Google Scholar] [CrossRef]
- Mitra, A.; Flynn, K.J.; Burkholder, J.M.; Berge, T.; Calbet, A.; Raven, J.A.; Granéli, E.; Glibert, P.M.; Hansen, P.J.; Stoecker, D.K.; et al. The role of mixotrophic protists in the biological carbon pump. Biogeosciences 2014, 11, 995–1005. [Google Scholar] [CrossRef]
- Krabberød, A.K.; Bjorbækmo, M.F.M.; Shalchian-Tabrizi, K.; Logares, R. Exploring the oceanic microeukaryotic interactome with metaomics approaches. Aquat. Microb. Ecol. 2017, 79, 1–12. [Google Scholar] [CrossRef]
- Ward, C.S.; Yung, C.M.; Davis, K.M.; Blinebry, S.K.; Williams, T.C.; Johnson, Z.I.; Hunt, D.E. Annual community patterns are driven by seasonal switching between closely related marine bacteria. ISME J. 2017, 11, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Needham, D.M.; Fuhrman, J.A. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat. Microbiol. 2016, 1, 16005. [Google Scholar] [CrossRef] [PubMed]
- Hansen, P.J.; Nielsen, L.T.; Johnson, M.; Berge, T.; Flynn, K.J. Acquired phototrophy in Mesodinium and Dinophysis—A review of cellular organization, prey selectivity, nutrient uptake and bioenergetics. Harmful Algae 2013, 28, 126–139. [Google Scholar] [CrossRef]
- Kim, M.; Nam, S.W.; Shin, W.; Coats, D.W.; Park, M.G. Fate of green plastids in Dinophysis caudata following ingestion of the benthic ciliate Mesodinium coatsi: Ultrastructure and psbA gene. Harmful Algae 2015, 43, 66–73. [Google Scholar] [CrossRef]
- Reguera, B.; Velo-Suárez, L.; Raine, R.; Park, M.G. Harmful dinophysis species: A review. Harmful Algae 2012, 14, 87–106. [Google Scholar] [CrossRef]
- Osterholz, H.; Singer, G.; Wemheuer, B.; Daniel, R.; Simon, M.; Niggemann, J.; Dittmar, T. Deciphering associations between dissolved organic molecules and bacterial communities in a pelagic marine system. ISME J. 2016, 10, 1717. [Google Scholar] [CrossRef] [PubMed]
- Lundgaard, A.S.B.; Treusch, A.H.; Stief, P.; Thamdrup, B.; Glud, R.N. Nitrogen cycling and bacterial community structure of sinking and aging diatom aggregates. Aquat. Microb. Ecol. 2017, 79, 85–99. [Google Scholar] [CrossRef]
- Lucas, I.A.; Vesk, M. The fine structure of two photosynthetic species of Dinophysis (Dinophysiales, Dinophyceae). J. Phycol. 1990, 26, 345–357. [Google Scholar] [CrossRef]
- Hansen, P.J.; Ojamae, K.; Berge, T.; Trampe, E.C.; Nielsen, L.T.; Lips, I.; Kuhl, M. Photoregulation in a Kleptochloroplastidic Dinoflagellate, Dinophysis acuta. Front. Microbiol. 2016, 7, 785. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kang, Y.G.; Kim, H.S.; Yih, W.; Coats, D.W.; Park, M.G. Growth and grazing responses of the mixotrophic dinoflagellate Dinophysis acuminata as functions of light intensity and prey concentration. Aquatic Microb. Ecol. 2008, 51, 301–310. [Google Scholar] [CrossRef]
- Seeyave, S.; Probyn, T.A.; Pitcher, G.C.; Lucas, M.I.; Purdie, D.A. Nitrogen nutrition in assemblages dominated by Pseudo-nitzschia spp., Alexandrium catenella and Dinophysis acuminata off the west coast of South Africa. Mar. Ecol. Prog. Ser. 2009, 379, 91–107. [Google Scholar] [CrossRef]
- Harred, L.B.; Campbell, L. Predicting harmful algal blooms: A case study with Dinophysis ovum in the Gulf of Mexico. J. Plankton Res. 2014, 36, 1434–1445. [Google Scholar] [CrossRef]
- Landa, M.; Cottrell, M.T.; Kirchman, D.L.; Blain, S.; Obernosterer, I. Changes in bacterial diversity in response to dissolved organic matter supply in a continuous culture experiment. Aquat. Microb. Ecol. 2013, 69, 157–168. [Google Scholar] [CrossRef]
- Buchan, A.; LeCleir, G.R.; Gulvik, C.A.; Gonzalez, J.M. Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 2014, 12, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Riemann, L.; Steward, G.F.; Azam, F. Dynamics of Bacterial Community Composition and Activity during a Mesocosm Diatom Bloom. Appl. Environ. Microbiol. 2000, 66, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, J.M.; Herndl, G.J. Changes in bacterial β-glucosidase diversity during a coastal phytoplankton bloom. Limnol. Oceanogr. 2002, 47, 594–599. [Google Scholar] [CrossRef]
- Hattenrath-Lehmann, T.K.; Marcoval, M.A.; Mittlesdorf, H.; Goleski, J.A.; Wang, Z.; Haynes, B.; Morton, S.L.; Gobler, C.J. Nitrogenous nutrients promote the growth and toxicity of Dinophysis acuminata during Estuarine bloom events. PLoS ONE 2015, 10, e0124148. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Flynn, K.J.; Tillmann, U.; Raven, J.A.; Caron, D.; Stoecker, D.K.; Not, F.; Hansen, P.J.; Hallegraeff, G.; Sanders, R.; et al. Defining Planktonic Protist Functional Groups on Mechanisms for Energy and Nutrient Acquisition: Incorporation of Diverse Mixotrophic Strategies. Protist 2016, 167, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, M.T.; Kirchman, D.L. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low-and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 2000, 66, 1692–1697. [Google Scholar] [CrossRef] [PubMed]
- Pearman, J.K.; Casas, L.; Merle, T.; Michell, C.; Irigoien, X. Bacterial and protist community changes during a phytoplankton bloom. Limnol. Oceanogr. 2016, 61, 198–213. [Google Scholar] [CrossRef]
- Bolch, C.J.; Subramanian, T.A.; Green, D.H. The Toxic Dinoflagellate Gymnodinium Catenatum (Dinophyceae) Requires Marine Bacteria for Growth. J. Phycol. 2011, 47, 1009–1022. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.-H.; Ramanan, R.; Kim, B.-H.; Lee, J.; Kim, S.; Yoo, C.; Choi, G.-G.; Oh, H.-M.; Kim, H.-S.; Lindell, D. Novel approach for the development of axenic microalgal cultures from environmental samples. J. Phycol. 2013, 49, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, F.; Escalera, L.; Reguera, B.; Rial, P.; Riobó, P.; da Silva, T.D.J. Morphological variability, toxinology and genetics of the dinoflagellate Dinophysis tripos (Dinophysiaceae, Dinophysiales). Harmful Algae 2012, 13, 26–33. [Google Scholar] [CrossRef]
- Smith, J.L.; Tong, M.; Fux, E.; Anderson, D.M. Toxin production, retention, and extracellular release by Dinophysis acuminata during extended stationary phase and culture decline. Harmful Algae 2012, 19, 125–132. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Reyon, D.; Tsai, S.Q.; Khayter, C.; Foden, J.A.; Sander, J.D.; Joung, J.K. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012, 30, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bittinger, K.; Charlson, E.S.; Hoffmann, C.; Lewis, J.; Wu, G.D.; Collman, R.G.; Bushman, F.D.; Li, H. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef] [PubMed]
- Pedrós-Alió, C. The rare bacterial biosphere. Ann. Rev. Mar. Sci. 2012, 4, 449–466. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Package ‘vegan’. Community Ecol. Package 2013, 2, 34–115. [Google Scholar]
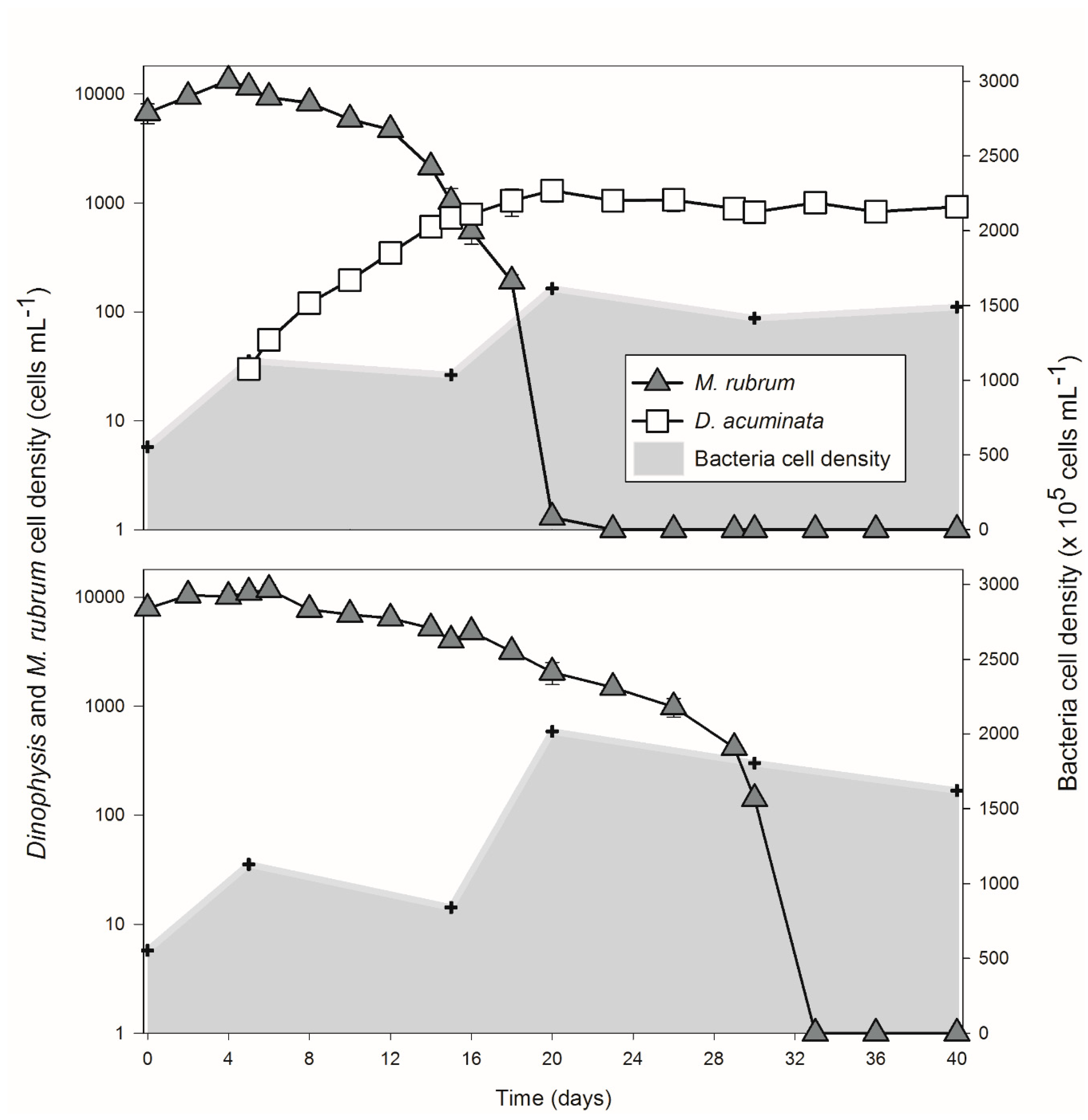


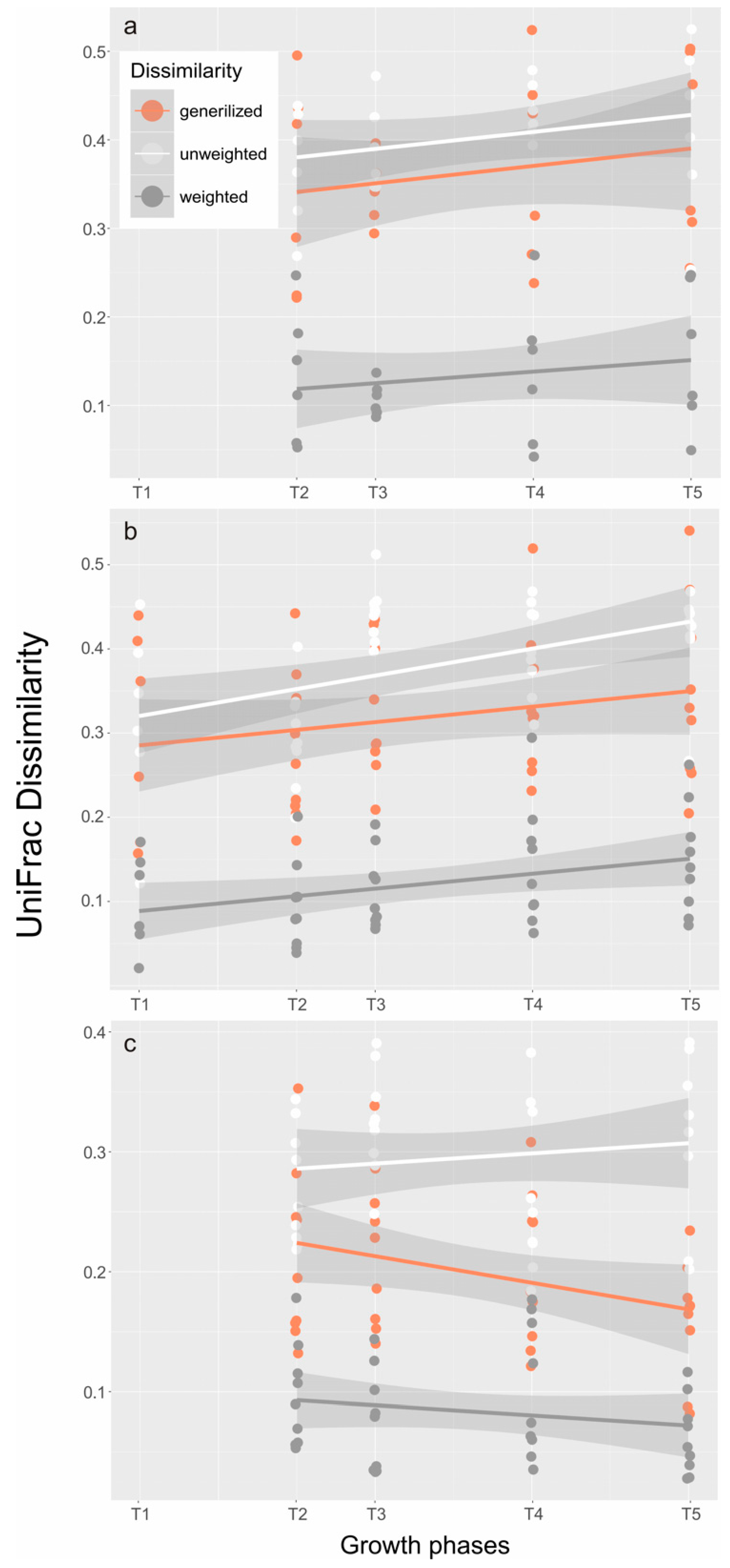
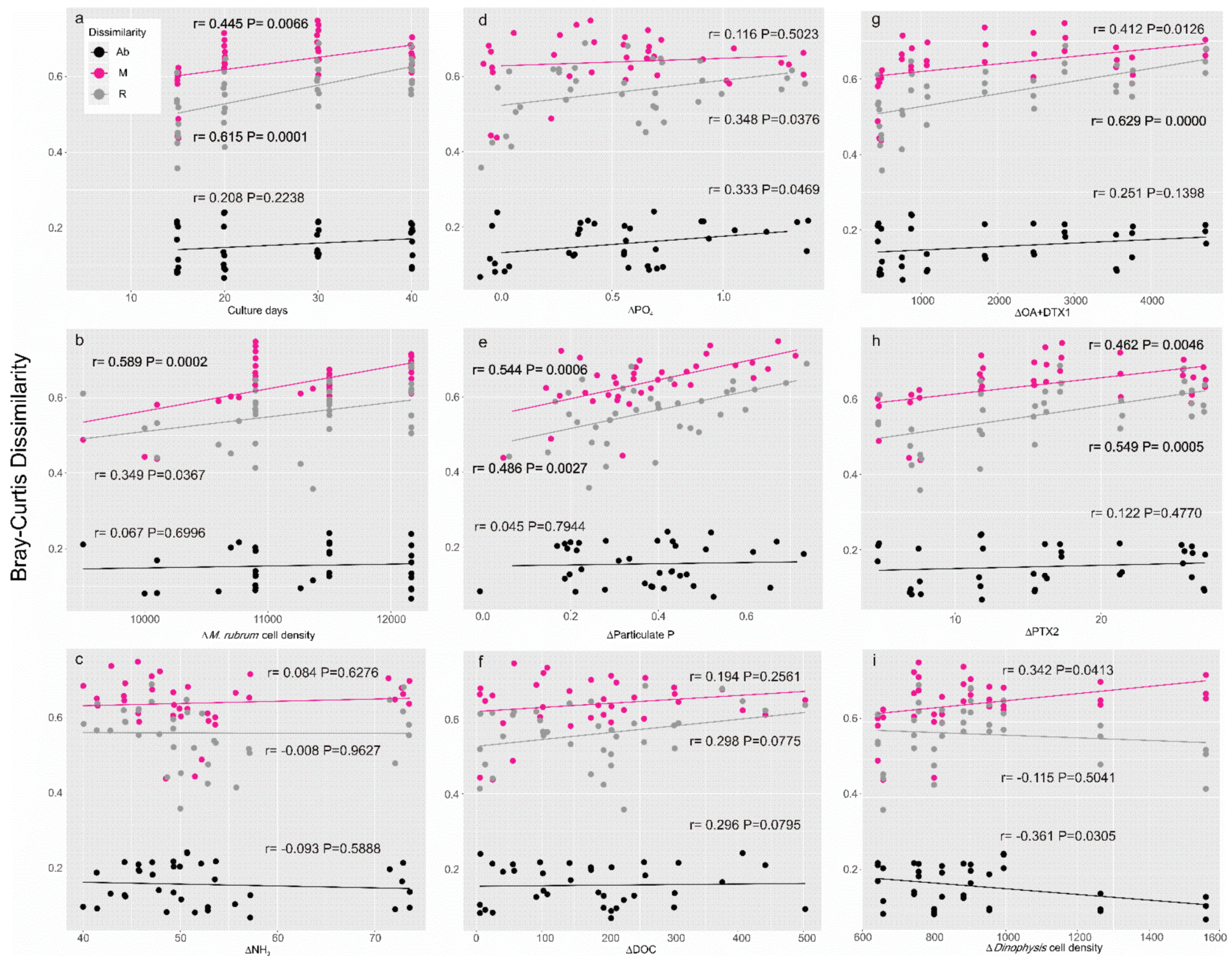
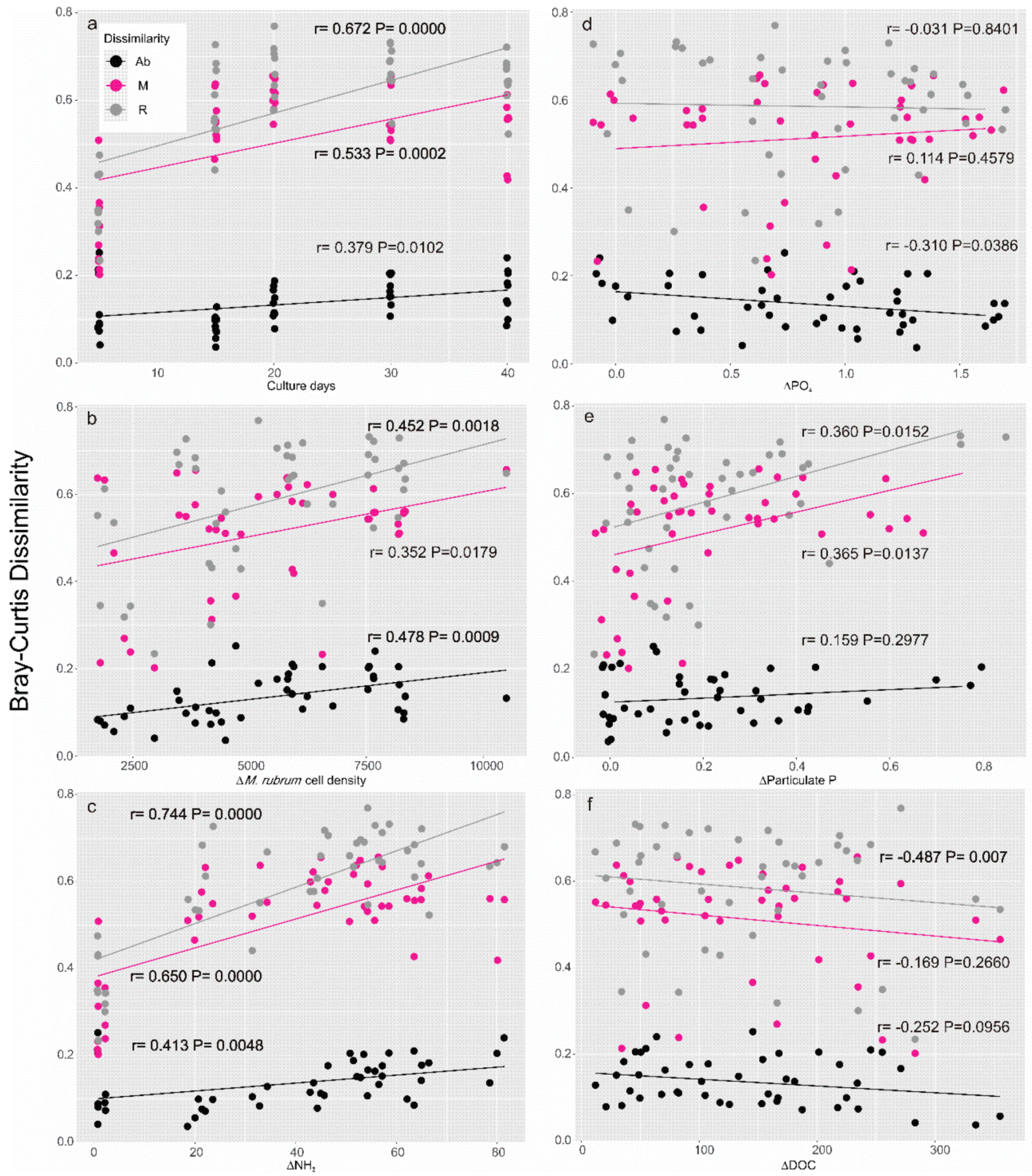

| Samples | POP (μM) | DIP (PO43−) (μM) | DIN (NH4+) (μM) | DIN (NO3−) (μM) | DOC (μM) | POC (μM) | OA + DTX1 (pg mL−1) | PTX2 (ng mL−1) |
|---|---|---|---|---|---|---|---|---|
| T0 | 1.21 ± 0.05 | 2.90 ± 0.85 | 2.86 ± 2.02 | 45.95 ± 5.77 | 633.61 ± 74.23 | 792.80 ± 133.67 | - | - |
| T1 | 1.26 ± 0.06 | 2.80 ± 0.37 | 2.62 ± 0.82 | 9.76 ± 5.77 | 977.85 ± 283.32 | 789.13 ± 124.75 | 99.33 ± 5.71 | 0.43 ± 0.03 |
| T2 (A) | 1.51 ± 0.12 | 2.47 ± 0.19 | 54.29 ± 1.89 | 2.50 ± 2.53 | 606.24 ± 211.48 | 666.22 ± 22.08 | 559.88 ± 23.09 | 6.87 ± 1.46 |
| T2 (B) | 1.31 ± 0.32 | 2.26 ± 0.32 | 30.36 ± 6.89 | 0.48 ± 0.41 | 463.64 ± 118.71 | 659.62 ± 30.02 | - | - |
| T3 (A) | 1.68 ± 0.03 | 2.37 ± 0.37 | 62.62 ± 11.79 | LOD | 693.88 ± 124.99 | 661.13 ± 23.72 | 1001.08 ± 163.95 | 13.45 ± 2.11 |
| T3 (B) | 1.45 ± 0.14 | 2.26 ± 0.32 | 50.24 ± 4.76 | 0.71 ± 0.71 | 764.90 ± 56.27 | 580.10 ± 16.55 | - | - |
| T4 (A) | 1.75 ± 0.17 | 2.15 ± 0.19 | 50.36 ± 2.51 | LOD | 614.06 ± 111.31 | 739.42 ± 53.55 | 2494.61 ± 526.41 | 18.71 ± 2.74 |
| T4 (B) | 1.65 ± 0.25 | 2.15 ± 0.19 | 57.86 ± 2.58 | 2.86 ± 2.02 | 636.20 ± 69.36 | 510.67 ± 56.38 | - | - |
| T5 (A) | 1.63 ± 0.19 | 2.04 ± 0.19 | 55.48 ± 17.04 | LOD | 391.74 ± 63.33 | 866.42 ± 70.63 | 4109.58 ± 621.79 | 26.74 ± 0.73 |
| T5 (B) | 1.35 ± 1.16 | 2.04 ± 0.19 | 72.38 ± 9.10 | LOD | 479.49 ± 36.14 | 409.98 ± 137.61 | - | - |
| Sample ID | Valid Tags | Valid% | Goods Coverage | OTU Counts | Simpson | Shannon Wiener | Chao1 |
|---|---|---|---|---|---|---|---|
| T0-1 | 35,860 | 91.37% | 0.9995 | 57 | 1.31 | 0.51 | 64.8 |
| T0-2 | 37,205 | 89.24% | 0.9995 | 79 | 1.75 | 0.56 | 83.4 |
| T0-3 | 36,596 | 91.55% | 0.9994 | 57 | 1.69 | 0.59 | 81.0 |
| T1-1 | 25,396 | 85.29% | 0.9996 | 81 | 1.93 | 0.57 | 85.0 |
| T1-2 | - | - | - | - | - | - | - |
| T1-3 | 38,153 | 92.66% | 0.9994 | 49 | 1.24 | 0.50 | 67.2 |
| T2-1 (A) | 38,611 | 92.30% | 0.9996 | 52 | 1.84 | 0.61 | 56.0 |
| T2-2 (A) | 34,985 | 90.78% | 0.9998 | 51 | 1.99 | 0.64 | 52.3 |
| T2-3 (A) | 33,591 | 87.98% | 0.9992 | 55 | 1.29 | 0.47 | 82.1 |
| T2-1 (B) | 34,373 | 89.63% | 0.9995 | 54 | 1.65 | 0.56 | 63.8 |
| T2-2 (B) | 36,081 | 91.48% | 0.9995 | 48 | 1.60 | 0.54 | 59.0 |
| T2-3 (B) | 35,202 | 87.22% | 0.9996 | 40 | 1.71 | 0.57 | 46.0 |
| T3-1 (A) | 38,304 | 92.79% | 0.9995 | 59 | 1.55 | 0.53 | 65.6 |
| T3-2 (A) | 35,826 | 91.63% | 0.9996 | 43 | 1.25 | 0.44 | 49.0 |
| T3-3 (A) | 36,145 | 92.71% | 0.9997 | 47 | 1.52 | 0.51 | 51.7 |
| T3-1 (B) | 37,516 | 91.71% | 0.9998 | 42 | 1.38 | 0.49 | 43.9 |
| T3-2 (B) | 35,153 | 90.96% | 0.9995 | 44 | 1.16 | 0.42 | 63.5 |
| T3-3 (B) | 36,480 | 91.28% | 0.9996 | 41 | 1.56 | 0.51 | 44.3 |
| T4-1 (A) | 35,231 | 91.41% | 0.9998 | 42 | 1.56 | 0.51 | 43.7 |
| T4-2 (A) | 37,869 | 90.60% | 0.9995 | 47 | 1.68 | 0.53 | 66.5 |
| T4-3 (A) | 36,610 | 92.61% | 0.9995 | 53 | 1.62 | 0.52 | 66.0 |
| T4-1 (B) | 38,995 | 93.12% | 0.9997 | 47 | 1.37 | 0.47 | 50.5 |
| T4-2 (B) | 35,561 | 87.96% | 0.9996 | 45 | 1.85 | 0.60 | 49.5 |
| T4-3 (B) | 34,364 | 90.38% | 0.9996 | 47 | 1.36 | 0.43 | 56.2 |
| T5-1 (A) | 33,756 | 88.31% | 0.9996 | 46 | 1.86 | 0.62 | 55.2 |
| T5-2 (A) | 37,201 | 88.99% | 0.9995 | 51 | 1.66 | 0.54 | 64.0 |
| T5-3 (A) | 36,041 | 89.82% | 0.9995 | 57 | 1.70 | 0.56 | 65.7 |
| T5-1 (B) | 36,348 | 91.57% | 0.9993 | 48 | 1.42 | 0.46 | 86.3 |
| T5-2 (B) | 36,231 | 90.09% | 0.9994 | 48 | 1.78 | 0.59 | 63.2 |
| T5-3 (B) | 29,843 | 86.83% | 0.9995 | 70 | 1.88 | 0.56 | 77.3 |
| Mantel Test | Pearson Correlation | Spearman Correlation | ||
|---|---|---|---|---|
| Statistic r | p Value | Statistic r | p Value | |
| Ab taxa | 0.361 | 0.002 | 0.363 | 0.001 |
| M taxa | 0.593 | 0.001 | 0.575 | 0.001 |
| R taxa | 0.520 | 0.001 | 0.542 | 0.001 |
| BIOENV | Pearson Correlation | Parameters in Best Model | Spearman Correlation | Parameters in Best Model |
|---|---|---|---|---|
| Ab taxa | 0.340 | DOC, OA + DTX1, PTX2, M. rubrum, Dinophysis | 0.287 | PO43−, DOC, OA + DTX1, M. rubrum, Dinophysis |
| M taxa | 0.756 | PO43−, NH4+, OA + DTX1, PTX2, M. rubrum, Dinophysis | 0.729 | PO43−, NH4+, OA + DTX1, PTX2, M. rubrum, Dinophysis |
| R taxa | 0.661 | POP, PO43−, NH4+, PTX2, M. rubrum | 0.670 | POP, PO43−, NH4+, PTX2, M. rubrum |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, H.; Hua, C.; Tong, M. Impact of Dinophysis acuminata Feeding Mesodinium rubrum on Nutrient Dynamics and Bacterial Composition in a Microcosm. Toxins 2018, 10, 443. https://doi.org/10.3390/toxins10110443
Gao H, Hua C, Tong M. Impact of Dinophysis acuminata Feeding Mesodinium rubrum on Nutrient Dynamics and Bacterial Composition in a Microcosm. Toxins. 2018; 10(11):443. https://doi.org/10.3390/toxins10110443
Chicago/Turabian StyleGao, Han, Chenfeng Hua, and Mengmeng Tong. 2018. "Impact of Dinophysis acuminata Feeding Mesodinium rubrum on Nutrient Dynamics and Bacterial Composition in a Microcosm" Toxins 10, no. 11: 443. https://doi.org/10.3390/toxins10110443
APA StyleGao, H., Hua, C., & Tong, M. (2018). Impact of Dinophysis acuminata Feeding Mesodinium rubrum on Nutrient Dynamics and Bacterial Composition in a Microcosm. Toxins, 10(11), 443. https://doi.org/10.3390/toxins10110443




