Repellency, Toxicity, Gene Expression Profiling and In Silico Studies to Explore Insecticidal Potential of Melaleuca alternifolia Essential Oil against Myzus persicae
Abstract
1. Introduction
2. Results and Discussion
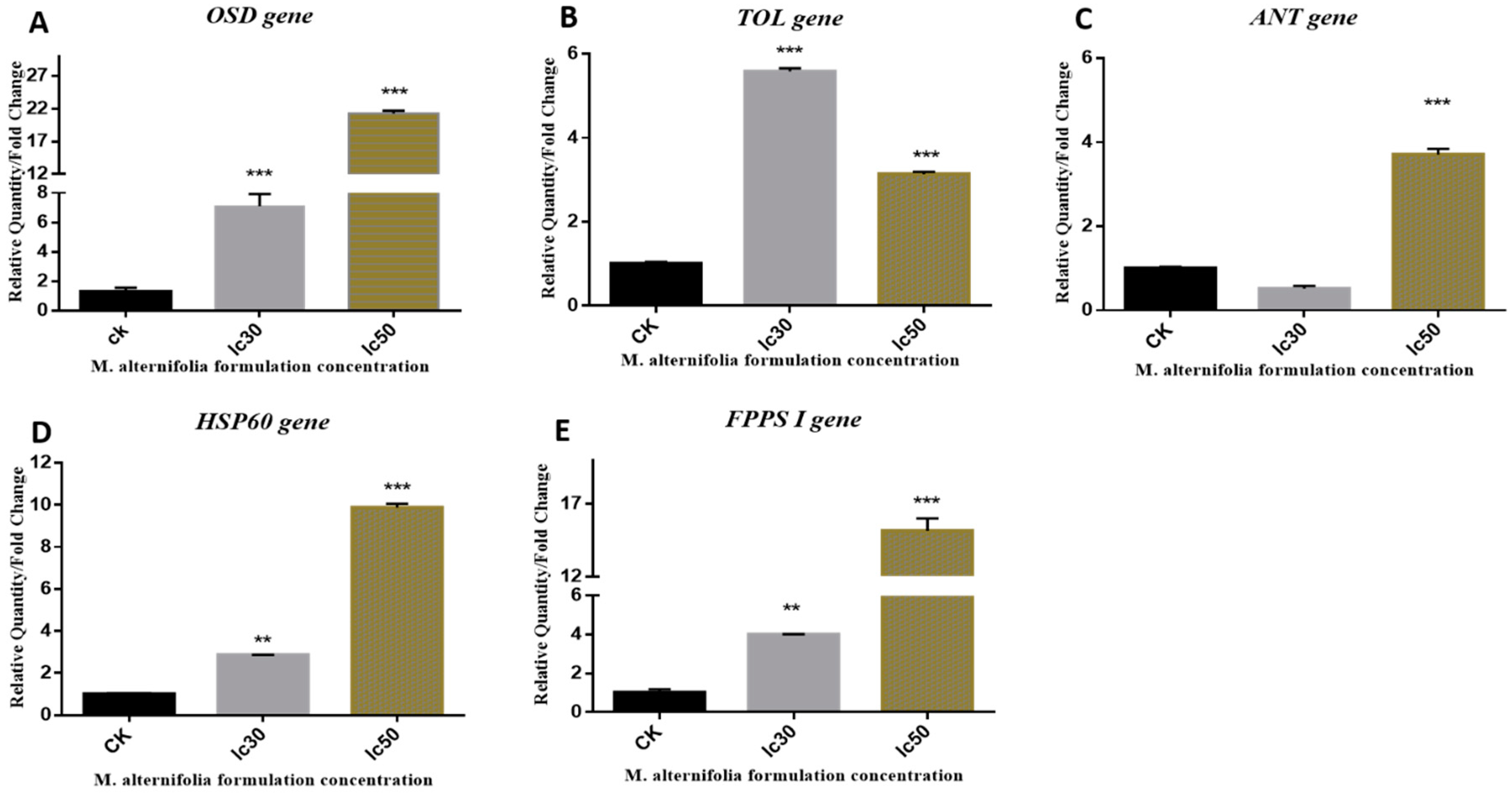
2.1. Gene Expression
2.1.1. OSD Gene
2.1.2. TOL Gene
2.1.3. ANT Gene
2.1.4. HSP 60 Gene
2.1.5. FPPS I Gene
2.2. In Silico Studies
2.2.1. Structural Description of the OSD, FPPS I, and HSP 60 3D Model
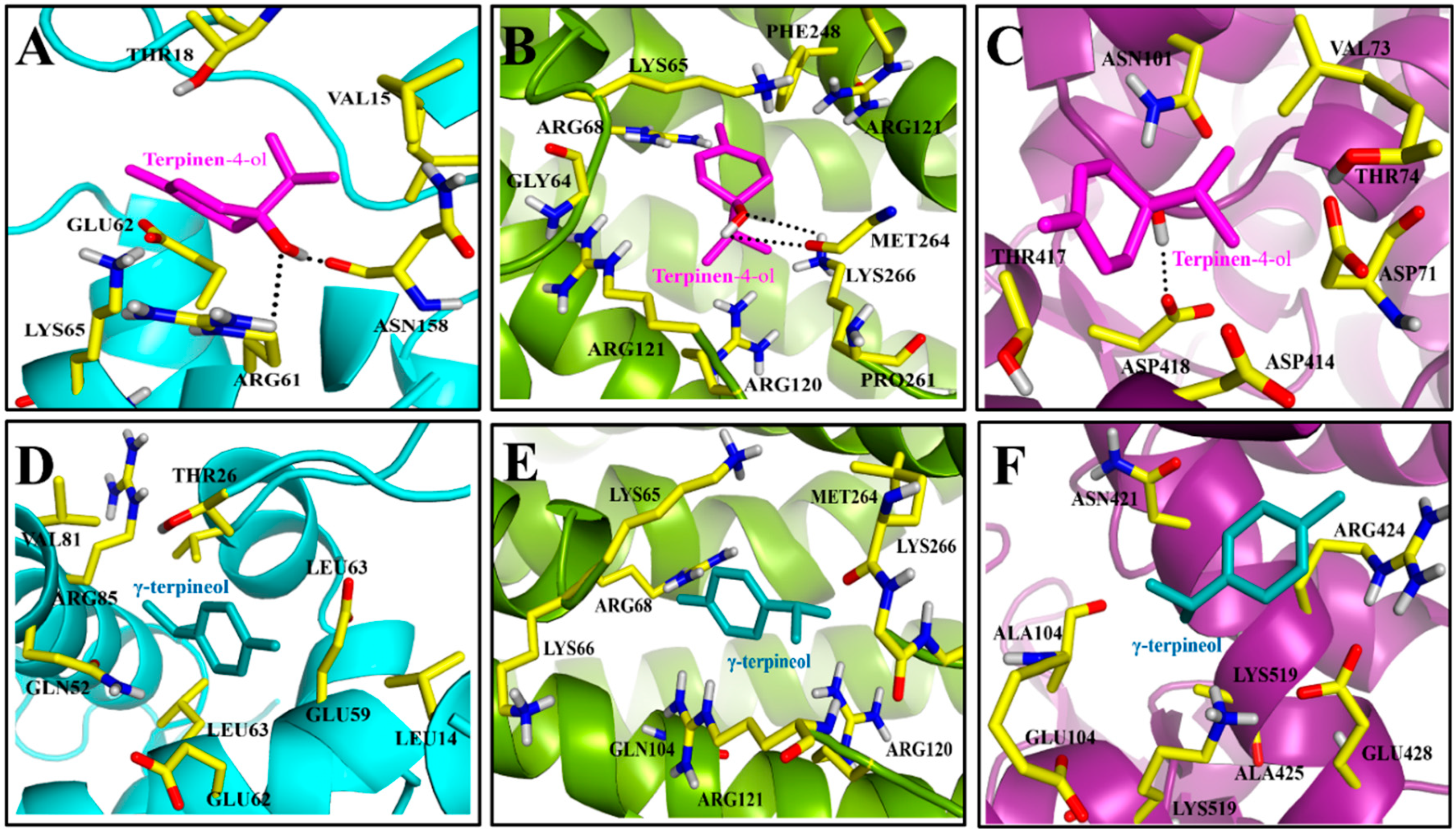
2.2.2. Molecular Docking and Molecular Dynamics Simulation
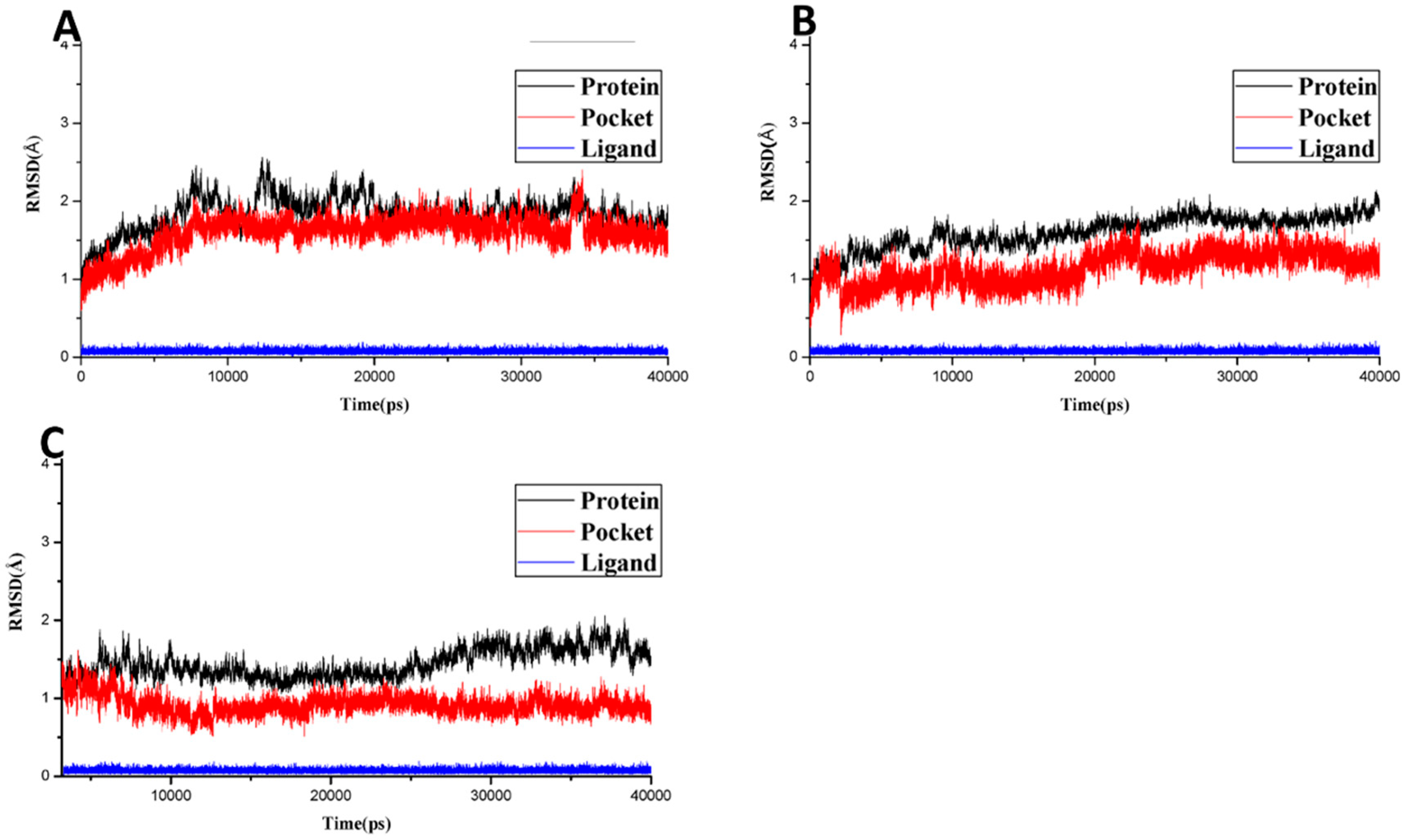
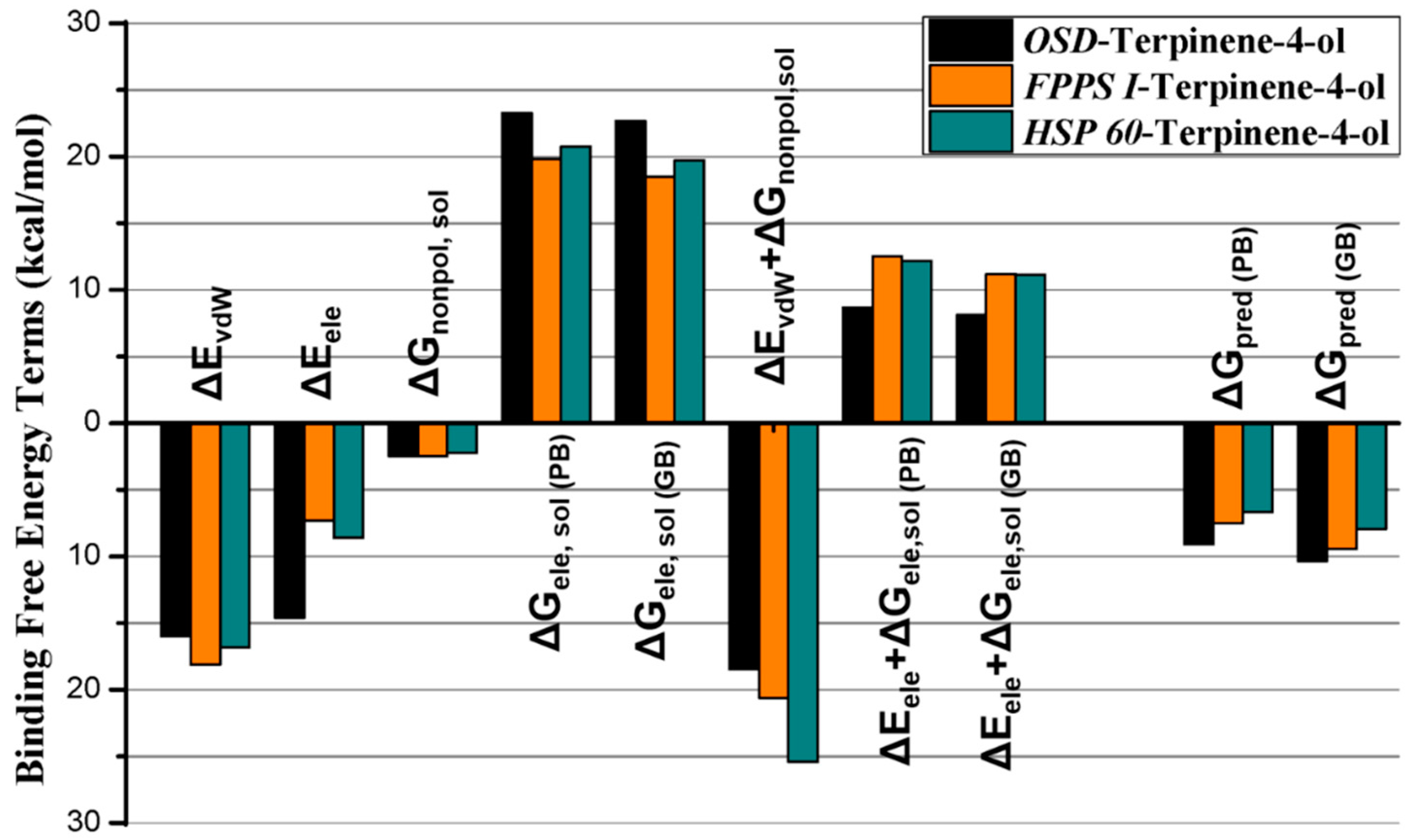
2.2.3. Molecular Dynamics Simulation, MMGB/PBSA
3. Significance of Study
4. Conclusions
5. Materials and Methods
5.1. 40% M. alternifolia Essential Oil Preparation
5.2. Myzus Persicae Culture
5.3. Bioassay of M. persicae
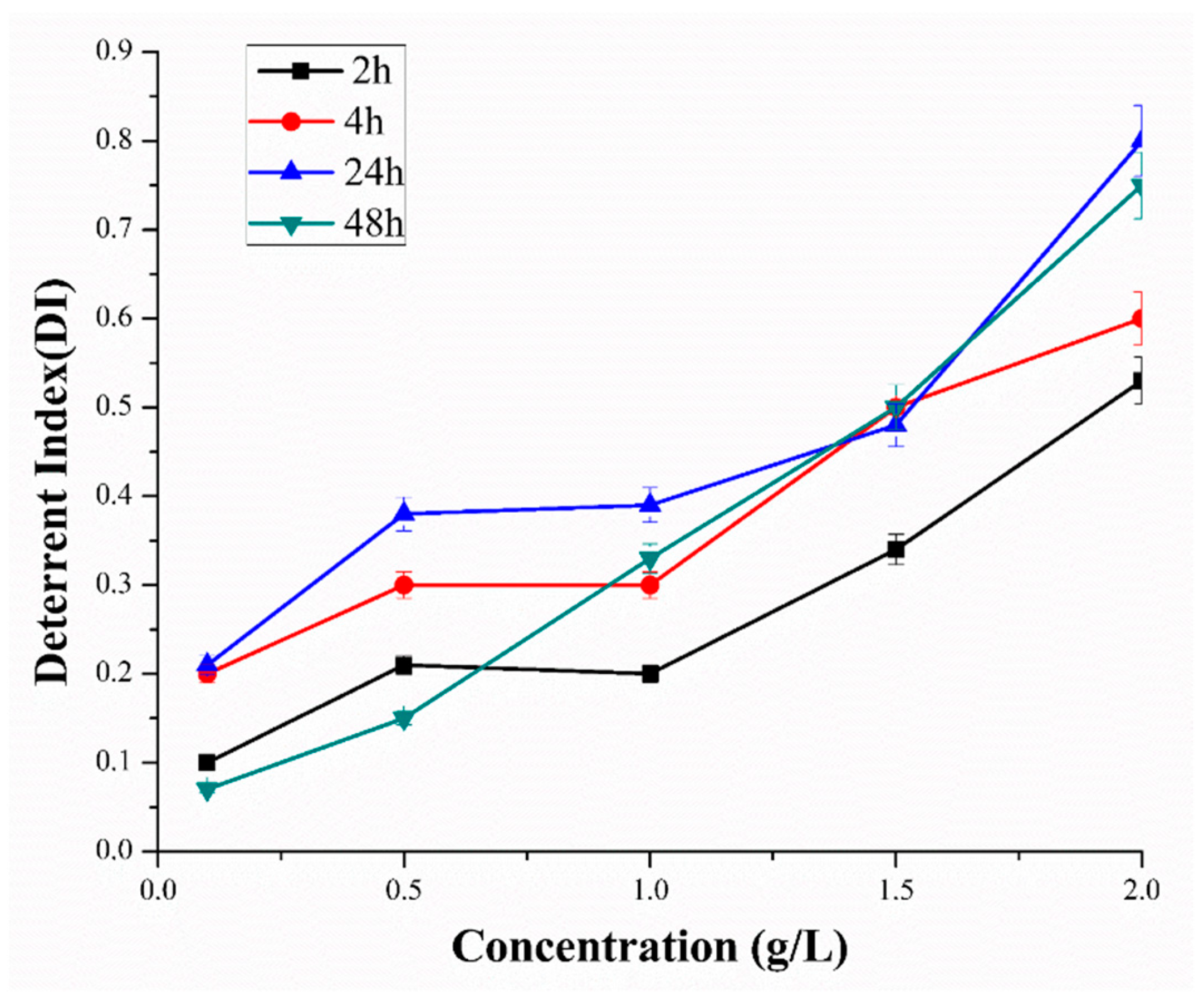
5.3.1. Deterrent Assay
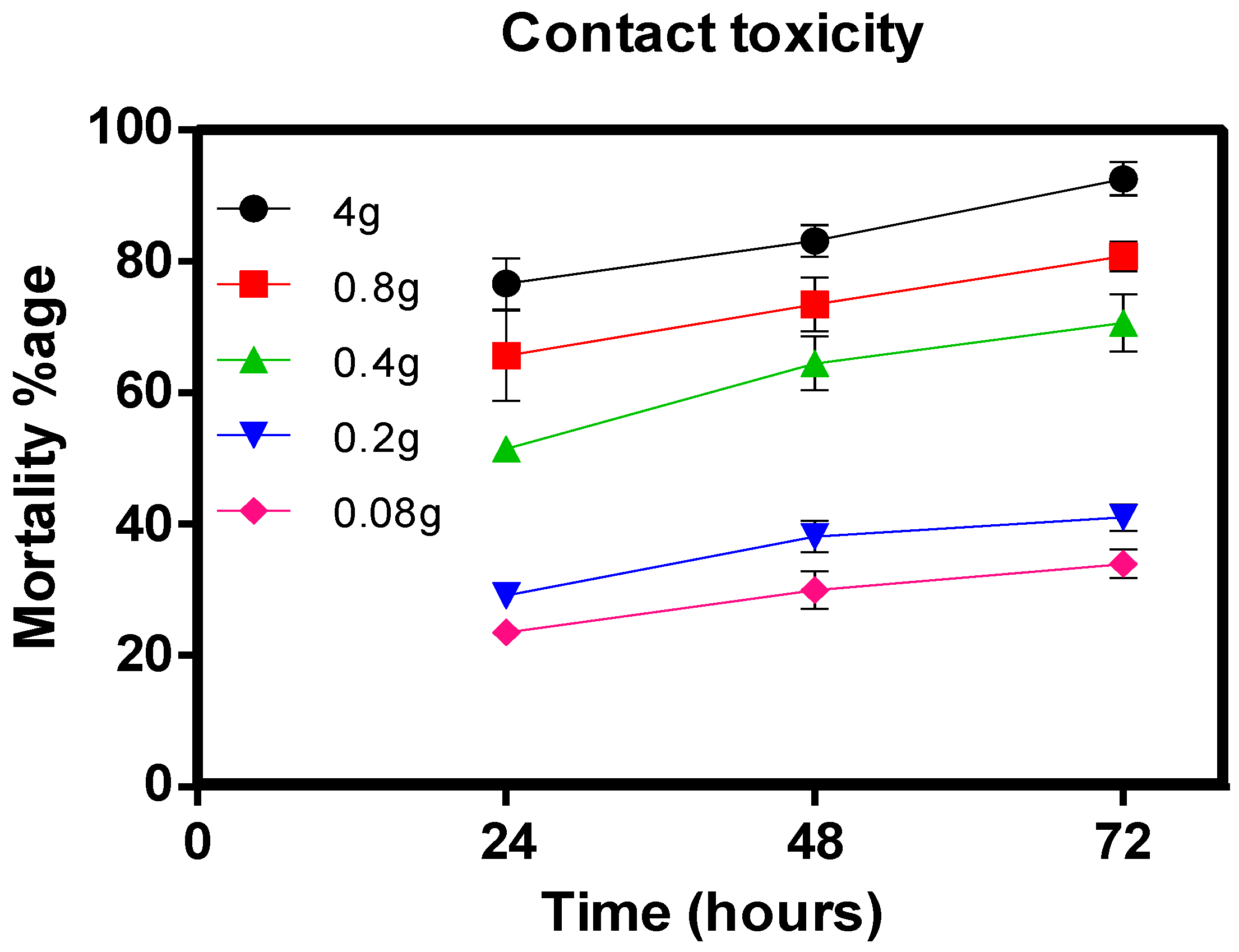
5.3.2. Leaf-Dip Bioassay
5.4. Real-Time Quantitative PCR (qRT-PCR)
5.5. Computation Methods
5.5.1. Sequence Analysis and Modeling
5.5.2. Ligand Preparation and Molecular Docking
5.5.3. Molecular Dynamics Simulations
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bakkali, F.; Averbeck, S.; Averbeck, D.; Idaomar, M. Biological effects of essential oils—A review. Food Chem. Toxicol. 2008, 46, 446–475. [Google Scholar] [CrossRef] [PubMed]
- Dedryver, C.-A.; Le Ralec, A.; Fabre, F. The conflicting relationships between aphids and men: A review of aphid damage and control strategies. C. R. Biol. 2010, 333, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Edwards, O.R.; Franzmann, B.; Thackray, D.; Micic, S. Insecticide resistance and implications for future aphid management in Australian grains and pastures: A review. Aust. J. Exp. Agric. 2008, 48, 1523–1530. [Google Scholar] [CrossRef]
- Hagenbucher, S.; Wäckers, F.L.; Romeis, J. Aphid honeydew quality as a food source for parasitoids is maintained in Bt cotton. PLoS ONE 2014, 9, e107806. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.X.; Jander, G.; Samaniego, H.; Ramsey, J.S.; Figueroa, C.C. Insecticide resistance mechanisms in the green peach aphid Myzus persicae (hemiptera: Aphididae) I: A transcriptomic survey. PLoS ONE 2012, 7, e36366. [Google Scholar] [CrossRef] [PubMed]
- Isman, M.B. Plant essential oils for pest and disease management. Crop Prot. 2000, 19, 603–608. [Google Scholar] [CrossRef]
- Khani, A.; Heydarian, M. Fumigant and repellent properties of sesquiterpene-rich essential oil from Teucrium polium subsp. Capitatum (L.). Asian Pac. J. Trop. Med. 2014, 7, 956–961. [Google Scholar] [CrossRef]
- Mossa, A.-T.H. Green pesticides: Essential oils as biopesticides in insect-pest management. J. Environ. Sci. Technol. 2016, 9, 354. [Google Scholar] [CrossRef]
- Nouri-Ganbalani, G.; Ebadollahi, A.; Nouri, A. Chemical composition of the essential oil of Eucalyptus procera dehnh. and its insecticidal effects against two stored product insects. J. Essent. Oil Bear. Plants 2016, 19, 1234–1242. [Google Scholar] [CrossRef]
- Carson, C.; Hammer, K.; Riley, T. Melaleuca alternifolia (tea tree) oil: A review of antimicrobial and other medicinal properties. Clin. Microbiol. Rev. 2006, 19, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, N.A.; Hammer, K.A.; Riley, T.V.; Van Belkum, A.; Carson, C.F. Effect of habituation to tea tree (Melaleuca alternifolia) oil on the subsequent susceptibility of Staphylococcus spp. to antimicrobials, triclosan, tea tree oil, terpinen-4-ol and carvacrol. Int. J. Antimicrob. Agents 2013, 41, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liao, M.; Yang, Q.; Xiao, J.; Hu, Z.; Zhou, L.; Cao, H. Transcriptome profiling reveals differential gene expression of detoxification enzymes in Sitophilus zeamais responding to terpinen-4-ol fumigation. Pestic. Biochem. Phys. 2018. [Google Scholar] [CrossRef] [PubMed]
- Gould, M.N. Cancer chemoprevention and therapy by monoterpenes. Environ. Health Perspect. 1997, 105, 977–979. [Google Scholar] [PubMed]
- Shelton, D.; Leach, D.; Baverstock, P.; Henry, R. Isolation of genes involved in secondary metabolism from Melaleuca alternifolia (Cheel) using expressed sequence tags (ESTs). Plant Sci. 2002, 162, 9–15. [Google Scholar] [CrossRef]
- Demusyak, A. International standards organization (ISO). Meas. Tech. 1967, 10, 651–655. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Y.; Su, Z.; Xian, J. Insecticidal and repellent action of pogostone against Myzus persicae (hemiptera: Aphididae). Fla. Entomol. 2017, 100, 346–349. [Google Scholar] [CrossRef][Green Version]
- Ebrahimi, M.; Safaralizade, M.H.; Valizadegan, O. Contact toxicity of Azadirachta indica (Adr. Juss.), Eucalyptus camaldulensis (Dehn.) and Laurus nobilis (L.) essential oils on mortality cotton aphids, aphis gossypii glover (hem.: Aphididae). Arch. Phytopathol. Plant Prot. 2013, 46, 2153–2162. [Google Scholar] [CrossRef]
- Ghanim, M.; Dombrovsky, A.; Raccah, B.; Sherman, A. A microarray approach identifies ANT, OS-D and takeout-like genes as differentially regulated in alate and apterous morphs of the green peach aphid Myzus persicae (Sulzer). Insect Biochem. Mol. Biol. 2006, 36, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Stanley, K.; Fenton, B. A member of the Hsp60 gene family from the peach potato aphid, Myzus persicae (Sulzer.). Insect Mol. Biol. 2000, 9, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Ayyanath, M.-M.; Cutler, G.C.; Scott-Dupree, C.D.; Prithiviraj, B.; Kandasamy, S.; Prithiviraj, K. Gene expression during imidacloprid-induced hormesis in green peach aphid. Dose-Response 2014, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Roote, J.; Brogna, S.; Davis, A.W.; Barbash, D.A.; Nash, D.; Ashburner, M. Stress sensitive B encodes an adenine nucleotide translocase in Drosophila melanogaster. Genetics 1999, 153, 891–903. [Google Scholar] [PubMed]
- Fujikawa, K.; Seno, K.; Ozaki, M. A novel takeout-like protein expressed in the taste and olfactory organs of the blowfly, Phormia regina. FEBS 2006, 273, 4311–4321. [Google Scholar] [CrossRef] [PubMed]
- Weil, T.; Korb, J.; Rehli, M. Comparison of queen-specific gene expression in related lower termite species. Mol. Biol. Evol. 2009, 26, 1841–1850. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bohbot, J.; Vogt, R.G. Antennal expressed genes of the yellow fever mosquito (Aedes aegypti L.); characterization of odorant-binding protein 10 and takeout. Insect Biochem. Mol. Biol. 2005, 35, 961–979. [Google Scholar] [CrossRef] [PubMed]
- Dauwalder, B.; Tsujimoto, S.; Moss, J.; Mattox, W. The drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 2002, 16, 2879–2892. [Google Scholar] [CrossRef] [PubMed]
- Parsell, D.; Lindquist, S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993, 27, 437–496. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-H.; Chen, B.; Kang, L. Impact of mild temperature hardening on thermotolerance, fecundity, and Hsp gene expression in Liriomyza huidobrensis. J. Insect Phys. 2007, 53, 1199–1205. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.; Prosser, I.; Mohib, A.; Field, L. Cloning and characterisation of a prenyltransferase from the aphid Myzus persicae with potential involvement in alarm pheromone biosynthesis. Insect Mol. Biol. 2008, 17, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Vandermoten, S.; Charloteaux, B.; Santini, S.; Sen, S.E.; Béliveau, C.; Vandenbol, M.; Francis, F.; Brasseur, R.; Cusson, M.; Haubruge, É. Characterization of a novel aphid prenyltransferase displaying dual geranyl/farnesyl diphosphate synthase activity. FEBS Lett. 2008, 582, 1928–1934. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Singhal, N.; Kumar, M.; Virdi, J.S. Molecular analysis of β-lactamase genes to understand their differential expression in strains of Yersinia enterocolitica biotype 1A. Sci. Rep. 2014, 4, 5270. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, G.H.I.M.; Indika, P.P.A.M.S.; Chandrasekharan, N.V.; Weerasinghe, M.S.S.; Wijesundera, R.L.C.; Wijesundera, W.S.S. Trichoderma virens β-glucosidase I (BGL I) gene; expression in Saccharomyces cerevisiae including docking and molecular dynamics studies. BMC Microbiol. 2017, 17, 137. [Google Scholar] [CrossRef] [PubMed]
- Smialowski, P.; Martin-Galiano, A.J.; Mikolajka, A.; Girschick, T.; Holak, T.A.; Frishman, D. Protein solubility: Sequence based prediction and experimental verification. Bioinformatics 2006, 23, 2536–2542. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.N. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003, 46, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Xiao, J.J.; Zhou, L.J.; Yao, X.; Tang, F.; Hua, R.M.; Wu, X.W.; Cao, H.Q. Chemical composition, insecticidal and biochemical effects of Melaleuca alternifolia essential oil on the Helicoverpa armigera. J. Appl. Entomol. 2017, 141, 721–728. [Google Scholar] [CrossRef]
- Cheng, C. The Antimicrobial Activity of Tea Tree Oil (Melaleuca alternifolia) Oil and Preparation of Its Emulsion in Water. Master’s Thesis, Anhui Agricultural University, Hefei, China, 2012. (In Chinese). [Google Scholar]
- You, C.X.; Jiang, H.Y.; Zhang, W.J.; Guo, S.S.; Yang, K.; Lei, N.; Ma, P.; Geng, Z.F.; Du, S.S. Contact toxicity and repellency of the main components from the essential oil of Clausena anisum-olens against two stored product insects. J. Insect Sci. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Moores, G.D.; Gao, X.; Denholm, I.; Devonshire, A.L. Characterisation of insensitive acetylcholinesterase in insecticide-resistant cotton aphids, Aphis gossypiiglover (homoptera: Aphididae). Pestic. Biochem. Phys. 1996, 56, 102–110. [Google Scholar] [CrossRef]
- Fong, D.K.; Kim, S.; Chen, Z.; DeSarbo, W.S. A bayesian multinomial probit model for the analysis of panel choice data. Psychometrika 2016, 81, 161–183. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-H.; Choi, W.-S.; Lee, S.-E.; Park, B.-S. Fumigant toxicity of essential oils and their constituent compounds towards the rice weevil, Sitophilus oryzae (L.). Crop Prot. 2001, 20, 317–320. [Google Scholar] [CrossRef]
- Rainen, L.; Oelmueller, U.; Jurgensen, S.; Wyrich, R.; Ballas, C.; Schram, J.; Herdman, C.; Bankaitis-Davis, D.; Nicholls, N.; Trollinger, D. Stabilization of mRNA expression in whole blood samples. Clin. Chem. 2002, 48, 1883–1890. [Google Scholar] [PubMed]
- Zhang, Y. I-Tasser server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Horio, Y.; Sugama, K.; Ito, S.; Liu, Y.; Fukui, H. Genomic cloning of the rat histamine H1 receptor. Biochem. Biophys. Res. Commun. 1993, 190, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U. Uniprot: A hub for protein information. Nucleic Acids Res. 2014, 43, D204–D212. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. Procheck: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. Prosa-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Ruppert, J.; Welch, W.; Jain, A.N. Automatic identification and representation of protein binding sites for molecular docking. Protein Sci. 1997, 6, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Chohan, T.A.; Chen, J.-J.; Qian, H.-Y.; Pan, Y.-L.; Chen, J.-Z. Molecular modeling studies to characterize N-phenylpyrimidin-2-amine selectivity for CDK2 and CDK4 through 3D-QSAR and molecular dynamics simulations. Mol. BioSyst. 2016, 12, 1250–1268. [Google Scholar] [CrossRef] [PubMed]
- Chohan, T.A.; Qian, H.-Y.; Pan, Y.-L.; Chen, J.-Z. Molecular simulation studies on the binding selectivity of 2-anilino-4-(thiazol-5-yl)-pyrimidines in complexes with CDK2 and CDK7. Mol. BioSyst. 2016, 12, 145–161. [Google Scholar] [CrossRef] [PubMed]
- Case, D.; Betz, R.; Botello-Smith, W.; Cerutti, D., III; Duke, R.; Giese, T.; Gohlke, H.; Goetz, A. Amber 16; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Rehman, K.; Chohan, T.A.; Waheed, I.; Gilani, Z.; Akash, M.S.H. Taxifolin prevents postprandial hyperglycemia by regulating the activity of α-amylase: Evidence from an in vivo and in silico studies. J. Cell. Biochem. 2018. [Google Scholar] [CrossRef] [PubMed]
| Compound | % (g/g) | Properties |
|---|---|---|
| M. alternifolia essential oil | 40 | Active ingredient |
| Ethylene glycol | 2.5 | Antifreeze |
| Epoxy ethane epoxide propane block polymer | 2.5 | Emulsifier |
| Polyoxyethylene Polyoxypropylene ether | 2.5 | Emulsifier |
| Silicone | 0.3 | Emulsifier |
| Gelatin | 0.2 | Thickener |
| Water | 52 | Deionized water |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chohan, T.A.; Chohan, T.A.; Zhou, L.; Yang, Q.; Min, L.; Cao, H. Repellency, Toxicity, Gene Expression Profiling and In Silico Studies to Explore Insecticidal Potential of Melaleuca alternifolia Essential Oil against Myzus persicae. Toxins 2018, 10, 425. https://doi.org/10.3390/toxins10110425
Chohan TA, Chohan TA, Zhou L, Yang Q, Min L, Cao H. Repellency, Toxicity, Gene Expression Profiling and In Silico Studies to Explore Insecticidal Potential of Melaleuca alternifolia Essential Oil against Myzus persicae. Toxins. 2018; 10(11):425. https://doi.org/10.3390/toxins10110425
Chicago/Turabian StyleChohan, Talha Ali, Tahir Ali Chohan, Lijun Zhou, Qianqian Yang, Liao Min, and Haiqun Cao. 2018. "Repellency, Toxicity, Gene Expression Profiling and In Silico Studies to Explore Insecticidal Potential of Melaleuca alternifolia Essential Oil against Myzus persicae" Toxins 10, no. 11: 425. https://doi.org/10.3390/toxins10110425
APA StyleChohan, T. A., Chohan, T. A., Zhou, L., Yang, Q., Min, L., & Cao, H. (2018). Repellency, Toxicity, Gene Expression Profiling and In Silico Studies to Explore Insecticidal Potential of Melaleuca alternifolia Essential Oil against Myzus persicae. Toxins, 10(11), 425. https://doi.org/10.3390/toxins10110425