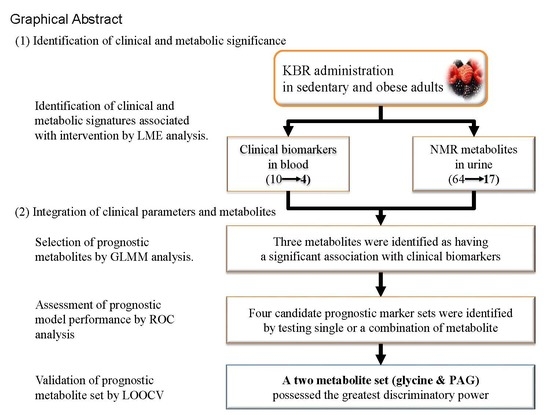
Integration of Traditional and Metabolomics Biomarkers Identifies Prognostic Metabolites for Predicting Responsiveness to Nutritional Intervention against Oxidative Stress and Inflammation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Materials
2.2. Participants
2.3. Experimental Design
2.4. Measurements of Traditional Biomarkers
2.5. 1H NMR Spectroscopy and Pre-Processing of NMR Spectra
2.6. Statistical Analysis
3. Results
3.1. Preliminary Study to Compare Oxidative Stress and Inflammation in the KBR and NAB Groups Using Traditional Biomarkers
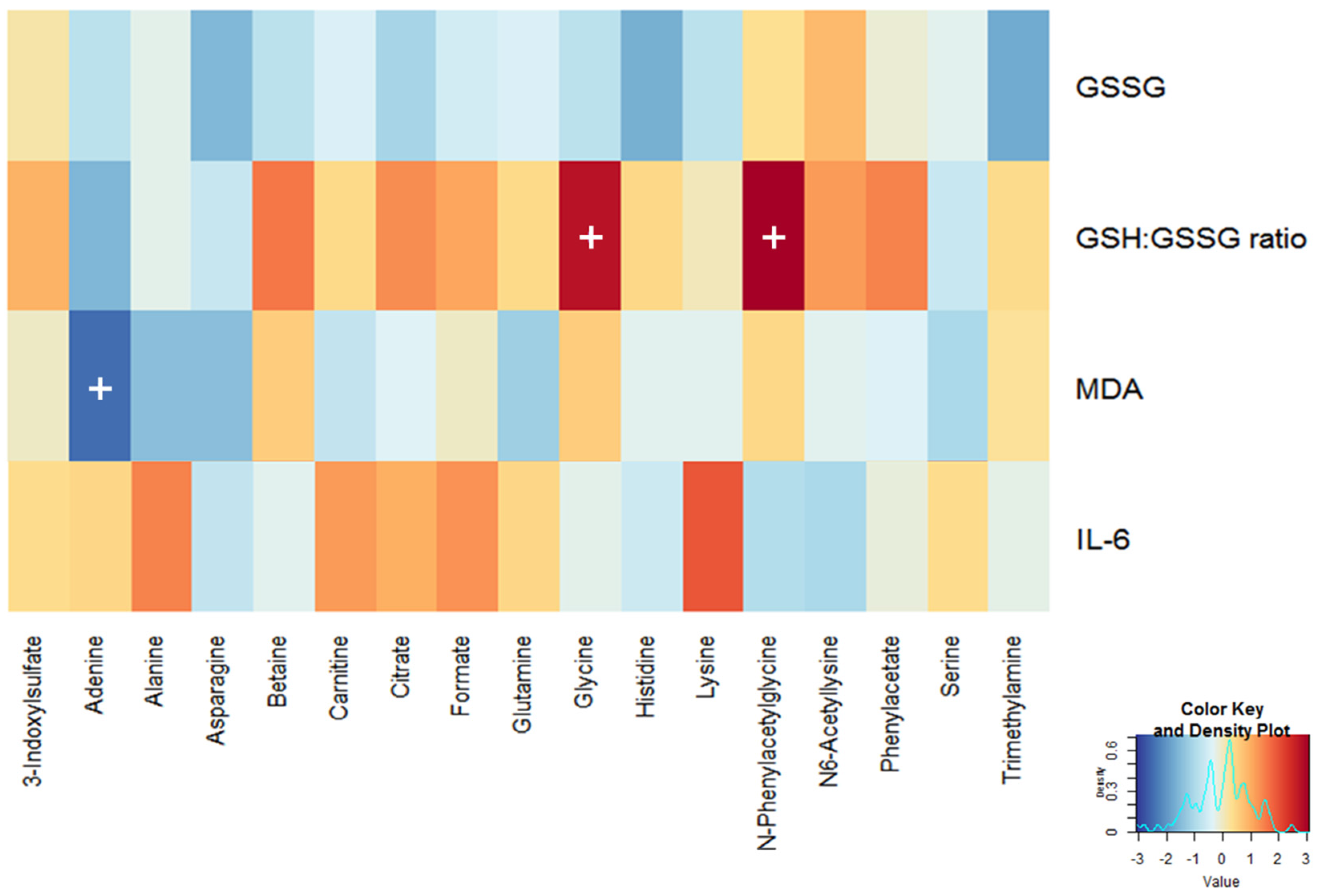
3.2. Selection of Traditional Biomarkers and Metabolites for Integration
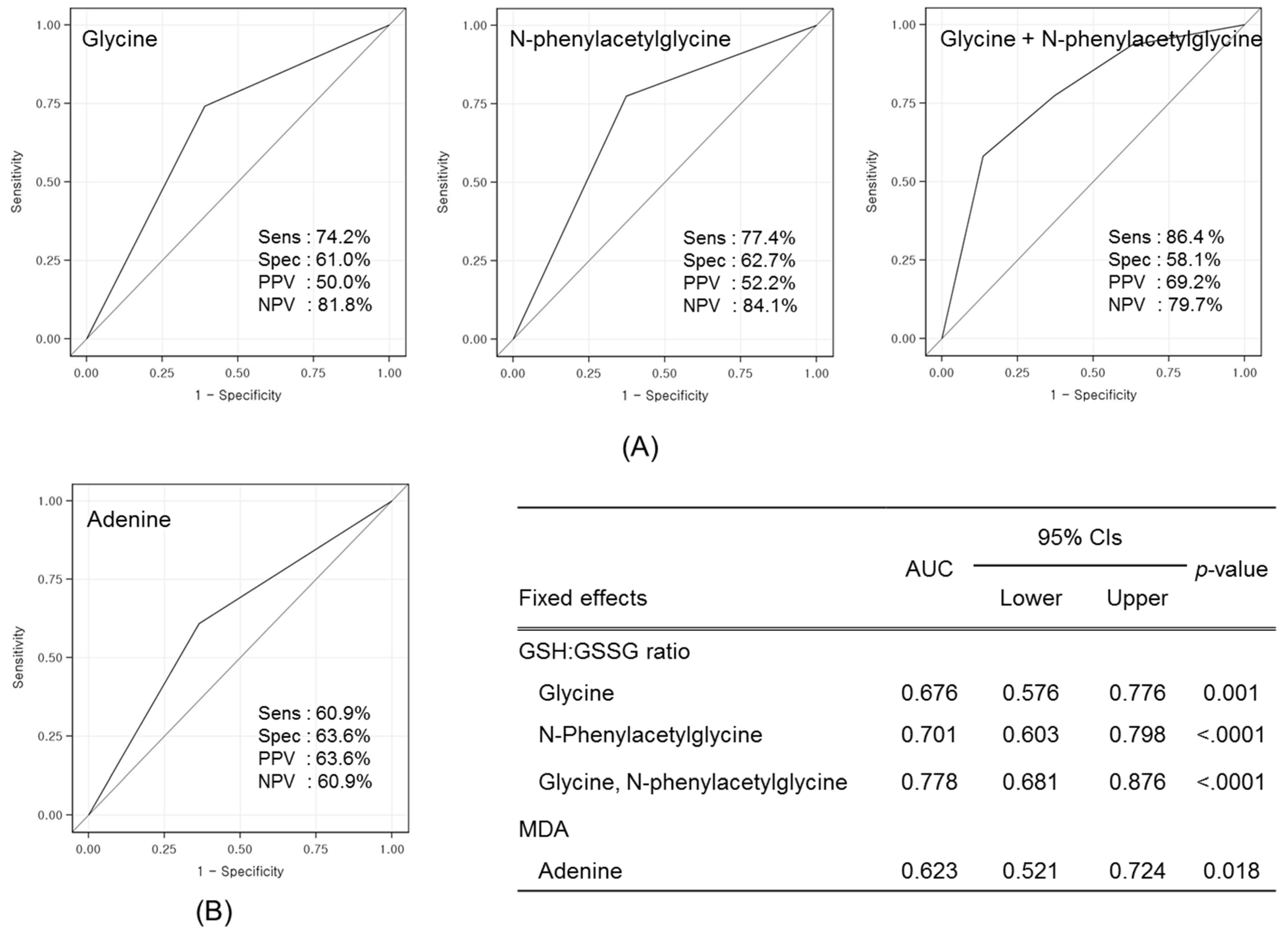
3.3. Identification of Candidate Prognostic Metabolites
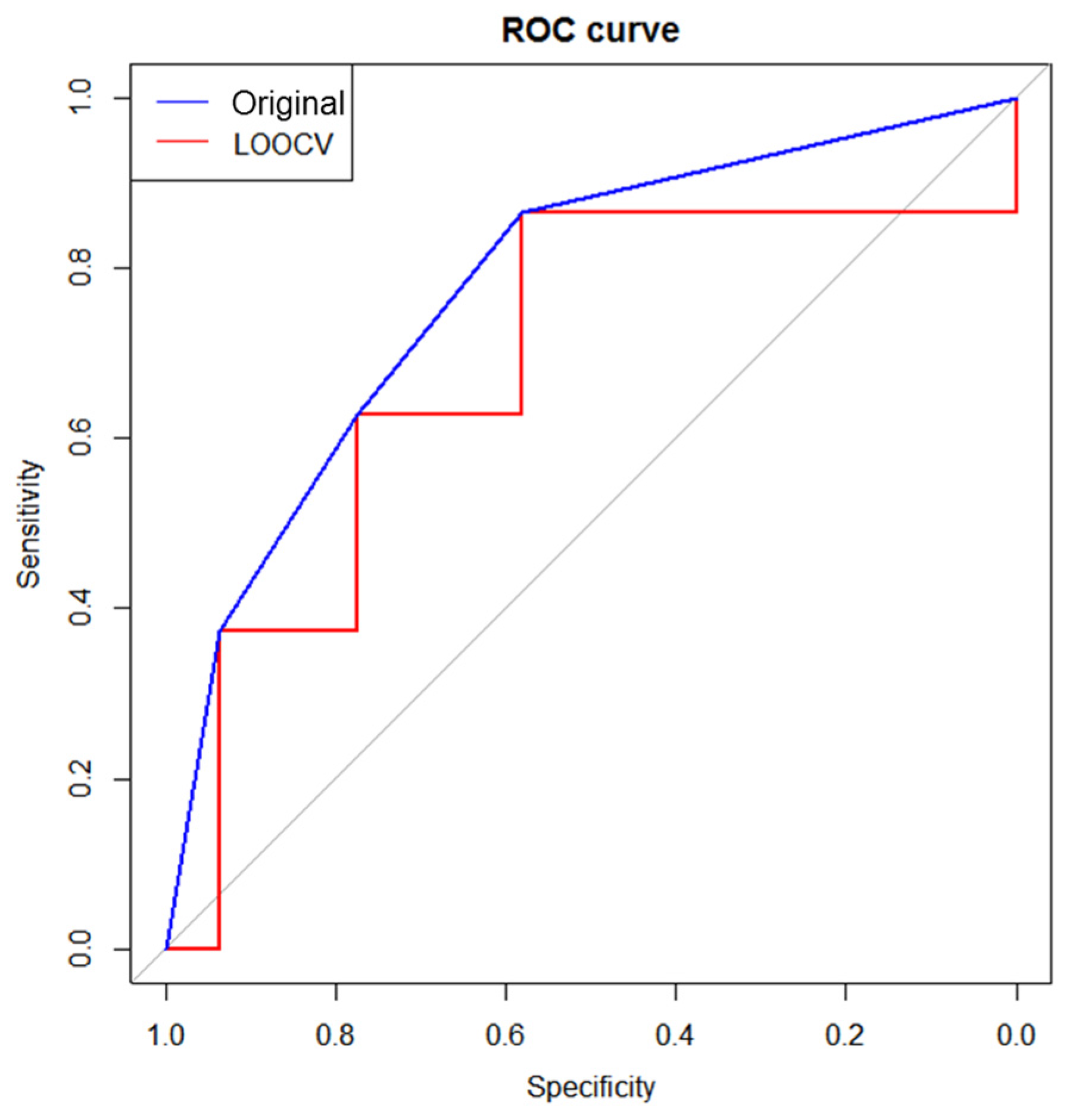
3.4. Validation of Prognostic Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sarkar, D.; Fisher, P.B. Molecular mechanisms of aging-associated inflammation. Cancer Lett. 2006, 236, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Holt, E.M.; Steffen, L.M.; Moran, A.; Basu, S.; Steinberger, J.; Ross, J.A.; Hong, C.P.; Sinaiko, A.R. Fruit and vegetable consumption and its relation to markers of inflammation and oxidative stress in adolescents. J. Am. Diet. Assoc. 2009, 109, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.H.; Lai, C.S.; Dushenkov, S.; Ho, C.T. Modulation of inflammatory genes by natural dietary bioactive compounds. J. Agric. Food Chem. 2009, 57, 4467–4477. [Google Scholar] [CrossRef] [PubMed]
- van Dorsten, F.A.; Grun, C.H.; van Velzen, E.J.; Jacobs, D.M.; Draijer, R.; van Duynhoven, J.P. The metabolic fate of red wine and grape juice polyphenols in humans assessed by metabolomics. Mol. Nutr. Food Res. 2010, 54, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, B.; Lee, S.; Erickson, D.; Mehta, S. Precision nutrition - review of methods for point-of-care assessment of nutritional status. Curr. Opin. Biotechnol. 2016, 44, 103–108. [Google Scholar] [CrossRef] [PubMed]
- van Ommen, B.; Keijer, J.; Kleemann, R.; Elliott, R.; Drevon, C.A.; McArdle, H.; Gibney, M.; Müller, M. The challenges for molecular nutrition research 2: Quantification of the nutritional phenotype. Genes Nutr. 2008, 3, 51–59. [Google Scholar] [CrossRef] [PubMed]
- van Ommen, B.; Keijer, J.; Heil, S.G.; Kaput, J. Challenging homeostasis to define biomarkers for nutrition related health. Mol. Nutr. Food Res. 2009, 53, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Gibney, M.J.; Walsh, M.; Brennan, L.; Roche, H.M.; German, B.; van Ommen, B. Metabolomics in human nutrition: Opportunities and challenges. Am. J. Clin. Nutr. 2005, 82, 497–503. [Google Scholar] [PubMed]
- Garcia-Aloy, M.; Llorach, R.; Urpi-Sarda, M.; Jauregui, O.; Corella, D.; Ruiz-Canela, M.; Salas-Salvado, J.; Fito, M.; Ros, E.; Estruch, R.; et al. A metabolomics-driven approach to predict cocoa product consumption by designing a multimetabolite biomarker model in free-living subjects from the predimed study. Mol. Nutr. Food Res. 2015, 59, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Heinzmann, S.S.; Brown, I.J.; Chan, Q.; Bictash, M.; Dumas, M.E.; Kochhar, S.; Stamler, J.; Holmes, E.; Elliott, P.; Nicholson, J.K. Metabolic profiling strategy for discovery of nutritional biomarkers: Proline betaine as a marker of citrus consumption. Am. J. Clin. Nutr. 2010, 92, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Ros, R.; Urpi-Sarda, M.; Lamuela-Raventos, R.M.; Estruch, R.; Martinez-Gonzalez, M.A.; Bullo, M.; Aros, F.; Cherubini, A.; Andres-Lacueva, C. Resveratrol metabolites in urine as a biomarker of wine intake in free-living subjects: The predimed study. Free Radic. Biol. Med. 2009, 46, 1562–1566. [Google Scholar] [CrossRef] [PubMed]
- Heinzmann, S.S.; Holmes, E.; Kochhar, S.; Nicholson, J.K.; Schmitt-Kopplin, P. 2-furoylglycine as a candidate biomarker of coffee consumption. J. Agric. Food chem. 2015, 63, 8615–8621. [Google Scholar] [CrossRef] [PubMed]
- McKeown, N.M.; Marklund, M.; Ma, J.; Ross, A.B.; Lichtenstein, A.H.; Livingston, K.A.; Jacques, P.F.; Rasmussen, H.M.; Blumberg, J.B.; Chen, C.Y. Comparison of plasma alkylresorcinols (ar) and urinary ar metabolites as biomarkers of compliance in a short-term, whole-grain intervention study. Eur. J. Nutr. 2015, 55, 1235–1244. [Google Scholar] [CrossRef] [PubMed]
- Hanhineva, K.; Lankinen, M.A.; Pedret, A.; Schwab, U.; Kolehmainen, M.; Paananen, J.; de Mello, V.; Sola, R.; Lehtonen, M.; Poutanen, K.; et al. Nontargeted metabolite profiling discriminates diet-specific biomarkers for consumption of whole grains, fatty fish, and bilberries in a randomized controlled trial. J. Nutr. 2015, 145, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Betts, J.; Gonzalez, J. Personalised nutrition: What makes you so special? Nutr. Bull. 2016, 41, 353–359. [Google Scholar] [CrossRef]
- Elnenaei, M.O.; Chandra, R.; Mangion, T.; Moniz, C. Genomic and metabolomic patterns segregate with responses to calcium and vitamin D supplementation. Br. J. Nutr. 2011, 105, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Kuehnbaum, N.L.; Gillen, J.B.; Gibala, M.J.; Britz-McKibbin, P. Personalized metabolomics for predicting glucose tolerance changes in sedentary women after high-intensity interval training. Scientific reports 2014, 4, 6166. [Google Scholar] [CrossRef] [PubMed]
- Hyun, T.K.; Lee, S.; Rim, Y.; Kumar, R.; Han, X.; Lee, S.Y.; Lee, C.H.; Kim, J.Y. De-novo rna sequencing and metabolite profiling to identify genes involved in anthocyanin biosynthesis in korean black raspberry (rubus coreanus miquel). PLoS ONE 2014, 9, e88292. [Google Scholar] [CrossRef] [PubMed]
- Finn, C.; Wennstrom, K.; Link, J.; Ridout, J. Evaluation of rubus leucodermis populations from the pacific northwest. HortScience 2003, 38, 1169–1172. [Google Scholar]
- Lee, J.; Dossett, M.; Finn, C.E. Anthocyanin fingerprinting of true bokbunja (rubus coreanus miq.) fruit. J. Funct. Foods 2013, 5, 1985–1990. [Google Scholar] [CrossRef]
- Park, S.J.; Hyun, S.-H.; Suh, H.W.; Lee, S.-Y.; Min, T.-S.; Auh, J.-H.; Lee, H.-J.; Kim, J.-H.; Cho, S.-M.; Choi, H.-K. Differentiation of black raspberry fruits according to species and geographic origins by genomic analysis and 1h-nmr-based metabolic profiling. J. Korean Soc. Appl. Biol. Chem. 2012, 55, 633–642. [Google Scholar] [CrossRef]
- Fisher-Wellman, K.; Bloomer, R.J. Acute exercise and oxidative stress: A 30 year history. Dyn. Med. DM 2009, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Park, S.J.; Hyun, S.-H.; Yang, S.-O.; Lee, J.; Auh, J.-H.; Kim, J.-H.; Cho, S.-M.; Marriott, P.J.; Choi, H.-K. Biochemical monitoring of black raspberry (rubus coreanus miquel) fruits according to maturation stage by 1h nmr using multiple solvent systems. Food Res. Int. 2011, 44, 1977–1987. [Google Scholar] [CrossRef]
- Kim, L.S.; Youn, S.H.; Kim, J.Y. Comparative study on antioxidant effects of extracts from rubus coreanus and rubus occidentalis. J. Korean Soc. Food Sci. Nutr. 2014, 43, 1357–1362. [Google Scholar] [CrossRef]
- Suh, H.W.; Kim, S.-H.; Park, S.J.; Hyun, S.-H.; Lee, S.-Y.; Auh, J.-H.; Lee, H.J.; Cho, S.-M.; Kim, J.-H.; Choi, H.-K. Effect of korean black raspberry (rubus coreanus miquel) fruit administration on DNA damage levels in smokers and screening biomarker investigation using 1h-nmr-based metabolic profiling. Food Res. Int. 2013, 54, 1255–1262. [Google Scholar] [CrossRef]
- Lee, J.E.; Park, E.; Lee, J.E.; Auh, J.H.; Choi, H.K.; Lee, J.; Cho, S.; Kim, J.H. Effects of a rubus coreanus miquel supplement on plasma antioxidant capacity in healthy Korean men. Nutr. Res. Pract. 2011, 5, 429–434. [Google Scholar] [CrossRef] [PubMed]
- WHO Expert Consultation. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet 2004, 363, 157–163. [Google Scholar]
- Food & Nutrition Information Center. Food & Nutrition Information Center. Physical Activity. In Dietary Reference Intakes; The National Academies Press: Washington, DC, USA, 2005; Chapter 12; pp. 880–935. [Google Scholar]
- Rahman, I.; Kode, A.; Biswas, S.K. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 2006, 1, 3159–3165. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, C.C.; Duarte, I.F.; Gomes, J.; Carrola, J.; Barros, A.S.; Gil, A.M.; Bousquet, J.; Bom, A.T.; Rocha, S.M. Urinary metabolomic changes as a predictive biomarker of asthma exacerbation. J. Allergy Clin. Immunol. 2014, 133, 261–263 e261–265. [Google Scholar] [CrossRef] [PubMed]
- Carrieri, M.; Trevisan, A.; Bartolucci, G.B. Adjustment to concentration-dilution of spot urine samples: Correlation between specific gravity and creatinine. Int. Arch. Occup. Environ. Health 2001, 74, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D. R Development Core Team. Nlme: Linear and Nonlinear Mixed Effects Models. 2015, 3.1-122. Available online: https://CRAN.R-project.org/package=nlme (accessed on 10 January 2016).
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S. Gplots: Various R Programming Tools for Plotting Data. R Package Version 2.12. 1. 2013. Available online: https://CRAN.R-project.org/package=gplots (accessed on 10 January 2016).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Muller, M. Proc: An open-source package for r and s+ to analyze and compare roc curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, P.; Giordan, M.; Wehrens, R. Multiple comparisons in mass-spectrometry-based-omics technologies. TrAC Trends Anal. Chem. 2013, 50, 11–21. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Zgoda-Pols, J.R.; Chowdhury, S.; Wirth, M.; Milburn, M.V.; Alexander, D.C.; Alton, K.B. Metabolomics analysis reveals elevation of 3-indoxyl sulfate in plasma and brain during chemically-induced acute kidney injury in mice: Investigation of nicotinic acid receptor agonists. Toxicol. Appl. Pharmacol. 2011, 255, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Vincent, H.K.; Innes, K.E.; Vincent, K.R. Oxidative stress and potential interventions to reduce oxidative stress in overweight and obesity. Diabetes Obes. Metab. 2007, 9, 813–839. [Google Scholar] [CrossRef] [PubMed]
- Lenz, E.M.; Bright, J.; Wilson, I.D.; Morgan, S.R.; Nash, A.F. A 1h nmr-based metabonomic study of urine and plasma samples obtained from healthy human subjects. J. Pharm. Biomed. Anal. 2003, 33, 1103–1115. [Google Scholar] [CrossRef]
- Stella, C.; Beckwith-Hall, B.; Cloarec, O.; Holmes, E.; Lindon, J.C.; Powell, J.; van der Ouderaa, F.; Bingham, S.; Cross, A.J.; Nicholson, J.K. Susceptibility of human metabolic phenotypes to dietary modulation. J. Proteome Res. 2006, 5, 2780–2788. [Google Scholar] [CrossRef] [PubMed]
- Pena, M.J.; Lambers Heerspink, H.J.; Hellemons, M.E.; Friedrich, T.; Dallmann, G.; Lajer, M.; Bakker, S.J.; Gansevoort, R.T.; Rossing, P.; de Zeeuw, D.; et al. Urine and plasma metabolites predict the development of diabetic nephropathy in individuals with type 2 diabetes mellitus. Diabet. Med. 2014, 31, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Scalbert, A.; Brennan, L.; Manach, C.; Andres-Lacueva, C.; Dragsted, L.O.; Draper, J.; Rappaport, S.M.; van der Hooft, J.J.; Wishart, D.S. The food metabolome: A window over dietary exposure. Am. J. Clin. Nutr. 2014, 99, 1286–1308. [Google Scholar] [CrossRef] [PubMed]
- Rezzi, S.; Ramadan, Z.; Fay, L.B.; Kochhar, S. Nutritional metabonomics: Applications and perspectives. J. Proteome Res. 2007, 6, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Hubert, J.; Llorach, R.; Scalbert, A. The complex links between dietary phytochemicals and human health deciphered by metabolomics. Mol. Nutr. Food Res. 2009, 53, 1303–1315. [Google Scholar] [CrossRef] [PubMed]
- Rechner, A.R.; Kuhnle, G.; Bremner, P.; Hubbard, G.P.; Moore, K.P.; Rice-Evans, C.A. The metabolic fate of dietary polyphenols in humans. Free Radic. Biol. Med. 2002, 33, 220–235. [Google Scholar] [CrossRef]
- Stephenson, A.J.; Smith, A.; Kattan, M.W.; Satagopan, J.; Reuter, V.E.; Scardino, P.T.; Gerald, W.L. Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy. Cancer 2005, 104, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Lin, J.C.; Chen, T.L.; Zhou, M.M.; Su, M.M.; Jia, W. Metabolic profiling reveals the protective effect of diammonium glycyrrhizinate on acute hepatic injury induced by carbon tetrachloride. Metabolomics 2011, 7, 226–236. [Google Scholar] [CrossRef]
- Bonvallot, N.; Tremblay-Franco, M.; Chevrier, C.; Canlet, C.; Warembourg, C.; Cravedi, J.P.; Cordier, S. Metabolomics tools for describing complex pesticide exposure in pregnant women in brittany (France). PLoS ONE 2013, 8, e64433. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Xiao, L.; Cao, W.; Fang, T.; Jia, G.; Chen, X.; Zhao, H.; Wu, C.; Wang, J. Changes in the metabolome of rats after exposure to arginine and N-carbamylglutamate in combination with diquat, a compound that causes oxidative stress, assessed by h nmr spectroscopy. Food Funct. 2016, 7, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhao, X.; Huang, C.; Lei, H.; Tang, H.; Wang, Y. Dynamic changes in metabolic profiles of rats subchronically exposed to mequindox. Mol. Biol. Syst. 2014, 10, 2914–2922. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Guardia, L.; Yubero-Serrano, E.M.; Delgado-Lista, J.; Perez-Martinez, P.; Garcia-Rios, A.; Marin, C.; Camargo, A.; Delgado-Casado, N.; Roche, H.M.; Perez-Jimenez, F.; et al. Effects of the mediterranean diet supplemented with coenzyme q10 on metabolomic profiles in elderly men and women. J. Gerontol. A Biol. Sci. Med. Sci. 2015, 70, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Kuo, C.H.; Tian, T.F.; Tsai, M.H.; Chiung, Y.M.; Hsiech, C.M.; Tsai, S.J.; Wang, S.Y.; Tsai, D.M.; Huang, C.C.; et al. Metabolomic characterization of laborers exposed to welding fumes. Chem. Res. Toxicol. 2012, 25, 676–686. [Google Scholar] [CrossRef] [PubMed]
- Ducena, K.; Abols, A.; Vilmanis, J.; Narbuts, Z.; Tars, J.; Andrejeva, D.; Line, A.; Pirags, V. Validity of multiplex biomarker model of 6 genes for the differential diagnosis of thyroid nodules. Thyroid Res. 2011, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.M.; Simon, R.; Pfeiffer, R.M. Prediction error estimation: A comparison of resampling methods. Bioinformatics 2005, 21, 3301–3307. [Google Scholar] [CrossRef] [PubMed]
- Ali, B.H.; Al-Husseni, I.; Beegam, S.; Al-Shukaili, A.; Nemmar, A.; Schierling, S.; Queisser, N.; Schupp, N. Effect of gum arabic on oxidative stress and inflammation in adenine-induced chronic renal failure in rats. PLoS ONE 2013, 8, e55242. [Google Scholar] [CrossRef] [PubMed]
- Belmonte, M.; Stasolla, C.; Loukanina, N.; Yeung, E.C.; Thorpe, T.A. Glutathione modulation of purine metabolism in cultured white spruce embryogenic tissue. Plant Sci. 2003, 165, 1377–1385. [Google Scholar] [CrossRef]
- Luan, H.; Liu, L.F.; Tang, Z.; Zhang, M.; Chua, K.K.; Song, J.X.; Mok, V.C.; Li, M.; Cai, Z. Comprehensive urinary metabolomic profiling and identification of potential noninvasive marker for idiopathic parkinson’s disease. Sci. Rep. 2015, 5, 13888. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.K.; Park, E.Y.; Kim, S.; Son, J.Y.; Kim, T.H.; Kang, W.G.; Jeong, T.C.; Kim, K.B.; Kwack, S.J.; Lee, J.; et al. Evaluation of cadmium-induced nephrotoxicity using urinary metabolomic profiles in sprague-dawley male rats. J. Toxicol. Environ. Health Part A 2014, 77, 1384–1398. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Fang, Y.Z.; Yang, S.; Lupton, J.R.; Turner, N.D. Glutathione metabolism and its implications for health. J. Nutr. 2004, 134, 489–492. [Google Scholar] [PubMed]
- Noctor, G.; Arisi, A.C.M.; Jouanin, L.; Valadier, M.H.; Roux, Y.; Foyer, C.H. The role of glycine in determining the rate of glutathione synthesis in poplar. Possible implications for glutathione production during stress. Physiol. Plant. 1997, 100, 255–263. [Google Scholar] [CrossRef]
- Higgins, P.; Dawson, J.; Walters, M. The potential for xanthine oxidase inhibition in the prevention and treatment of cardiovascular and cerebrovascular disease. Cardiovasc. Psychiatry Neurol. 2009, 2009, 282059. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.N.; Chatterjee, S.; Rana, A.R.S. Electrochemical sensor based on oxidation of 2,8-dihydroxyadenine to monitor DNA damage in calf thymus DNA. Electroanalysis 2011, 23, 1383–1390. [Google Scholar] [CrossRef]

| Variables | Placebo | KBR | β 2 | q-Value 3 | ||
|---|---|---|---|---|---|---|
| Baseline | Delta Change | Baseline | Delta Change | |||
| Traditional biomarkers | ||||||
| GSSG (µM/g Hb) | 12.9 ± 0.6 4 | 1.8 ± 0.3 | 12.9 ± 0.7 | 0.3 ± 0.4 | −1.117 | 0.027 |
| GSH:GSSG ratio | 3.0 ± 0.2 | −0.3 ± 0.1 | 2.8 ± 0.2 | 0.2 ± 0.2 | 0.045 | 0.039 |
| MDA (nM) | 14.5 ± 1.5 | 0.0 ± 0.4 | 16.4 ± 1.6 | −2.5 ± 0.6 | −0.058 | 0.006 |
| IL-6 (pg/mL) | 196.1 ± 22.8 | 51.4 ± 31.8 | 182.7 ± 24.4 | −52.1 ± 17.9 | −0.199 | 0.006 |
| Urinary metabolites (μM) | ||||||
| 3-Indoxylsulfate | 2.31 ± 0.16 | 0.11 ± 0.21 | 2.16 ± 0.18 | 0.81 ± 0.2 | 0.399 | 0.009 |
| Adenine | 1.26 ± 0.11 | 0.09 ± 0.18 | 1.64 ± 0.21 | −0.6 ± 0.2 | −0.286 | 0.041 |
| Alanine | 2.79 ± 0.18 | −0.14 ± 0.17 | 2.49 ± 0.16 | 0.35 ± 0.13 | 0.194 | 0.021 |
| Asparagine | 1.62 ± 0.09 | 0.16 ± 0.13 | 1.55 ± 0.09 | 0.47 ± 0.12 | 0.199 | 0.041 |
| Betaine | 1.63 ± 0.12 | −0.07 ± 0.13 | 1.55 ± 0.14 | 0.31 ± 0.15 | 0.295 | 0.024 |
| Carnitine | 0.95 ± 0.11 | 0.22 ± 0.14 | 0.91 ± 0.09 | −0.13 ± 0.11 | −0.555 | 0.009 |
| Citrate | 11.49 ± 0.78 | −1.15 ± 0.5 | 11.19 ± 1.16 | 0.53 ± 0.68 | 0.029 | 0.037 |
| Formate | 3.22 ± 0.28 | 0.01 ± 0.32 | 2.58 ± 0.19 | 1.31 ± 0.48 | 0.314 | 0.034 |
| Glutamine | 5.29 ± 0.29 | −0.26 ± 0.24 | 4.61 ± 0.22 | 0.95 ± 0.22 | 0.220 | 0.0001 |
| Glycine | 10.51 ± 1.29 | −0.83 ± 0.81 | 8.63 ± 0.75 | 1.18 ± 0.56 | 0.200 | 0.021 |
| Histidine | 4.33 ± 0.4 | −0.55 ± 0.4 | 3.76 ± 0.34 | 1.25 ± 0.38 | 0.018 | 0.013 |
| Lysine | 1.83 ± 0.25 | −0.27 ± 0.2 | 1.11 ± 0.1 | 0.3 ± 0.12 | 0.329 | 0.021 |
| N-Phenylacetylglycine | 3.05 ± 0.15 | −0.16 ± 0.2 | 2.62 ± 0.13 | 0.53 ± 0.14 | 0.007 | 0.016 |
| N6-Acetyllysine | 1.04 ± 0.03 | 0.02 ± 0.04 | 0.99 ± 0.03 | 0.14 ± 0.04 | 0.001 | 0.028 |
| Phenylacetate | 1.04 ± 0.04 | 0.01 ± 0.05 | 1.07 ± 0.06 | 0.21 ± 0.06 | 0.002 | 0.021 |
| Serine | 5.19 ± 0.31 | 0.42 ± 0.35 | 4.05 ± 0.18 | 1.51 ± 0.31 | 0.248 | 0.021 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.J.; Huh, I.; Kim, J.Y.; Park, S.; Ryu, S.H.; Kim, K.-B.; Kim, S.; Park, T.; Kwon, O. Integration of Traditional and Metabolomics Biomarkers Identifies Prognostic Metabolites for Predicting Responsiveness to Nutritional Intervention against Oxidative Stress and Inflammation. Nutrients 2017, 9, 233. https://doi.org/10.3390/nu9030233
Kim YJ, Huh I, Kim JY, Park S, Ryu SH, Kim K-B, Kim S, Park T, Kwon O. Integration of Traditional and Metabolomics Biomarkers Identifies Prognostic Metabolites for Predicting Responsiveness to Nutritional Intervention against Oxidative Stress and Inflammation. Nutrients. 2017; 9(3):233. https://doi.org/10.3390/nu9030233
Chicago/Turabian StyleKim, You Jin, Iksoo Huh, Ji Yeon Kim, Saejong Park, Sung Ha Ryu, Kyu-Bong Kim, Suhkmann Kim, Taesung Park, and Oran Kwon. 2017. "Integration of Traditional and Metabolomics Biomarkers Identifies Prognostic Metabolites for Predicting Responsiveness to Nutritional Intervention against Oxidative Stress and Inflammation" Nutrients 9, no. 3: 233. https://doi.org/10.3390/nu9030233
APA StyleKim, Y. J., Huh, I., Kim, J. Y., Park, S., Ryu, S. H., Kim, K.-B., Kim, S., Park, T., & Kwon, O. (2017). Integration of Traditional and Metabolomics Biomarkers Identifies Prognostic Metabolites for Predicting Responsiveness to Nutritional Intervention against Oxidative Stress and Inflammation. Nutrients, 9(3), 233. https://doi.org/10.3390/nu9030233