Dietary Gluten-Induced Gut Dysbiosis Is Accompanied by Selective Upregulation of microRNAs with Intestinal Tight Junction and Bacteria-Binding Motifs in Rhesus Macaque Model of Celiac Disease
Abstract
:1. Introduction
2. Experimental Section
2.1. Ethics Approval
2.2. Rhesus Macaques, Diets and Samples Collected
2.3. DNA Extraction, Library Preparation and Profiling
2.4. Operational Taxonomic Unit (OTU) Selection
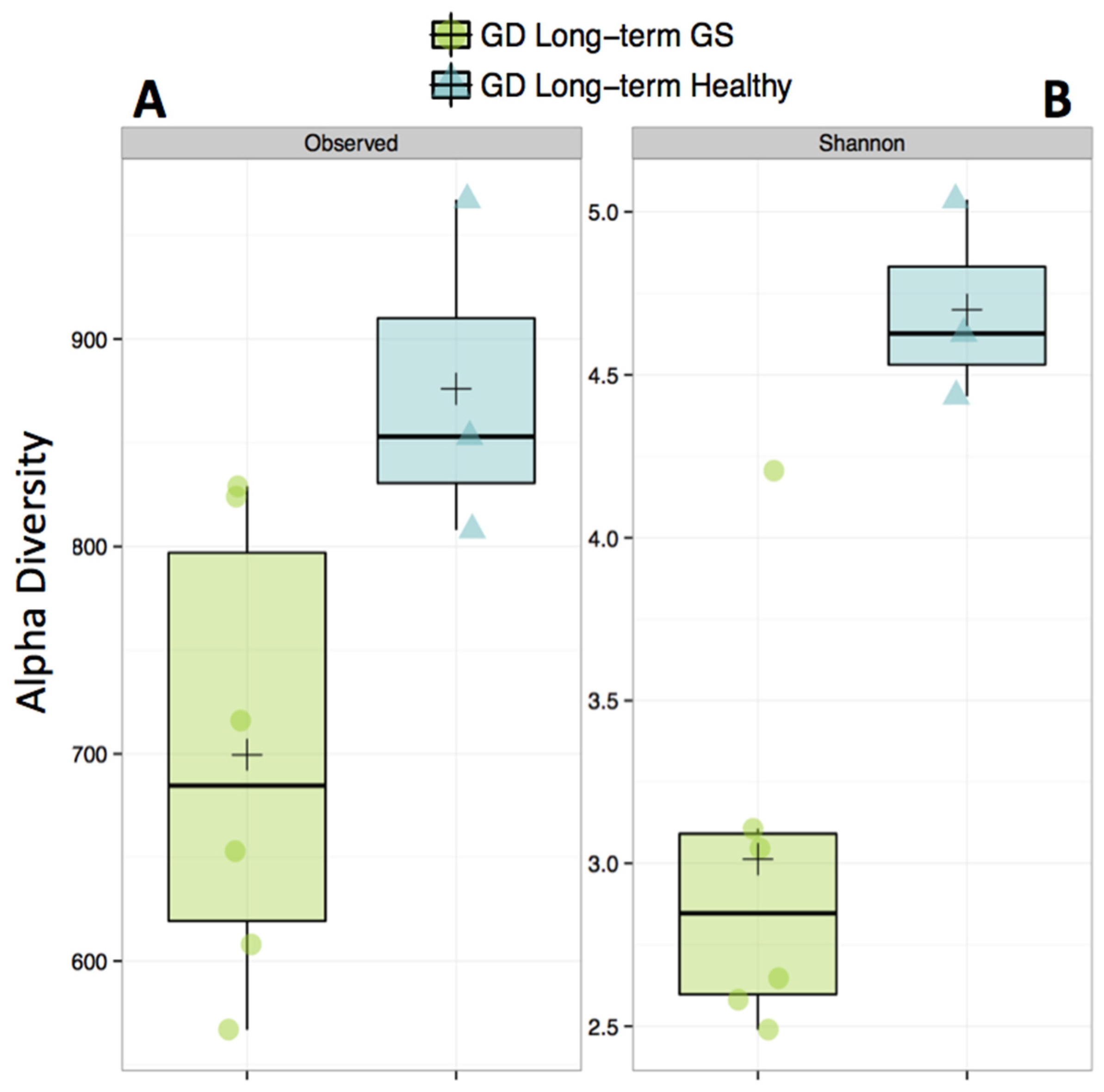
2.5. Alpha- (within Sample) and Beta- (between Samples) Diversity
2.6. Ordination and Clustering
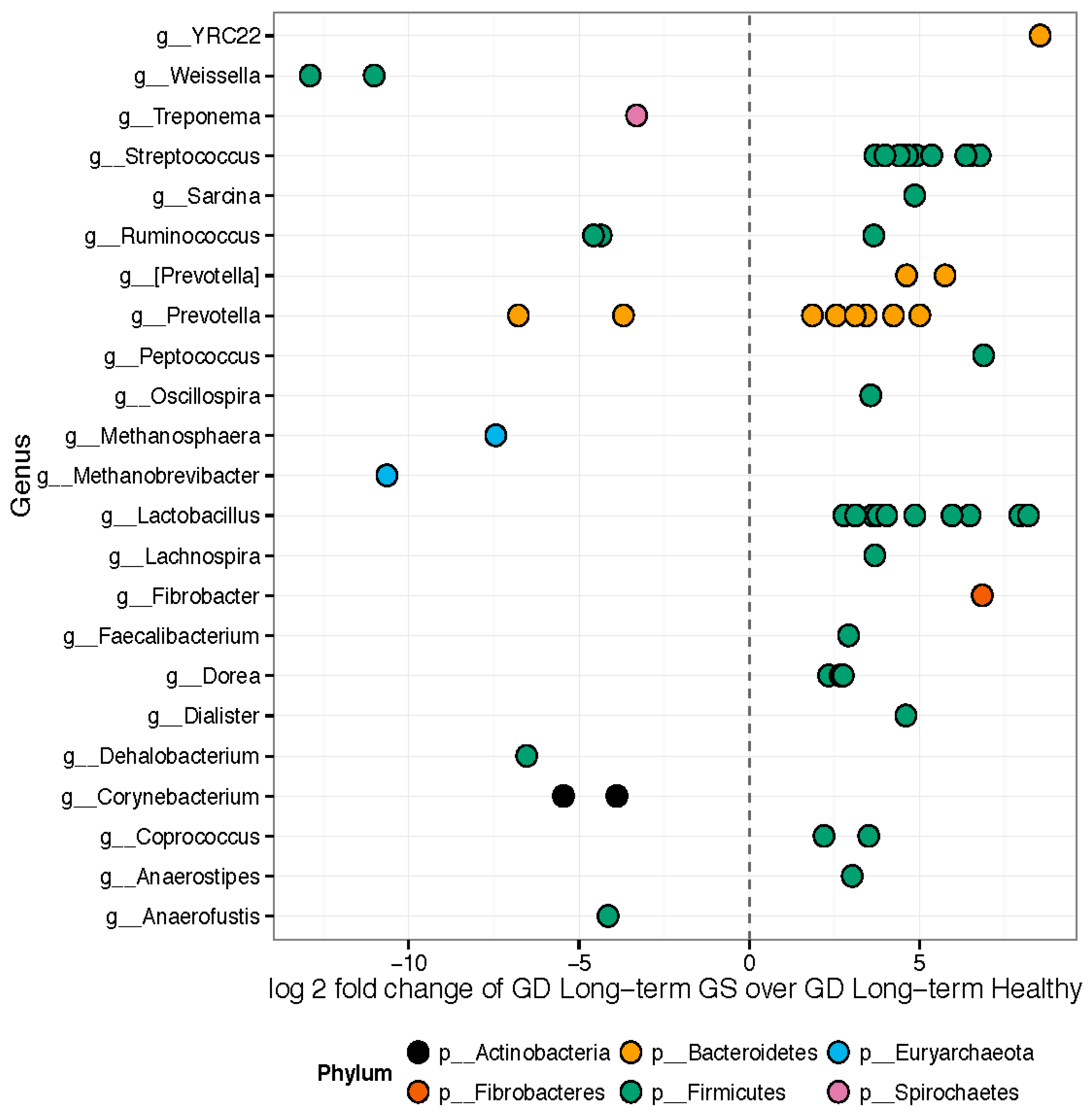
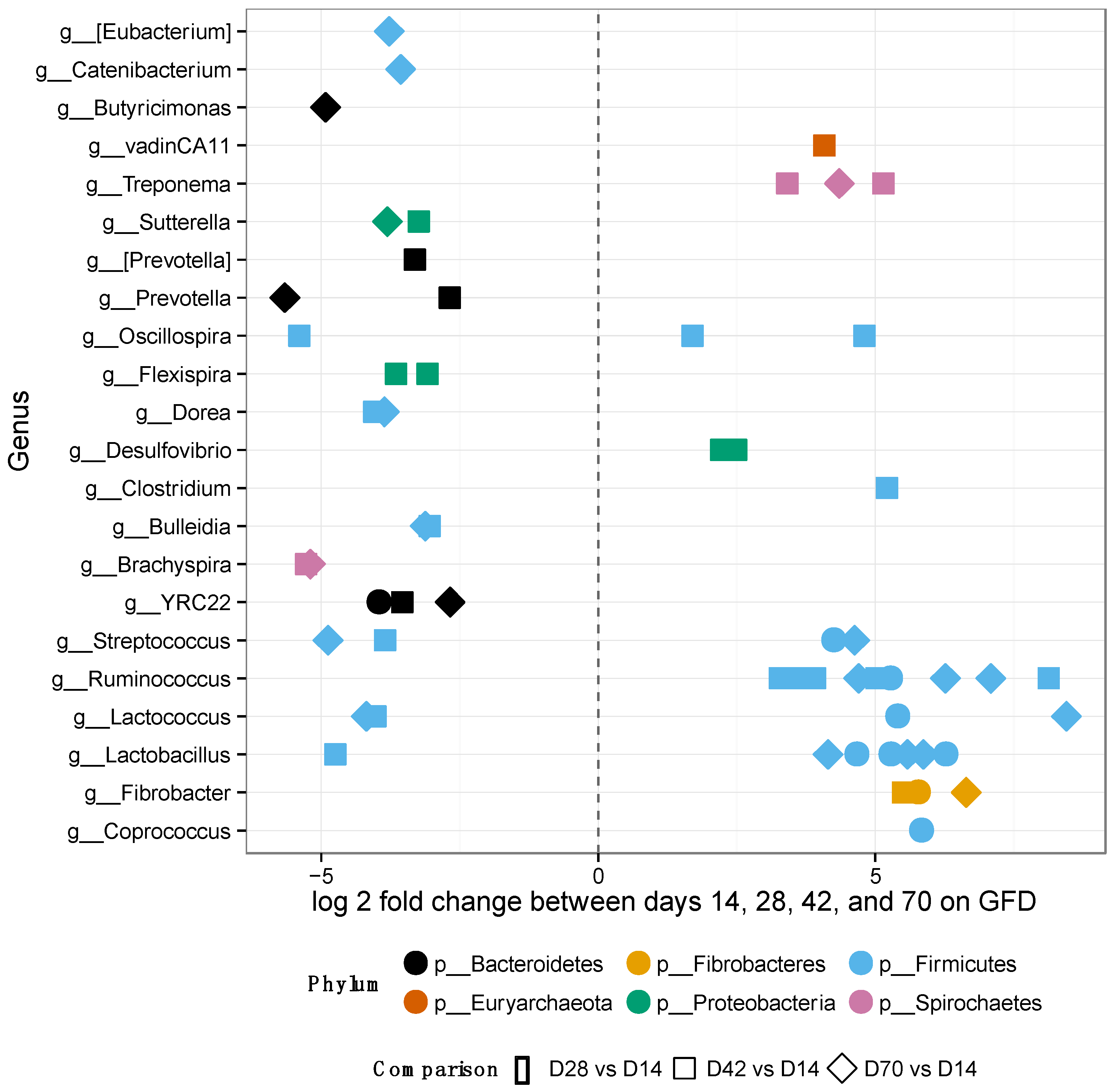
2.7. Whole Microbiome and Taxon Significance Testing
2.8. miRNA Profiling, Real Time qRT-PCR and Confocal Microscopy
2.9. Data Analysis
3. Results
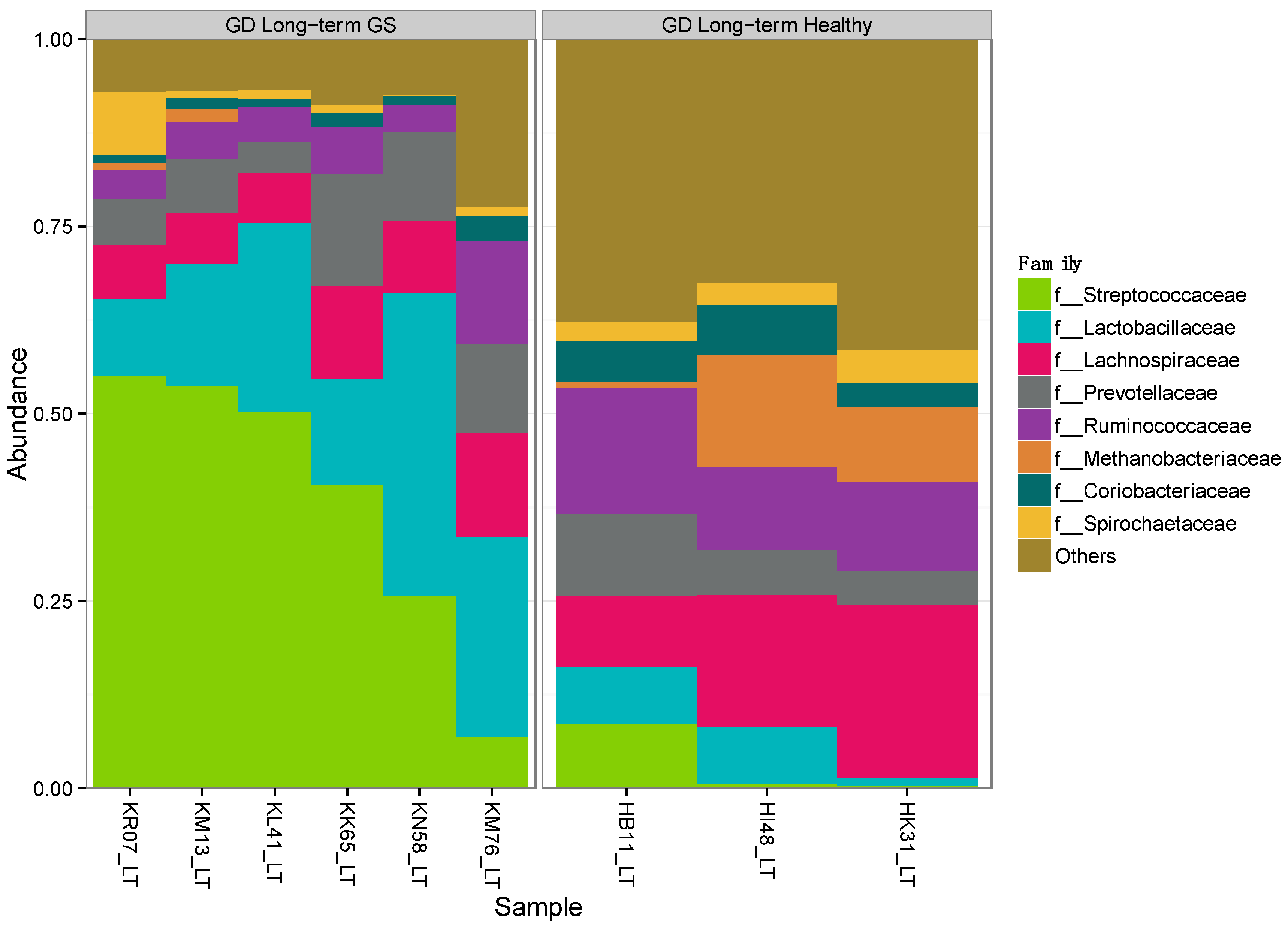
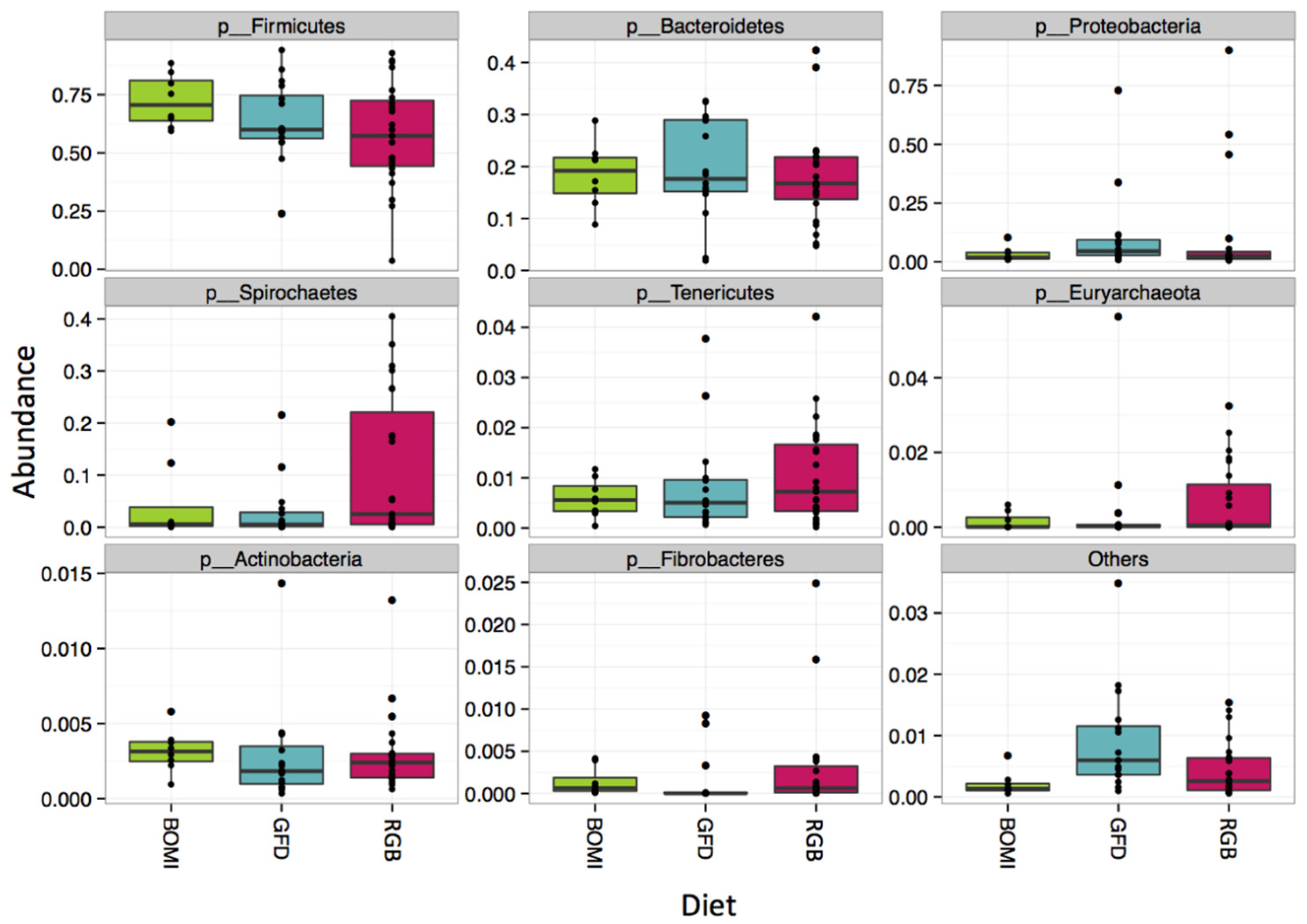
3.1. Gut Microbiomes Differ Significantly between Healthy and GS Macaques
3.2. Gut Microbiomes of GS Macaques Are Influenced by GFD
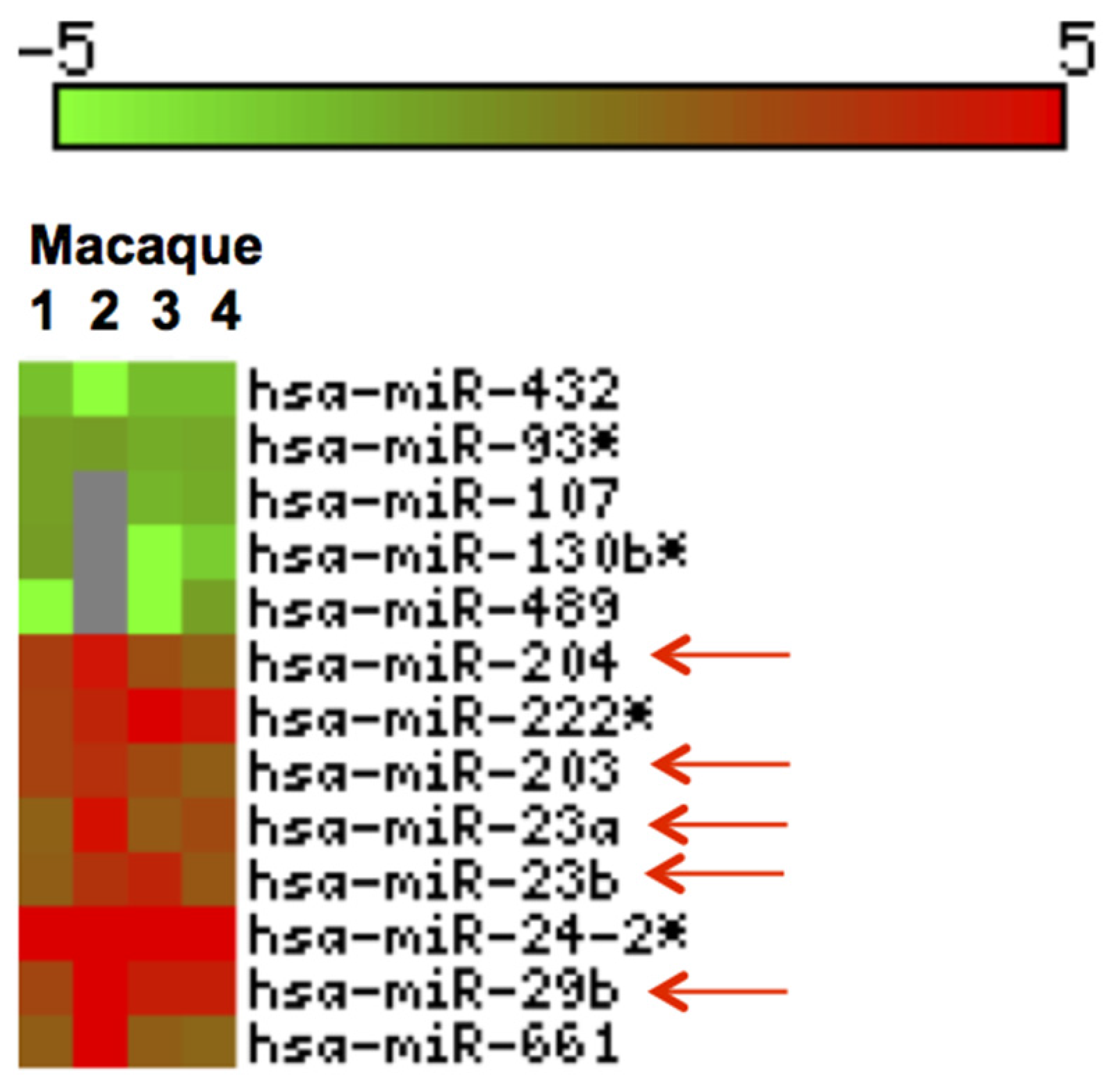
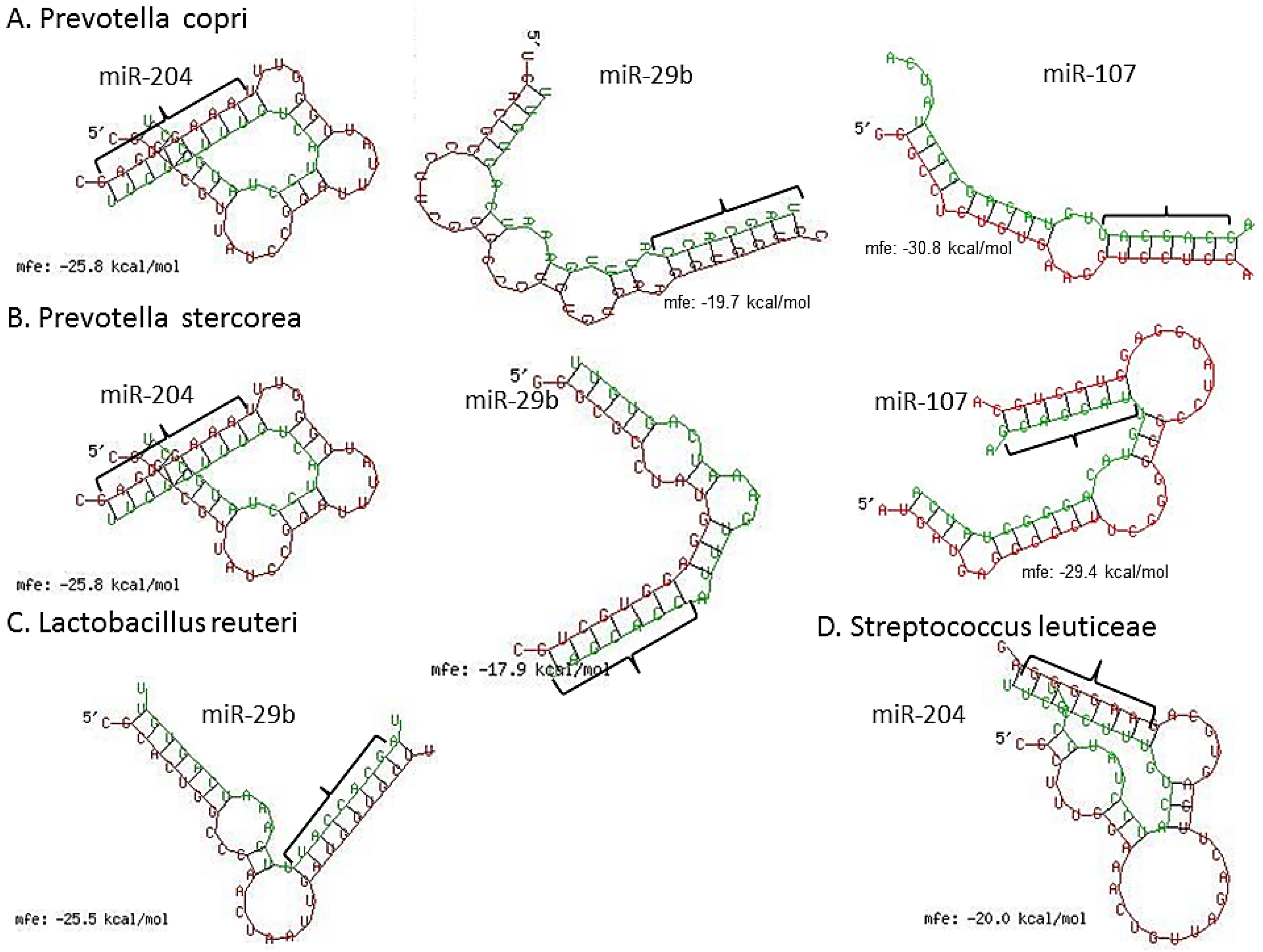
3.3. Inflammation-Associated miRNAs Are Upregulated in Jejunum of GS Macaques and Have Predicted Binding Sites on Bacterial 16S Ribosomal RNA
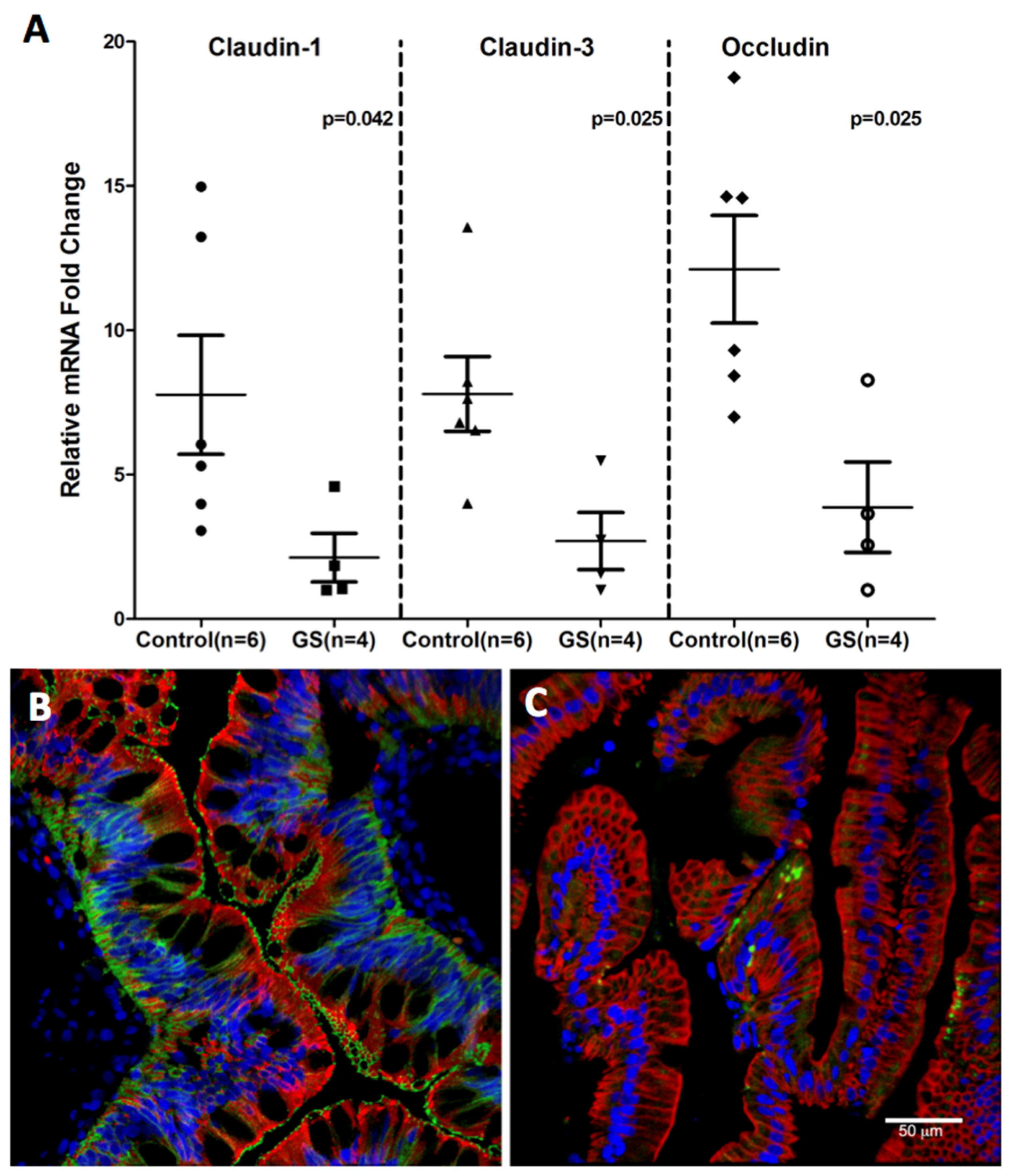
3.4. Claudin-1, an Epithelial Tight Junction Protein and A Validated Target of miR-29b Is Significantly Downregulated in Jejunum of GS Macaques
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Atarashi, K.; Honda, K. Microbiota in autoimmunity and tolerance. Curr. Opin. Immunol. 2011, 23, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Helander, H.F.; Fandriks, L. Surface area of the digestive tract-revisited. Scand. J. Gastroenterol. 2014, 49, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Lederberg, J. Infectious history. Science 2000, 288, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.; Knight, R.; Gordon, J.I. The human microbiome project: Exploring the microbial part of ourselves in a changing world. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, B.; Zhang, M.; Rantalainen, M.; Wang, S.; Zhou, H.; Zhang, Y.; Shen, J.; Pang, X.; Zhang, M.; et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. USA 2008, 105, 2117–2122. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; McFall-Ngai, M.; Relman, D.A. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 2007, 449, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
- Zoetendal, E.; Rajilić-Stojanović, M.; De Vos, W. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 2008, 57, 1605–1615. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Tyakht, A.V.; Kostryukova, E.S.; Popenko, A.S.; Belenikin, M.S.; Pavlenko, A.V.; Larin, A.K.; Karpova, I.Y.; Selezneva, O.V.; Semashko, T.A.; Ospanova, E.A.; et al. Human gut microbiota community structures in urban and rural populations in Russia. Nat. Commun. 2013. [Google Scholar] [CrossRef] [PubMed]
- Bergström, A.; Skov, T.H.; Bahl, M.I.; Roager, H.M.; Christensen, L.B.; Ejlerskov, K.T.; Mølgaard, C.; Michaelsen, K.F.; Licht, T.R. Establishment of intestinal microbiota during early life: A longitudinal, explorative study of a large cohort of Danish infants. Appl. Environ. Microbiol. 2014, 80, 2889–2900. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Prince, A.L.; Bader, D.; Hu, M.; Ganu, R.; Baquero, K.; Blundell, P.; Harris, R.A.; Frias, A.E.; Grove, K.L.; et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Schnorr, S.L.; Candela, M.; Rampelli, S.; Centanni, M.; Consolandi, C.; Basaglia, G.; Turroni, S.; Biagi, E.; Peano, C.; Severgnini, M.; et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014, 5, 3654. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Materna, A.C.; Friedman, J.; Campos-Baptista, M.I.; Blackburn, M.C.; Perrotta, A.; Erdman, S.E.; Alm, E.J. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014, 15, R8. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.E.; Bhatnagar, S.; George, M.D.; Paster, B.J.; Canfield, D.R.; Eisen, J.A.; Solnick, J.V. The impact of Helicobacter pylori infection on the gastric microbiota of the rhesus macaque. PLoS ONE 2013, 8, e76375. [Google Scholar] [CrossRef] [PubMed]
- McKenna, P.; Hoffmann, C.; Minkah, N.; Aye, P.P.; Lackner, A.; Liu, Z.; Lozupone, C.A.; Hamady, M.; Knight, R.; Bushman, F.D. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008, 4, e20. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; Carbonero, F.; Zoetendal, E.G.; DeLany, J.P.; Wang, M.; Newton, K.; Gaskins, H.R.; O’Keefe, S.J. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr. 2013, 98, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Ardeshir, A.; Narayan, N.R.; Méndez-Lagares, G.; Lu, D.; Rauch, M.; Huang, Y.; Van Rompay, K.K.; Lynch, S.V.; Hartigan-O’Connor, D.J. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci. Transl. Med. 2014, 6, 252ra120. [Google Scholar] [CrossRef] [PubMed]
- Broadhurst, M.J.; Ardeshir, A.; Kanwar, B.; Mirpuri, J.; Gundra, U.M.; Leung, J.M.; Wiens, K.E.; Vujkovic-Cvijin, I.; Kim, C.C.; Yarovinsky, F.; et al. Therapeutic helminth infection of macaques with idiopathic chronic diarrhea alters the inflammatory signature and mucosal microbiota of the colon. PLoS Pathog. 2012, 8, e1003000. [Google Scholar] [CrossRef] [PubMed]
- Ardeshir, A.; Sankaran, S.; Oslund, K.; Hartigan-O’Connor, D.; Lerche, N.; Hyde, D.; Dandekar, S. Inulin treatment leads to changes in intestinal microbiota and resolution of idiopathic chronic diarrhea in rhesus macaques. Ann. Am. Thorac. Soc. 2014, 11, S75–S75. [Google Scholar] [CrossRef]
- Lupp, C.; Robertson, M.L.; Wickham, M.E.; Sekirov, I.; Champion, O.L.; Gaynor, E.C.; Finlay, B.B. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007, 2, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Seksik, P. The intestinal microbiota in inflammatory bowel diseases: Time to connect with the host. Curr. Opin. Gastroenterol. 2010, 26, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Li, X.; Parfrey, L.W.; Roth, B.; Ippoliti, A.; Wei, B.; Borneman, J.; McGovern, D.P.; Frank, D.N.; Li, E.; et al. A modular organization of the human intestinal mucosal microbiota and its association with Inflammatory Bowel Disease. PLoS ONE 2013, 8, e80702. [Google Scholar] [CrossRef] [PubMed]
- Walujkar, S.A.; Dhotre, D.P.; Marathe, N.P.; Lawate, P.S.; Bharadwaj, R.S.; Shouche, Y.S. Characterization of bacterial community shift in human Ulcerative Colitis patients revealed by Illumina based 16S rRNA gene amplicon sequencing. Gut Path. 2014, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Lucke, K.; Miehlke, S.; Jacobs, E.; Schuppler, M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J. Med. Microbiol. 2006, 55, 617–624. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Nadal, I.; Collado, M.C.; Sanz, Y. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects. Br. J. Nutr. 2009, 102, 1154–1160. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Capilla, A.; Nova, E.; Castillejo, G.; Varea, V.; Pozo, T.; Garrote, J.A.; Polanco, I.; López, A.; Ribes-Koninckx, C.; et al. Influence of milk-feeding type and genetic risk of developing coeliac disease on intestinal microbiota of infants: The PROFICEL study. PLoS ONE 2012, 7, e30791. [Google Scholar] [CrossRef] [PubMed]
- Sellitto, M.; Bai, G.; Serena, G.; Fricke, W.F.; Sturgeon, C.; Gajer, P.; White, J.R.; Koenig, S.S.; Sakamoto, J.; Boothe, D.; et al. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS ONE 2012, 7, e33387. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Kalliomäki, M.; Heilig, H.G.; Palva, A.; Lähteenoja, H.; de Vos, W.M.; Salojärvi, J.; Satokari, R. Duodenal microbiota composition and mucosal homeostasis in pediatric celiac disease. BMC Gastroenterol. 2013, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, E.; Donat, E.; Ribes-Koninckx, C.; Fernández-Murga, M.L.; Sanz, Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl. Environ. Microbiol. 2013, 79, 5472–5479. [Google Scholar] [CrossRef] [PubMed]
- Ivarsson, A.; Persson, L.Å.; Nyström, L.; Ascher, H.; Cavell, B.; Danielsson, L.; Dannaeus, A.; Lindberg, T.; Lindquist, B.; Stenhammar, L.; et al. Epidemic of coeliac disease in Swedish children. Acta Paediatr. 2000, 89, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Ou, G.; Hedberg, M.; Hörstedt, P.; Baranov, V.; Forsberg, G.; Drobni, M.; Sandström, O.; Wai, S.N.; Johansson, I.; Hammarström, M.L.; et al. Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. Am. J. Gastroenterol. 2009, 104, 3058–3067. [Google Scholar] [CrossRef] [PubMed]
- Sjöberg, V.; Sandström, O.; Hedberg, M.; Hammarström, S.; Hernell, O.; Hammarström, M.L. Intestinal T-cell responses in celiac disease—Impact of celiac disease associated bacteria. PLoS ONE 2013, 8, e53414. [Google Scholar] [CrossRef] [PubMed]
- Caminero, A.; Herrán, A.R.; Nistal, E.; Pérez-Andrés, J.; Vaquero, L.; Vivas, S.; de Morales, J.M.; Albillos, S.M.; Casqueiro, J. Diversity of the cultivable human gut microbiome involved in gluten metabolism: Isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol. Ecol. 2014, 88, 309–319. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Casaburi, G.; Precone, V.; Pagliuca, C.; Colicchio, R.; Sarnataro, D.; Discepolo, V.; Kim, S.M.; Russo, I.; Blanco, G.D.; et al. Metagenomics reveals dysbiosis and a potentially pathogenic N. flavescens strain in duodenum of adult celiac patients. Am. J. Gastroenterol. 2016, 111, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Feely, S.L.; Wang, X.; Liu, D.X.; Borda, J.T.; Dufour, J.; Li, W.; Aye, P.P.; Doxiadis, G.G.; Khosla, C.; et al. Gluten-sensitive enteropathy coincides with decreased capability of intestinal T cells to secrete IL-17 and IL-22 in a macaque model for celiac disease. Clin. Immunol. 2013, 147, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; da Cunha, A.P.; Rezende, R.M.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.E.; Gandhi, R.; Weiner, H.L. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe 2016, 19, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Sestak, K.; Thwin, H.; Dufour, J.; Aye, P.P.; Liu, D.X.; Moehs, C.P. The effects of reduced gluten barley diet on humoral and cell-mediated systemic immune responses of gluten-sensitive rhesus macaques. Nutrients 2015, 7, 1657–1671. [Google Scholar] [CrossRef] [PubMed]
- Sestak, K.; Merritt, C.K.; Borda, J.; Saylor, E.; Schwamberger, S.R.; Cogswell, F.; Didier, E.S.; Didier, P.J.; Plauche, G.; Bohm, R.P.; et al. Infectious agent and immune response characteristics of chronic enterocolitis in captive rhesus macaques. Infect. Immun. 2003, 71, 4079–4086. [Google Scholar] [CrossRef] [PubMed]
- Sestak, K.; Thwin, H.; Dufour, J.; Liu, D.X.; Alvarez, X.; Laine, D.; Clarke, A.; Doyle, A.; Aye, P.P.; Blanchard, J.; et al. Supplementation of reduced gluten barley diet with oral prolyl endopeptidase effectively abrogates enteropathy-associated changes in gluten-sensitive macaques. Nutrients 2016, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, N.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Aust. Ecol. 2001, 26, 32–46. [Google Scholar]
- Love, M.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [PubMed]
- Mohan, M.; Kumar, V.; Lackner, A.A.; Alvarez, X. Dysregulated miR-34a-SIRT1-acetyl p65 axis is a potential mediator of immune activation in the colon during chronic simian immunodeficiency virus infection of rhesus macaques. J. Immunol. 2015, 194, 291–306. [Google Scholar] [CrossRef] [PubMed]
- Chandra, L.C.; Kumar, V.; Torben, W.; Stouwe, C.V.; Winsauer, P.; Amedee, A.; Molina, P.E.; Mohan, M. Chronic administration of Δ9-tetrahydrocannabinol induces intestinal anti-inflammatory microRNA expression during acute SIV infection of rhesus macaques. J. Virol. 2015, 89, 1168–1181. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Costinean, S.; Croce, C.M.; Brasier, A.R.; Merwat, S.; Larson, S.A.; Basra, S.; Verne, G.N. MicroRNA 29 targets nuclear factor-κB-repressing factor and Claudin 1 to increase intestinal permeability. Gastroenterology 2015, 148, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.F.; Fang, J.; Wu, X.N.; Yu, C.H. MicroRNA-203 accelerates apoptosis in LPS-stimulated alveolar epithelial cells by targeting PIK3CA. Biochem. Biophys. Res. Commun. 2014, 450, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Peck, B.C.; Weiser, M.; Lee, S.E.; Gipson, G.R.; Iyer, V.B.; Sartor, R.B.; Herfarth, H.H.; Long, M.D.; Hansen, J.J.; Isaacs, K.L.; et al. MicroRNAs Classify Different Disease Behavior Phenotypes of Crohn’s Disease and May Have Prognostic Utility. Inflamm. Bowel Dis. 2015, 21, 2178–2187. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Luna, C.; Qiu, J.; Epstein, D.L.; Gonzalez, P. Role of miR-204 in the regulation of apoptosis, endoplasmic reticulum stress response, and inflammation in human trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 2011, 52, 2999–3007. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, S.Q.; Li, C.; Lykken, E.; Jiang, S.; Wong, E.; Gong, Z.; Tao, Z.; Zhu, B.; Wan, Y.; et al. MicroRNA-23a Curbs Necrosis during Early T Cell Activation by Enforcing Intracellular Reactive Oxygen Species Equilibrium. Immunity 2016, 44, 568–581. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Pan, W.; Song, X.; Liu, Y.; Shao, X.; Tang, Y.; Liang, D.; He, D.; Wang, H.; Liu, W.; et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat. Med. 2012, 18, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Chapman, C.G.; Pekow, J. The emerging role of miRNAs in inflammatory bowel disease: A review. Ther. Adv. Gastroenterol. 2015, 8, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucl. Acids Res. 2006, 34, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Cichon, C.; Sabharwal, H.; Rüter, C.; Schmidt, M.A. MicroRNAs regulate tight junction proteins and modulate epithelial/endothelial barrier functions. Tissue Barriers 2014, 2, e944446. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Rao, J.N.; Wang, J.Y. Posttranscriptional Regulation of Intestinal Epithelial Tight Junction Barrier by RNA-binding Proteins and microRNAs. Tissue Barriers 2014, 2, e28320. [Google Scholar] [CrossRef] [PubMed]
- Bressan, W. Biological control of maize seed pathogenic fungi by use of actinomycetes. BioControl 2003, 48, 233–240. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Pound, L.D.; Kievit, P.; Grove, K.L. The nonhuman primate as a model for type 2 diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 2014, 21, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Sui, Y.; Gordon, S.; Franchini, G.; Berzofsky, J.A. Nonhuman primate models for HIV/AIDS vaccine development. Curr. Protoc. Immunol. 2013, 102, 12–14. [Google Scholar]
- Beaudoin-Gobert, M.; Sgambato-Faure, V. Serotonergic pharmacology in animal models: From behavioral disorders to dyskinesia. Neuropharmacology 2014, 81, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, K.; Oh, K.; Ren, B.; Tickle, T.L.; Franzosa, E.A.; Wachtman, L.M.; Miller, A.D.; Westmoreland, S.V.; Mansfield, K.G.; Vallender, E.J.; et al. Biogeography of the intestinal mucosal and luminal microbiome in the rhesus macaque. Cell Host Microbe 2015, 17, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Dassanayake, R.P.; Zhou, Y.; Hinkley, S.; Stryker, C.J.; Plauche, G.; Borda, J.T.; Sestak, K.; Duhamel, G.E. Characterization of cytolethal distending toxin of Campylobacter species isolated from captive macaque monkeys. J. Clin. Microbiol. 2005, 43, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Ardeshir, A.; Oslund, K.L.; Ventimiglia, F.; Yee, J.; Lerche, N.W.; Hyde, D.M. Idiopathic microscopic colitis of rhesus macaques: Quantitative assessment of colonic mucosa. Anat. Rec. 2013, 296, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Zeller, J.; Takeuchi, A. Infection of the colon of the rhesus monkey by spiral-shaped organisms. Vet. Pathol. Suppl. 1982, 7, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Littman, D.R. The microbiome in infectious disease and inflammation. Ann. Rev. Immunol. 2012, 30, 759–795. [Google Scholar] [CrossRef] [PubMed]
- Caminero, A.; Galipeau, H.J.; McCarville, J.L.; Johnston, C.W.; Bernier, S.P.; Russell, A.K.; Jury, J.; Herran, A.R.; Casqueiro, J.; Tye-Din, J.A.; et al. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology 2016, 151, 670–683. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, G.B.; Balachandran, L. Antibacterial agents from actinomycetes—A Review. Front. Biosci. 2012, 4, 240–253. [Google Scholar] [CrossRef]
- Barcia, G.; Posar, A.; Santucci, M.; Parmeggiani, A. Autism and coeliac disease. J. Autism Dev. Disord. 2008, 38, 407–408. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Krajmalnik-Brown, R.; Porazinska, D.L.; Weiss, S.J.; Knight, R. Toward effective probiotics for Autism and other neurodevelopmental disorders. Cell 2013, 155, 1446–1448. [Google Scholar] [CrossRef] [PubMed]
- Scheperjans, F.; Aho, V.; Pereira, P.A.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M.; et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2014, 30, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; LaBaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef] [PubMed]
- Sestak, K.; Conroy, L.; Aye, P.P.; Mehra, S.; Doxiadis, G.G.; Kaushal, D. Improved xenobiotic metabolism and reduced susceptibility to cancer in gluten-sensitive macaques upon introduction of a gluten-free diet. PLoS ONE 2011, 6, e18648. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Guo, S.; Al-Sadi, R.; Ma, T.Y. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology 2011, 141, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Wacklin, P.; Laurikka, P.; Lindfors, K.; Collin, P.; Salmi, T.; Lähdeaho, M.L.; Saavalainen, P.; Mäki, M.; Mättö, J.; Kurppa, K.; et al. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am. J. Gastroenterol. 2014, 109, 1933–1941. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohan, M.; Chow, C.-E.T.; Ryan, C.N.; Chan, L.S.; Dufour, J.; Aye, P.P.; Blanchard, J.; Moehs, C.P.; Sestak, K. Dietary Gluten-Induced Gut Dysbiosis Is Accompanied by Selective Upregulation of microRNAs with Intestinal Tight Junction and Bacteria-Binding Motifs in Rhesus Macaque Model of Celiac Disease. Nutrients 2016, 8, 684. https://doi.org/10.3390/nu8110684
Mohan M, Chow C-ET, Ryan CN, Chan LS, Dufour J, Aye PP, Blanchard J, Moehs CP, Sestak K. Dietary Gluten-Induced Gut Dysbiosis Is Accompanied by Selective Upregulation of microRNAs with Intestinal Tight Junction and Bacteria-Binding Motifs in Rhesus Macaque Model of Celiac Disease. Nutrients. 2016; 8(11):684. https://doi.org/10.3390/nu8110684
Chicago/Turabian StyleMohan, Mahesh, Cheryl-Emiliane T. Chow, Caitlin N. Ryan, Luisa S. Chan, Jason Dufour, Pyone P. Aye, James Blanchard, Charles P. Moehs, and Karol Sestak. 2016. "Dietary Gluten-Induced Gut Dysbiosis Is Accompanied by Selective Upregulation of microRNAs with Intestinal Tight Junction and Bacteria-Binding Motifs in Rhesus Macaque Model of Celiac Disease" Nutrients 8, no. 11: 684. https://doi.org/10.3390/nu8110684
APA StyleMohan, M., Chow, C.-E. T., Ryan, C. N., Chan, L. S., Dufour, J., Aye, P. P., Blanchard, J., Moehs, C. P., & Sestak, K. (2016). Dietary Gluten-Induced Gut Dysbiosis Is Accompanied by Selective Upregulation of microRNAs with Intestinal Tight Junction and Bacteria-Binding Motifs in Rhesus Macaque Model of Celiac Disease. Nutrients, 8(11), 684. https://doi.org/10.3390/nu8110684




