Properties of Gluten Intolerance: Gluten Structure, Evolution, Pathogenicity and Detoxification Capabilities
Abstract
:1. Introduction
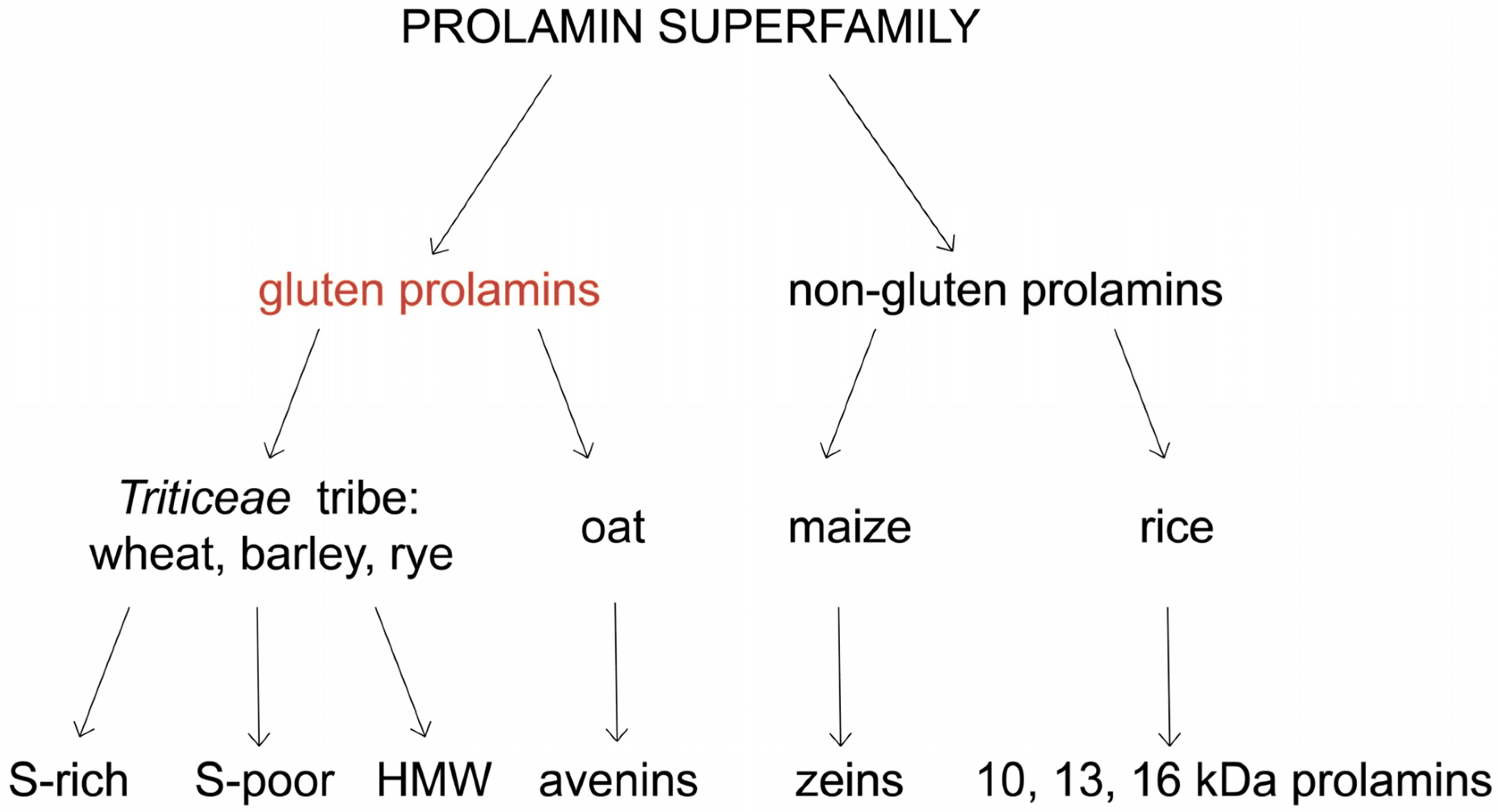
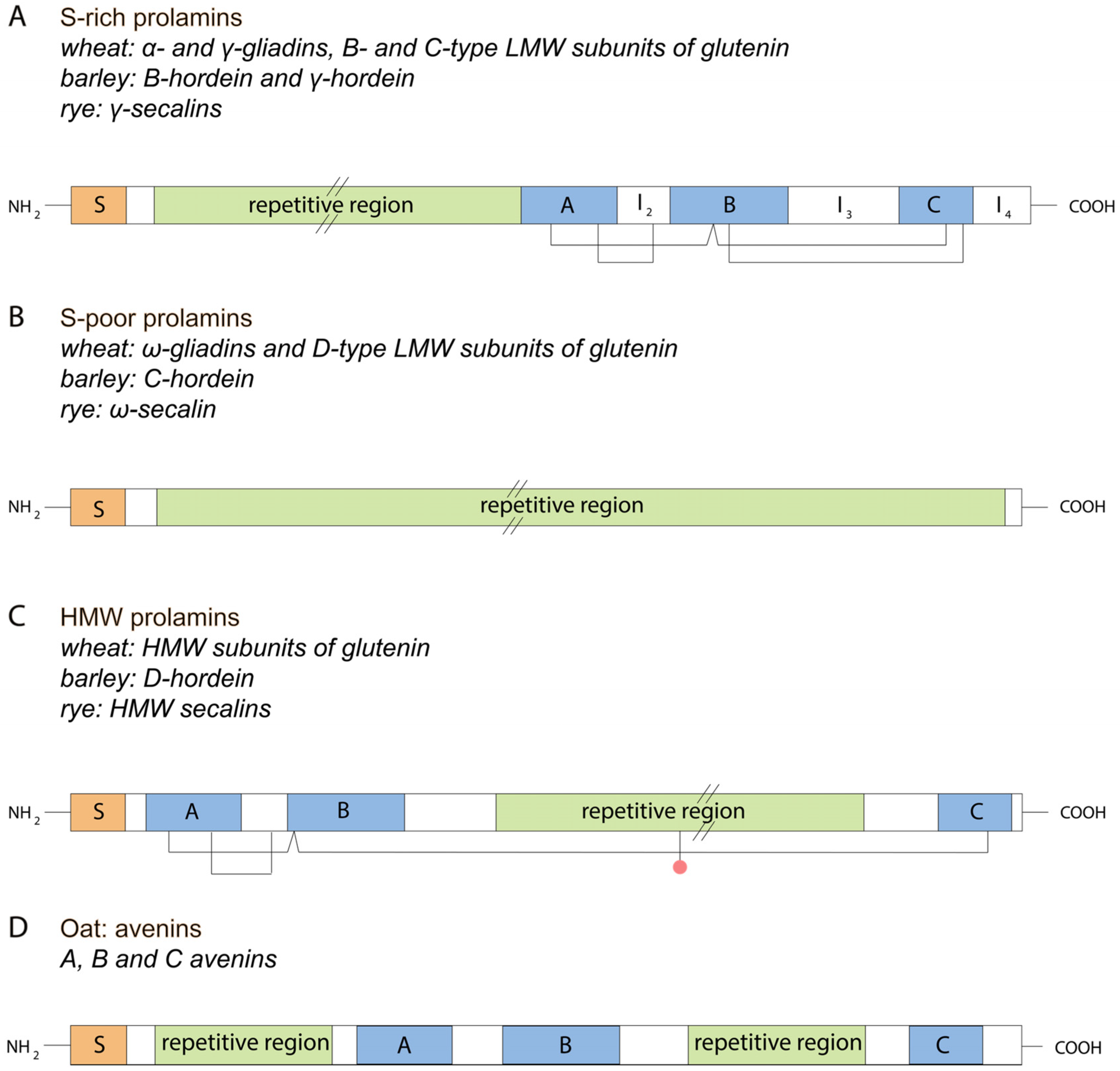
2. Classification and Structure of Gluten Proteins
2.1. Wheat
2.2. Barley
2.3. Rye
2.4. Triticale
2.5. Oat
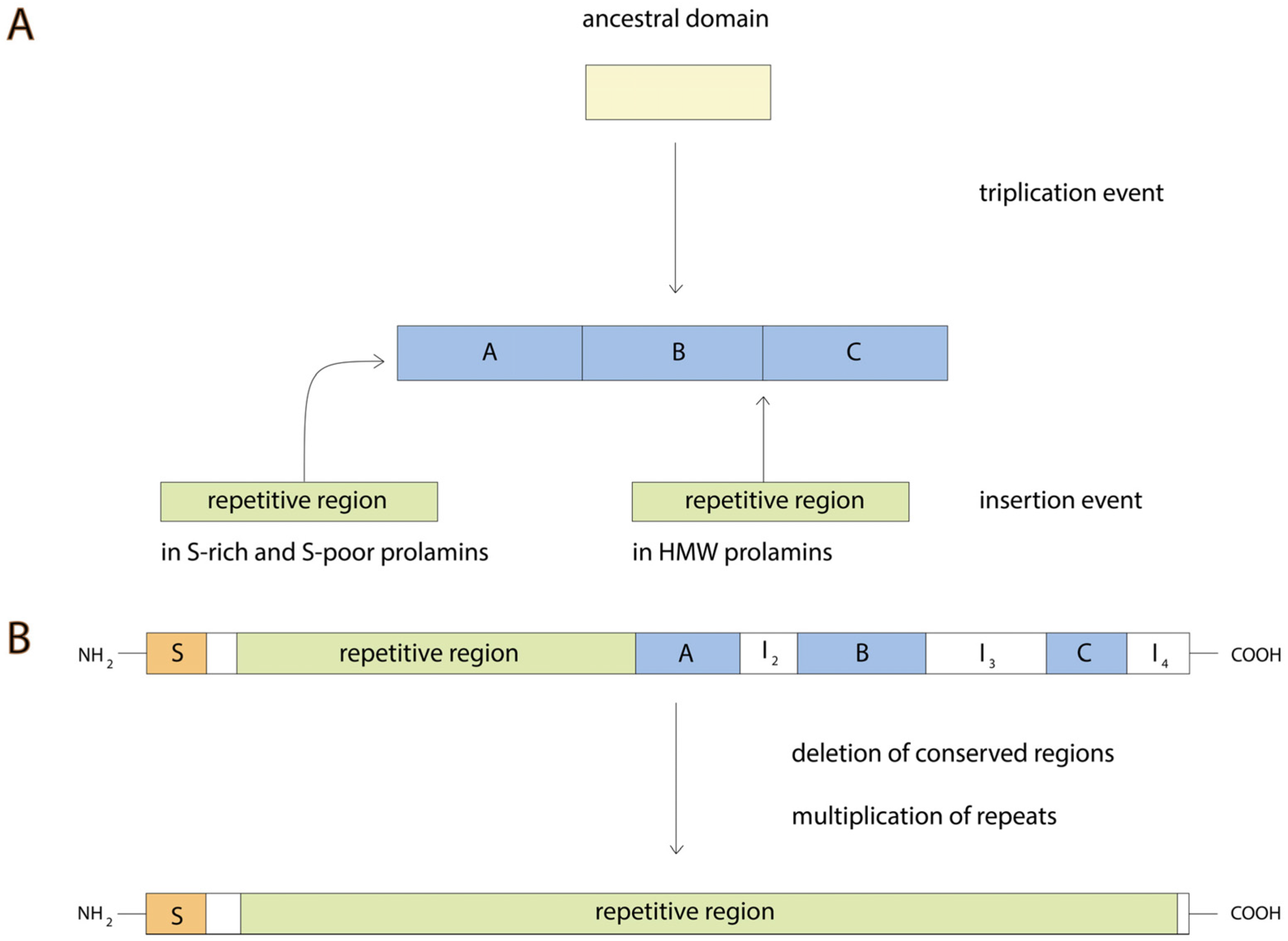
3. Gluten Evolution
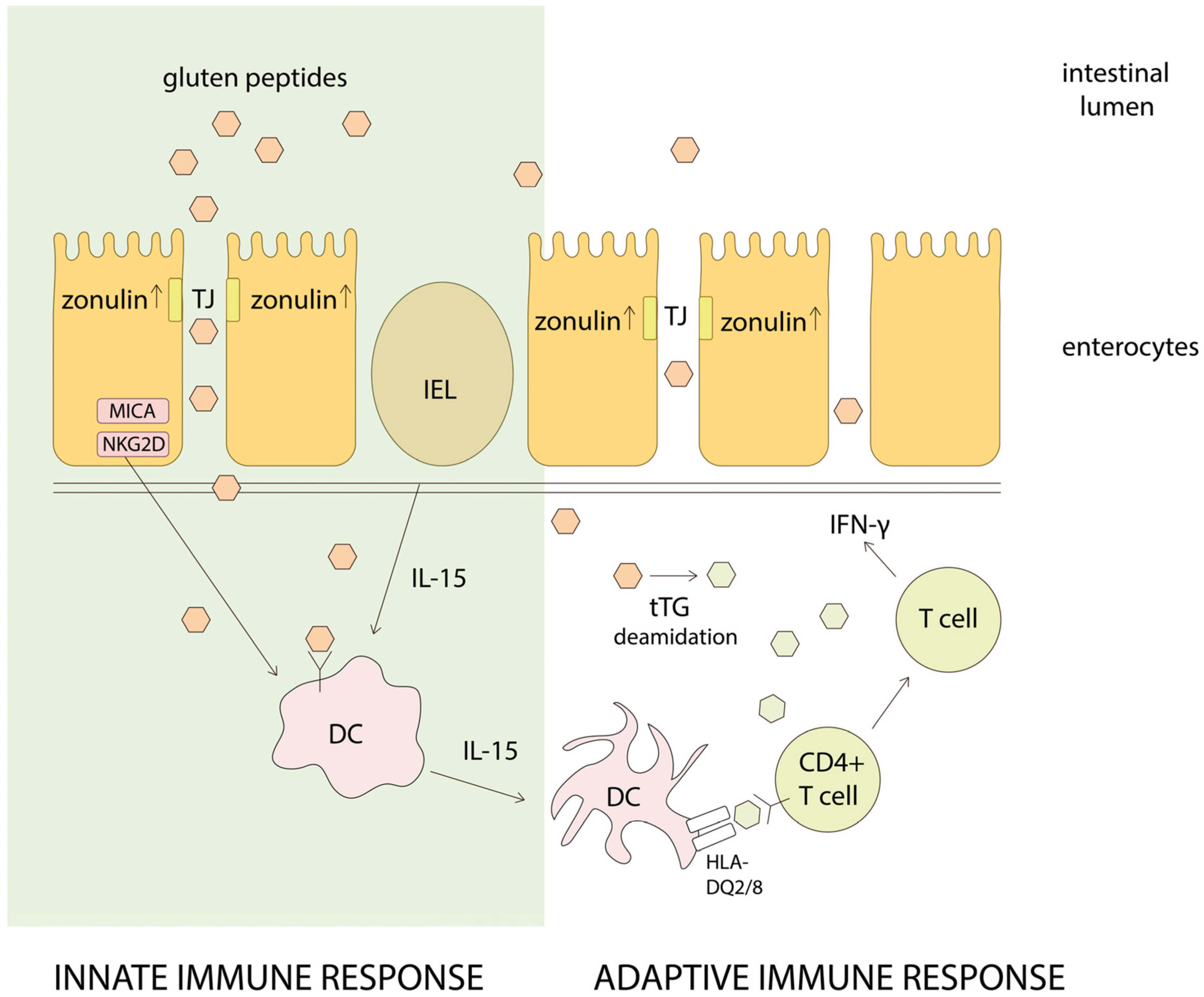
4. Gluten Intolerance Pathogenesis
4.1. Cross-Reactivity between Gluten Proteins
4.2. Celiac Disease
4.3. Wheat Allergy
4.4. Non-Celiac Gluten Sensitivity (NCGS)
5. Gluten Detoxification Strategies
5.1. Gluten Free Diet (GFD)
5.2. Detoxification of Gluten Proteins with Enzymatic Therapy
5.3. Modified Grains
5.4. Corrections of Gluten Pathogenicity Pathways
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CD | celiac disease |
| NCGS | non-celiac gluten sensitivity |
| HLA | human leukocyte antigen |
| MHC | major histocompatibility complex |
| tTG | tissue transglutaminase |
| IgA (IgG, IgE) | immunoglobulin A (G, E) |
| IEL | intraepithelial lymphocyte |
| TJ | tight junction |
| DC | dendritic cell |
| GA | gluten ataxia |
| HMW | high molecular weight |
| LMW | low molecular weight |
| HMW-GS | high molecular weight glutenin subunits |
| LMW-GS | low molecular weight glutenin subunits |
| HMW-SS | high molecular weight secalin subunits |
| SDS-PAGE | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| ER | endoplasmic reticulum |
| DNA | deoxyribonucleic acid |
| RNA | ribonucleic acid |
| FISH | fluorescence in situ hybridization |
| HPLC | high-performance liquid chromatography |
| PDB | protein data bank |
| NGS | next-generation sequencing |
| IFN-γ | interferon-γ |
| IL | interleukin |
| TCL | T cell line |
| TCC | T cell clone |
| PBMC | peripheral blood mononuclear cells |
| WDEIA | wheat-dependent exercise-induced anaphylaxis |
| GFD | gluten-free diet |
| IBS | irritable bowel syndrome |
| GSRS | gastrointestinal symptom rating scale |
| NRS | numerical rating scale |
| BMD | bone mineral density |
References
- Ferguson, A.; MacDonald, T.T.; McClure, J.P.; Holden, R.J. Cell-mediated immunity to gliadin within the small-intestinal mucosa in coeliac disease. Lancet 1975, 1, 895–897. [Google Scholar] [CrossRef]
- Czaja-Bulsa, G. Non coeliac gluten sensitivity—A new disease with gluten intolerance. Clin. Nutr. 2015, 34, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Elli, L.; Bonaz, B.; Bouma, G.; Carroccio, A.; Castillejo, G.; Cellier, C.; Cristofori, F.; de Magistris, L.; Dolinsek, J.; et al. Diagnosis of Non-Celiac Gluten Sensitivity (NCGS): The Salerno Experts’ Criteria. Nutrients 2015, 7, 4966–4977. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Gatti, S.; Lionetti, E. World perspective and celiac disease epidemiology. Dig. Dis. 2015, 33, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Husby, S.; Koletzko, S.; Korponay-Szabo, I.R.; Mearin, M.L.; Phillips, A.; Shamir, R.; Troncone, R.; Giersiepen, K.; Branski, D.; Catassi, C.; et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J. Pediatr. Gastroenterol. Nutr. 2012, 54, 136–160. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A.; Catassi, C. Clinical practice. Celiac disease. N. Engl. J. Med. 2012, 367, 2419–2426. [Google Scholar] [CrossRef] [PubMed]
- Leffler, D.A.; Green, P.H.; Fasano, A. Extraintestinal manifestations of coeliac disease. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Van Heel, D.A.; West, J. Recent advances in coeliac disease. Gut 2006, 55, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Uzzau, S.; Goldblum, S.E.; Fasano, A. Human zonulin, a potential modulator of intestinal tight junctions. J. Cell Sci. 2000, 113, 4435–4440. [Google Scholar] [PubMed]
- Schuppan, D. Current concepts of celiac disease pathogenesis. Gastroenterology 2000, 119, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, E.M.; Jahnsen, F.L.; Lundin, K.E.; Johansen, F.E.; Fausa, O.; Sollid, L.M.; Jahnsen, J.; Scott, H.; Brandtzaeg, P. Gluten induces an intestinal cytokine response strongly dominated by interferon gamma in patients with celiac disease. Gastroenterology 1998, 115, 551–563. [Google Scholar] [CrossRef]
- Dewar, D.; Pereira, S.P.; Ciclitira, P.J. The pathogenesis of coeliac disease. Int. J. Biochem. Cell Biol. 2004, 36, 17–24. [Google Scholar] [CrossRef]
- Jacquenet, S.; Morisset, M.; Battais, F.; Denery-Papini, S.; Croizier, A.; Baudouin, E.; Bihain, B.; Moneret-Vautrin, D.A. Interest of ImmunoCAP system to recombinant omega-5 gliadin for the diagnosis of exercise-induced wheat allergy. Int. Arch. Allergy Immunol. 2009, 149, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Battais, F.; Mothes, T.; Moneret-Vautrin, D.A.; Pineau, F.; Kanny, G.; Popineau, Y.; Bodinier, M.; Denery-Papini, S. Identification of IgE-binding epitopes on gliadins for patients with food allergy to wheat. Allergy 2005, 60, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, L.; Hernandez-Lahoz, C.; Lauret, E.; Rodriguez-Pelaez, M.; Soucek, M.; Ciccocioppo, R.; Kruzliak, P. Gluten ataxia is better classified as non-celiac gluten sensitivity than as celiac disease: A comparative clinical study. Immunol. Res. 2016, 64, 558. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Halford, N.G. Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 2002, 53, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Kreis, M.; Forde, B.G.; Rahman, S.; Miflin, B.J.; Shewry, P.R. Molecular evolution of the seed storage proteins of barley, rye and wheat. J. Mol. Biol. 1985, 183, 499–502. [Google Scholar] [CrossRef]
- Wieser, H. Chemistry of gluten proteins. Food Microbiol. 2007, 24, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; MacRitchie, F. Application of polymer science to properties of gluten. J. Cereal Sci. 2001, 33, 231–243. [Google Scholar] [CrossRef]
- Muccilli, V.; Cunsolo, V.; Saletti, R.; Foti, S.; Masci, S.; Lafiandra, D. Characterization of B- and C-type low molecular weight glutenin subunits by electrospray ionization mass spectrometry and matrix-assisted laser desorption/ionization mass spectrometry. Proteomics 2005, 5, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Zilic, S.; Barac, M.; Pesic, M.; Dodig, D.; Ignjatovic-Micic, D. Characterization of proteins from grain of different bread and durum wheat genotypes. Int. J. Mol. Sci. 2011, 12, 5878–5894. [Google Scholar] [CrossRef] [PubMed]
- Wrigley, C.W.; Shepherd, K.W. Electrofocusing of grain proteins from wheat genotypes. Ann. N. Y. Acad. Sci. 1973, 209, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Ferranti, P.; Mamone, G.; Picariello, G.; Addeo, F. Mass spectrometry analysis of gliadins in celiac disease. J. Mass Spectrom. 2007, 42, 1531–1548. [Google Scholar] [CrossRef] [PubMed]
- Tatham, A.S.; Drake, A.F.; Shewry, P.R. A conformational study of a glutamine- and proline-rich cereal seed protein, C hordein. Biochem. J. 1985, 226, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Schofield, J.D.; Bottomley, R.C.; Timms, M.F.; Booth, M.R. The effect of heat on wheat gluten and the involvement of sulphydryl-disulphide interchange reactions. J. Cereal Sci. 1983, 1, 241–253. [Google Scholar] [CrossRef]
- Rombouts, I.; Lagrain, B.; Brijs, K.; Delcour, J.A. β-Elimination reactions and formation of covalent cross-links in gliadin during heating at alkaline pH. J. Cereal Sci. 2010, 52, 362–367. [Google Scholar] [CrossRef]
- Lagrain, B.; Rombouts, I.; Brijs, K.; Delcour, J.A. Kinetics of heat-induced polymerization of gliadin. J. Agric. Food Chem. 2011, 59, 2034–2039. [Google Scholar] [CrossRef] [PubMed]
- Barak, S.; Mudgil, D.; Khatkar, B.S. Biochemical and functional properties of wheat gliadins: A review. Crit. Rev. Food Sci. Nutr. 2015, 55, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Hamer, R.J.; van Vliet, T. Understanding the structure and properties of gluten: An overview. In Wheat Gluten; Shewry, P.R., Tatham, A.S., Eds.; Royal Society of Chemistry: Cambrige, UK, 2000; pp. 125–131. [Google Scholar]
- Ozuna, C.V.; Iehisa, J.C.; Gimenez, M.J.; Alvarez, J.B.; Sousa, C.; Barro, F. Diversification of the celiac disease alpha-gliadin complex in wheat: A 33-mer peptide with six overlapping epitopes, evolved following polyploidization. Plant J. 2015, 82, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Tatham, A.S.; Marsh, M.N.; Wieser, H.; Shewry, P.R. Conformational studies of peptides corresponding to the coeliac-activating regions of wheat alpha-gliadin. Biochem. J. 1990, 270, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.G.; Benedini, L.A.; Lonez, C.; Schilardi, P.L.; Hellweg, T.; Ruysschaert, J.M.; Dodero, V.I. Self-assembly of 33-mer gliadin peptide oligomers. Soft Matter 2015, 11, 8648–8660. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Matsumiya, A.; Higashino, Y.; Funaki, S.; Kitao, Y.; Oba, Y.; Inoue, R.; Arisaka, F.; Sugiyama, M.; Urade, R. Molecular assembly of wheat gliadins into nanostructures: A small-angle X-ray scattering study of gliadins in distilled water over a wide concentration range. J. Agric. Food Chem. 2015, 63, 8715–8721. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.G.; Veuthey, T.V.; Dodero, V.I. Self-organization of gliadin in aqueous media under physiological digestive pHs. Colloids Surf. B Biointerfaces 2016, 141, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Banc, A.; Desbat, B.; Renard, D.; Popineau, Y.; Mangavel, C.; Navailles, L. Exploring the interactions of gliadins with model membranes: Effect of confined geometry and interfaces. Biopolymers 2009, 91, 610–622. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Napier, J.A.; Tatham, A.S. Seed storage proteins: Structures and biosynthesis. Plant Cell 1995, 7, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Wellner, N.; Mills, E.N.; Brownsey, G.; Wilson, R.H.; Brown, N.; Freeman, J.; Halford, N.G.; Shewry, P.R.; Belton, P.S. Changes in protein secondary structure during gluten deformation studied by dynamic fourier transform infrared spectroscopy. Biomacromolecules 2005, 6, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, G.J.; Shepherd, K.W. Chromosomal location of genes controlling seed proteins in species related to wheat. Theor. Appl. Genet. 1981, 59, 25–31. [Google Scholar] [PubMed]
- Shewry, P.R.; Halford, N.G.; Tatham, A.S.; Popineau, Y.; Lafiandra, D.; Belton, P.S. The high molecular weight subunits of wheat glutenin and their role in determining wheat processing properties. Adv. Food Nutr. Res. 2003, 45, 219–302. [Google Scholar] [PubMed]
- Payne, P.I.; Holt, L.M.; Jarvis, G.; Jackson, E.A. Two-dimensional fractionation of the endosperm proteins of bread wheat (Triticum aestivum): Biochemical and genetic studies. Cereal Chem. 1985, 62, 319–326. [Google Scholar]
- Tosi, P.; D’Ovidio, R.; Napier, J.A.; Bekes, F.; Shewry, P.R. Expression of epitope-tagged LMW glutenin subunits in the starchy endosperm of transgenic wheat and their incorporation into glutenin polymers. Theor. Appl. Genet. 2004, 108, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Masci, S.; Egorov, T.A.; Ronchi, C.; Kuzmicky, D.D.; Kasarda, D.D.; Lafiandra, D. Evidence for the presence of only one cysteine residue in the D-type low molecular weight subunits of wheat glutenin. J. Cereal Sci. 1999, 29, 17–25. [Google Scholar] [CrossRef]
- Gorg, A.; Postel, W.; Baumer, M.; Weiss, W. Two-dimensional polyacrylamide gel electrophoresis, with immobilized pH gradients in the first dimension, of barley seed proteins: Discrimination of cultivars with different malting grades. Electrophoresis 1992, 13, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.; Jorgensen, J.H.; Jensen, H.P.; Giese, H.; Doll, H. Linkage of the hordein loci Hor1 and Hor2 with the powdery mildew resistance loci Ml-k and Ml-a on Barley chromosome 5. Theor. Appl. Genet. 1980, 58, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G.; Kreis, M.; Williamson, M.S.; Fry, R.P.; Pywell, J.; Shewry, P.R.; Bunce, N.; Miflin, B.J. Short tandem repeats shared by B- and C-hordein cDNAs suggest a common evolutionary origin for two groups of cereal storage protein genes. EMBO J. 1985, 4, 9–15. [Google Scholar] [PubMed]
- Anderson, O.D. The B-hordein prolamin family of barley. Genome 2013, 56, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Rechinger, K.B.; Simpson, D.J.; Svendsen, I.; Cameron-Mills, V. A role for gamma 3 hordein in the transport and targeting of prolamin polypeptides to the vacuole of developing barley endosperm. Plant J. 1993, 4, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.; Bishop, D.H.L.; Tatham, A.S.; Shewry, P.R. Exploring disulphide bond formation in a low molecular weight subunit of glutenin using a baculovirus expression system. Gluten Proteins 1994, 345–355. [Google Scholar]
- Faulks, A.J.; Shewry, P.R.; Miflin, B.J. The polymorphism and structural homology of storage polypeptides (hordein) coded by the Hor-2 locus in barley (Hordeum vulgare L.). Biochem. Genet. 1981, 19, 841–858. [Google Scholar] [CrossRef] [PubMed]
- Tatham, A.S.; Shewry, P.R.; Belton, P.S. 13C-n.m.r. study of C hordein. Biochem. J. 1985, 232, 617–620. [Google Scholar] [PubMed]
- I’Anson, K.J.; Morris, V.J.; Shewry, P.R.; Tatham, A.S. Small-angle X-ray-scattering studies of the C hordeins of barley (Hordeum vulgare). Biochem. J. 1992, 287, 183–185. [Google Scholar] [CrossRef] [PubMed]
- Cameron-Mills, V.; von Wettstein, D. Protein body formation in the developing barley endosperm. Carlsberg Res. Commun. 1980, 45, 577–594. [Google Scholar] [CrossRef]
- Halford, N.G.; Tatham, A.S.; Sui, E.; Daroda, L.; Dreyer, T.; Shewry, P.R. Identification of a novel beta-turn-rich repeat motif in the D hordeins of barley. Biochim. Biophys. Acta 1992, 1122, 118–122. [Google Scholar] [CrossRef]
- Hull, G.; Sabelli, P.A.; Shewry, P.R. Restriction fragment analysis of the secalin loci of rye. Biochem. Genet. 1992, 30, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Salmanowicz, B.P.; Langner, M.; Kubicka-Matusiewicz, H. Variation of high-molecular-weight secalin subunit composition in Rye (Secale cereale L.) inbred lines. J. Agric. Food Chem. 2014, 62, 10535–10541. [Google Scholar] [CrossRef] [PubMed]
- Miles, M.J.; Carr, H.J.; McMaster, T.; Belton, P.S.; Morris, V.J.; Field, J.M.; Shewry, P.R.; Tatham, A.S. Scanning tunnelling microscopy of a wheat gluten protein reveals details of a spiral supersecondary structure. Proc. Natl. Acad. Sci. USA 1991, 88, 68. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.K.; Shepherd, K.W.; McIntosh, R.A. Linkage mapping of genes for resistance to leaf, stem and stripe rusts and omega-secalins on the short arm of rye chromosome 1R. Theor. Appl. Genet. 1990, 80, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Clarke, B.C.; Mukai, Y.; Appels, R. The Sec-1 locus on the short arm of chromosome 1R of rye (Secale cereale). Chromosoma 1996, 105, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Mukai, Y. High-resolution physical mapping of the secalin-1 locus of rye on extended DNA fibers. Cytogenet. Genome Res. 2005, 109, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.T.; Wei, Y.M.; Andre, L.; Lu, Z.X.; Pu, Z.E.; Peng, Y.Y.; Zheng, Y.L. Characterization of ω-secalin genes from rye, triticale, and a wheat 1BL/1RS translocation line. J. Appl. Genet. 2010, 51, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Dennett, A.L.; Cooper, K.V.; Trethowan, R.M. The genotypic and phenotypic interaction of wheat and rye storage proteins in primary triticale. Euphytica 2013, 194, 235–242. [Google Scholar] [CrossRef]
- Shewry, P.R. Plant storage proteins. Biol. Rev. Camb. Philos. Soc. 1995, 70, 375–426. [Google Scholar] [CrossRef] [PubMed]
- Chesnut, R.S.; Shotwell, M.A.; Boyer, S.K.; Larkins, B.A. Analysis of avenin proteins and the expression of their mRNAs in developing oat seeds. Plant Cell 1989, 1, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Lapvetelainen, A.; Bietz, J.; Huebner, F. Reversed-phase high-performance liquid chromatography of oat proteins: Application of cultivar comparison and analysis of the effect of wet processing. Cereal Chem. 1995, 72, 259–264. [Google Scholar]
- Egorov, T.A.; Musolyamov, A.K.; Andersen, J.S.; Roepstorff, P. The complete amino acid sequence and disulphide bond arrangement of oat alcohol-soluble avenin-3. Eur. J. Biochem. 1994, 224, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Real, A.; Comino, I.; de Lorenzo, L.; Merchan, F.; Gil-Humanes, J.; Gimenez, M.J.; Lopez-Casado, M.A.; Torres, M.I.; Cebolla, A.; Sousa, C.; et al. Molecular and immunological characterization of gluten proteins isolated from oat cultivars that differ in toxicity for celiac disease. PLoS ONE 2012, 7, e48365. [Google Scholar]
- Lending, C.R.; Chesnut, R.S.; Shaw, K.L.; Larkins, B.A. Immunolocalization of avenin and globulin storage proteins in developing endosperm of Avena sativa L. Planta 1989, 178, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.F.; Le, C.X.; Wang, Z.; Liu, Y.B.; Chen, Q.; Wei, Z.Z.; Xu, B.J.; Wei, Z.Y.; Dai, S.F.; Wei, Y.M.; et al. The gamma-gliadin-like gamma-prolamin genes in the tribe Triticeae. J. Genet. 2014, 93, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Vader, W.; Kooy, Y.; Van Veelen, P.; De Ru, A.; Harris, D.; Benckhuijsen, W.; Pena, S.; Mearin, L.; Drijfhout, J.W.; Koning, F. The gluten response in children with celiac disease is directed toward multiple gliadin and glutenin peptides. Gastroenterology 2002, 122, 1729–1737. [Google Scholar] [CrossRef] [PubMed]
- Bushara, K.O. Neurologic presentation of celiac disease. Gastroenterology 2005, 128, S92–S97. [Google Scholar] [CrossRef] [PubMed]
- Green, P.H.; Alaedini, A.; Sander, H.W.; Brannagan, T.H., 3rd; Latov, N.; Chin, R.L. Mechanisms underlying celiac disease and its neurologic manifestations. Cell. Mol. Life Sci. 2005, 62, 791–799. [Google Scholar]
- Alaedini, A.; Okamoto, H.; Briani, C.; Wollenberg, K.; Shill, H.A.; Bushara, K.O.; Sander, H.W.; Green, P.H.; Hallett, M.; Latov, N. Immune cross-reactivity in celiac disease: Anti-gliadin antibodies bind to neuronal synapsin I. J. Immunol. 2007, 178, 6590–6595. [Google Scholar] [CrossRef] [PubMed]
- Palosuo, K.; Alenius, H.; Varjonen, E.; Kalkkinen, N.; Reunala, T. Rye gamma-70 and gamma-35 secalins and barley gamma-3 hordein cross-react with omega-5 gliadin, a major allergen in wheat-dependent, exercise-induced anaphylaxis. Clin. Exp. Allergy 2001, 31, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Festenstein, G.W.; Hay, F.C.; Shewry, P.R. Immunochemical relationships of the prolamin storage proteins of barley, wheat, rye and oat. Biochim. Biophys. Acta 1987, 912, 371–383. [Google Scholar] [CrossRef]
- Vader, L.W.; Stepniak, D.T.; Bunnik, E.M.; Kooy, Y.M.; de Haan, W.; Drijfhout, J.W.; Van Veelen, P.A.; Koning, F. Characterization of cereal toxicity for celiac disease patients based on protein homology in grains. Gastroenterology 2003, 125, 1105–1113. [Google Scholar] [CrossRef]
- Hogberg, L.; Laurin, P.; Falth-Magnusson, K.; Grant, C.; Grodzinsky, E.; Jansson, G.; Ascher, H.; Browaldh, L.; Hammersjo, J.A.; Lindberg, E.; et al. Oats to children with newly diagnosed coeliac disease: A randomised double blind study. Gut 2004, 53, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Gatti, S.; Caporelli, N.; Galeazzi, T.; Francavilla, R.; Barbato, M.; Roggero, P.; Malamisura, B.; Iacono, G.; Budelli, A.; Gesuita, R.; et al. Oats in the diet of children with celiac disease: Preliminary results of a double-blind, randomized, placebo-controlled multicenter Italian study. Nutrients 2013, 5, 4653–4664. [Google Scholar] [CrossRef] [PubMed]
- Wieser, H. Relation between gliadin structure and coeliac toxicity. Acta Paediatr. 1996, 85, 3–9. [Google Scholar] [CrossRef]
- Hardy, M.Y.; Tye-Din, J.A.; Stewart, J.A.; Schmitz, F.; Dudek, N.L.; Hanchapola, I.; Purcell, A.W.; Anderson, R.P. Ingestion of oats and barley in patients with celiac disease mobilizes cross-reactive T cells activated by avenin peptides and immuno-dominant hordein peptides. J. Autoimmun. 2015, 56, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Hovhannisyan, Z.; Weiss, A.; Martin, A.; Wiesner, M.; Tollefsen, S.; Yoshida, K.; Ciszewski, C.; Curran, S.A.; Murray, J.A.; David, C.S.; et al. The role of HLA-DQ8 beta57 polymorphism in the anti-gluten T-cell response in coeliac disease. Nature 2008, 456, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.T.; Mosekilde, L.; Duan, Y.; Zhang, X.Z.; Tornvig, L.; Thomsen, J.S.; Seeman, E. The structural and hormonal basis of sex differences in peak appendicular bone strength in rats. J. Bone Miner. Res. 2003, 18, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Ciccocioppo, R.; Di Sabatino, A.; Corazza, G.R. The immune recognition of gluten in coeliac disease. Clin. Exp. Immunol. 2005, 140, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Van de Wal, Y.; Kooy, Y.M.C.; van Veelen, P.; Vader, W.; August, S.A.; Drijfhout, J.W.; Pena, S.A.; Koning, F. Glutenin is involved in the glutendriven mucosal T cell response. Eur. J. Immunol. 1999, 29, 3133–3139. [Google Scholar] [CrossRef]
- Barone, M.V.; Zimmer, K.P. Endocytosis and transcytosis of gliadin peptides. Mol. Cell. Pediatr. 2016, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.G.; Zamarreno, F.; Costabel, M.; Ritacco, H.; Hutten, A.; Sewald, N.; Dodero, V.I. Circular dichroism and electron microscopy studies in vitro of 33-mer gliadin peptide revealed secondary structure transition and supramolecular organization. Biopolymers 2014, 101, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Molberg, O.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.M.; Sollid, L.M.; Khosla, C. Structural basis for gluten intolerance in celiac sprue. Science 2002, 297, 2275–2279. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, C.; Menard, S.; Abed, J.; Moura, I.C.; Coppo, R.; Dugave, C.; Monteiro, R.C.; Fricot, A.; Traore, M.G.; Griffin, M.; et al. Interactions among secretory immunoglobulin A, CD71, and transglutaminase-2 affect permeability of intestinal epithelial cells to gliadin peptides. Gastroenterology 2012, 143, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, K.; Alvarez, X.; Borda, J.T.; Dufour, J.; Martin, E.; Bethune, M.T.; Khosla, C.; Sestak, K. Visualization of transepithelial passage of the immunogenic 33-residue peptide from alpha-2 gliadin in gluten-sensitive macaques. PLoS ONE 2010, 5, e10228. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Viljanen, M.K.; Hinkkanen, A.E.; Arvilommi, H.; Mertsola, J.; Viander, M. No evidence of cross-reactivity of human antibodies to a 33-mer peptide of the alpha-gliadin component of gluten with Bordetella pertussis pertactin. Vaccine 2005, 23, 3336–3340. [Google Scholar] [CrossRef] [PubMed]
- Vartdal, F.; Johansen, B.H.; Friede, T.; Thorpe, C.J.; Stevanovic, S.; Eriksen, J.E.; Sletten, K.; Thorsby, E.; Rammensee, H.G.; Sollid, L.M. The peptide binding motif of the disease associated HLA-DQ (alpha 1* 0501, beta 1* 0201) molecule. Eur. J. Immunol. 1996, 26, 2764–2772. [Google Scholar] [CrossRef] [PubMed]
- Arentz-Hansen, H.; McAdam, S.N.; Molberg, O.; Fleckenstein, B.; Lundin, K.E.; Jorgensen, T.J.; Jung, G.; Roepstorff, P.; Sollid, L.M. Celiac lesion T cells recognize epitopes that cluster in regions of gliadins rich in proline residues. Gastroenterology 2002, 123, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.P.; Degano, P.; Godkin, A.J.; Jewell, D.P.; Hill, A.V. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat. Med. 2000, 6, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Tye-Din, J.A.; Stewart, J.A.; Dromey, J.A.; Beissbarth, T.; van Heel, D.A.; Tatham, A.; Henderson, K.; Mannering, S.I.; Gianfrani, C.; Jewell, D.P.; et al. Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci. Transl. Med. 2010, 2, 41ra51. [Google Scholar] [CrossRef] [PubMed]
- Beissbarth, T.; Tye-Din, J.A.; Smyth, G.K.; Speed, T.P.; Anderson, R.P. A systematic approach for comprehensive T-cell epitope discovery using peptide libraries. Bioinformatics 2005, 21 (Suppl. 1), i29–i37. [Google Scholar] [CrossRef] [PubMed]
- Camarca, A.; Del Mastro, A.; Gianfrani, C. Repertoire of gluten peptides active in celiac disease patients: Perspectives for translational therapeutic applications. Endocr. Metab. Immune Disord. Drug Targets 2012, 12, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Comino, I.; Bernardo, D.; Bancel, E.; de Lourdes Moreno, M.; Sanchez, B.; Barro, F.; Suligoj, T.; Ciclitira, P.J.; Cebolla, A.; Knight, S.C.; et al. Identification and molecular characterization of oat peptides implicated on coeliac immune response. Food Nutr. Res. 2016, 60, 30324. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.Y.; Volcheck, G.W. Food allergy: Common causes, diagnosis, and treatment. Mayo Clin. Proc. 2015, 90, 1411–1419. [Google Scholar] [CrossRef] [PubMed]
- Chehade, M.; Mayer, L. Oral tolerance and its relation to food hypersensitivities. J. Allergy Clin. Immunol. 2005, 115, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, H.; Yokooji, T.; Taogoshi, T. Common food allergens and their IgE-binding epitopes. Allergol. Int. 2015, 64, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Tatham, A.S.; Shewry, P.R. Allergens to wheat and related cereals. Clin. Exp. Allergy 2008, 38, 1712–1726. [Google Scholar] [PubMed]
- Beyer, K.; Chung, D.; Schulz, G.; Mishoe, M.; Niggemann, B.; Wahn, U.; Sampson, H.A. The role of wheat omega-5 gliadin IgE antibodies as a diagnostic tool for wheat allergy in childhood. J. Allergy Clin. Immunol. 2008, 122, 419–421. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, S. IgE-binding abilities of pentapeptides, QQPFP and PQQPF, in wheat gliadin. J. Nutr. Sci. Vitaminol. (Tokyo) 2004, 50, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, S.; Arai, S.; Yanagihara, Y.; Mita, H.; Takahashi, K.; Watanabe, M. A major wheat allergen has a Gln-Gln-Gln-Pro-Pro motif identified as an IgE-binding epitope. Biochem. Biophys. Res. Commun. 1996, 219, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Volta, U.; Bardella, M.T.; Calabro, A.; Troncone, R.; Corazza, G.R.; The Study Group for Non-Celiac Gluten Sensitivity. An Italian prospective multicenter survey on patients suspected of having non-celiac gluten sensitivity. BMC Med. 2014, 12, 85. [Google Scholar] [PubMed]
- Aziz, I.; Lewis, N.R.; Hadjivassiliou, M.; Winfield, S.N.; Rugg, N.; Kelsall, A.; Newrick, L.; Sanders, D.S. A UK study assessing the population prevalence of self-reported gluten sensitivity and referral characteristics to secondary care. Eur. J. Gastroenterol. Hepatol. 2014, 26, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Sapone, A.; Lammers, K.M.; Casolaro, V.; Cammarota, M.; Giuliano, M.T.; De Rosa, M.; Stefanile, R.; Mazzarella, G.; Tolone, C.; Russo, M.I.; et al. Divergence of gut permeability and mucosal immune gene expression in two gluten-associated conditions: Celiac disease and gluten sensitivity. BMC Med. 2011, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Bucci, C.; Zingone, F.; Russo, I.; Morra, I.; Tortora, R.; Pogna, N.; Scalia, G.; Iovino, P.; Ciacci, C. Gliadin does not induce mucosal inflammation or basophil activation in patients with nonceliac gluten sensitivity. Clin. Gastroenterol. Hepatol. 2013, 11, 1294–1299.e1. [Google Scholar] [CrossRef] [PubMed]
- Aziz, I.; Hadjivassiliou, M.; Sanders, D.S. The spectrum of noncoeliac gluten sensitivity. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.; Eaton, W.; Cascella, N.; Fasano, A.; Santora, D.; Sullivan, K.; Feldman, S.; Raley, H.; McMahon, R.P.; Carpenter, W.T., Jr.; et al. Gluten sensitivity and relationship to psychiatric symptoms in people with schizophrenia. Schizophr. Res. 2014, 159, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, E.; Leonardi, S.; Franzonello, C.; Mancardi, M.; Ruggieri, M.; Catassi, C. Gluten Psychosis: Confirmation of a New Clinical Entity. Nutrients 2015, 7, 5532–5539. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.S.; Aziz, I. Non-celiac wheat sensitivity: Separating the wheat from the chat! Am. J. Gastroenterol. 2012, 107, 1908–1912. [Google Scholar] [CrossRef] [PubMed]
- Makharia, A.; Catassi, C.; Makharia, G.K. The overlap between irritable bowel syndrome and non-celiac gluten sensitivity: A clinical dilemma. Nutrients 2015, 7, 10417–10426. [Google Scholar] [CrossRef] [PubMed]
- Leonard, M.M.; Camhi, S.; Huedo-Medina, T.B.; Fasano, A. Celiac disease genomic, environmental, microbiome, and metabolomic (CDGEMM) study design: Approach to the future of personalized prevention of celiac disease. Nutrients 2015, 7, 9325–9336. [Google Scholar] [CrossRef] [PubMed]
- Leonard, M.M.; Fasano, A. The microbiome as a possible target to prevent celiac disease. Expert Rev. Gastroenterol. Hepatol. 2016, 10, 555–556. [Google Scholar] [CrossRef] [PubMed]
- Hischenhuber, C.; Crevel, R.; Jarry, B.; Maki, M.; Moneret-Vautrin, D.A.; Romano, A.; Troncone, R.; Ward, R. Review article: Safe amounts of gluten for patients with wheat allergy or coeliac disease. Aliment. Pharmacol. Ther. 2006, 23, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Penagini, F.; Dilillo, D.; Meneghin, F.; Mameli, C.; Fabiano, V.; Zuccotti, G.V. Gluten-free diet in children: An approach to a nutritionally adequate and balanced diet. Nutrients 2013, 5, 4553–4565. [Google Scholar] [CrossRef] [PubMed]
- Saturni, L.; Ferretti, G.; Bacchetti, T. The gluten-free diet: Safety and nutritional quality. Nutrients 2010, 2, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Hallert, C.; Grant, C.; Grehn, S.; Granno, C.; Hulten, S.; Midhagen, G.; Strom, M.; Svensson, H.; Valdimarsson, T. Evidence of poor vitamin status in coeliac patients on a gluten-free diet for 10 years. Aliment. Pharmacol. Ther. 2002, 16, 1333–1339. [Google Scholar] [CrossRef] [PubMed]
- Hallert, C.; Svensson, M.; Tholstrup, J.; Hultberg, B. Clinical trial: B vitamins improve health in patients with coeliac disease living on a gluten-free diet. Aliment. Pharmacol. Ther. 2009, 29, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Pantaleoni, S.; Luchino, M.; Adriani, A.; Pellicano, R.; Stradella, D.; Ribaldone, D.G.; Sapone, N.; Isaia, G.C.; Di Stefano, M.; Astegiano, M. Bone mineral density at diagnosis of celiac disease and after 1 year of gluten-free diet. Sci. World J. 2014, 2014, 173082. [Google Scholar] [CrossRef] [PubMed]
- Caruso, R.; Pallone, F.; Stasi, E.; Romeo, S.; Monteleone, G. Appropriate nutrient supplementation in celiac disease. Ann. Med. 2013, 45, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Vici, G.; Belli, L.; Biondi, M.; Polzonetti, V. Gluten free diet and nutrient deficiencies: A review. Clin. Nutr. 2016, in press. [Google Scholar] [CrossRef] [PubMed]
- Lamacchia, C.; Camarca, A.; Picascia, S.; Di Luccia, A.; Gianfrani, C. Cereal-based gluten-free food: How to reconcile nutritional and technological properties of wheat proteins with safety for celiac disease patients. Nutrients 2014, 6, 575–590. [Google Scholar] [CrossRef] [PubMed]
- Szajewska, H.; Shamir, R.; Mearin, L.; Ribes-Koninckx, C.; Catassi, C.; Domellof, M.; Fewtrell, M.S.; Husby, S.; Papadopoulou, A.; Vandenplas, Y.; et al. Gluten introduction and the risk of coeliac disease: A position paper by the european society for pediatric gastroenterology, hepatology, and nutrition. J. Pediatr. Gastroenterol. Nutr. 2016, 62, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A. Novel therapeutic/integrative approaches for celiac disease and dermatitis herpetiformis. Clin. Dev. Immunol. 2012, 2012, 959061. [Google Scholar] [CrossRef] [PubMed]
- Zamyatnin, A.A., Jr. Plant proteases involved in regulated cell death. Biochemistry 2015, 80, 1701–1715. [Google Scholar] [CrossRef] [PubMed]
- Yike, I. Fungal proteases and their pathophysiological effects. Mycopathologia 2011, 171, 299–323. [Google Scholar] [CrossRef] [PubMed]
- Kantyka, T.; Shaw, L.N.; Potempa, J. Papain-like proteases of Staphylococcus aureus. Adv. Exp. Med. Biol. 2011, 712, 1–14. [Google Scholar] [PubMed]
- Bethune, M.T.; Khosla, C. Oral enzyme therapy for celiac sprue. Methods Enzymol. 2012, 502, 241–271. [Google Scholar] [PubMed]
- Wieser, H.; Koehler, P. Detoxification of gluten by means of enzymatic treatment. J. AOAC Int. 2012, 95, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Savvateeva, L.V.; Zamyatnin, A.A. Prospects of Developing Medicinal Therapeutic strategies and pharmaceutical design for effective gluten intolerance treatment. Curr. Pharm. Des. 2016, 22, 2439–2449. [Google Scholar] [CrossRef] [PubMed]
- Savvateeva, L.V.; Gorokhovets, N.V.; Makarov, V.A.; Serebryakova, M.V.; Solovyev, A.G.; Morozov, S.Y.; Reddy, V.P.; Zernii, E.Y.; Zamyatnin, A.A., Jr.; Aliev, G. Glutenase and collagenase activities of wheat cysteine protease Triticain-alpha: Feasibility for enzymatic therapy assays. Int. J. Biochem. Cell Biol. 2015, 62, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Bethune, M.T.; Strop, P.; Tang, Y.; Sollid, L.M.; Khosla, C. Heterologous expression, purification, refolding, and structural-functional characterization of EP-B2, a self-activating barley cysteine endoprotease. Chem. Biol. 2006, 13, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Marti, T.; Sollid, L.M.; Gray, G.M.; Khosla, C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: Implications for coeliac sprue. Biochem. J. 2004, 383, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Goptar, I.A.; Semashko, T.A.; Danilenko, S.A.; Lysogorskaya, E.N.; Oksenoit, E.S.; Zhuzhikov, D.P.; Belozersky, M.A.; Dunaevsky, Y.E.; Oppert, B.; Filippova, I.Y.; et al. Cysteine digestive peptidases function as post-glutamine cleaving enzymes in tenebrionid stored-product pests. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012, 161, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Lahdeaho, M.L.; Kaukinen, K.; Laurila, K.; Vuotikka, P.; Koivurova, O.P.; Karja-Lahdensuu, T.; Marcantonio, A.; Adelman, D.C.; Maki, M. Glutenase ALV003 attenuates gluten-induced mucosal injury in patients with celiac disease. Gastroenterology 2014, 146, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Tye-Din, J.A.; Anderson, R.P.; Ffrench, R.A.; Brown, G.J.; Hodsman, P.; Siegel, M.; Botwick, W.; Shreeniwas, R. The effects of ALV003 pre-digestion of gluten on immune response and symptoms in celiac disease in vivo. Clin. Immunol. 2010, 134, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; De Angelis, M.; Auricchio, S.; Greco, L.; Clarke, C.; De Vincenzi, M.; Giovannini, C.; D’Archivio, M.; Landolfo, F.; Parrilli, G.; et al. Sourdough bread made from wheat and nontoxic flours and started with selected lactobacilli is tolerated in celiac sprue patients. Appl. Environ. Microbiol. 2004, 70, 1088–1096. [Google Scholar] [CrossRef] [PubMed]
- Altenbach, S.B.; Allen, P.V. Transformation of the US bread wheat ‘Butte 86‘ and silencing of omega-5 gliadin genes. GM Crops 2011, 2, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Gil-Humanes, J.; Piston, F.; Tollefsen, S.; Sollid, L.M.; Barro, F. Effective shutdown in the expression of celiac disease-related wheat gliadin T-cell epitopes by RNA interference. Proc. Natl. Acad. Sci. USA 2010, 107, 17023–17028. [Google Scholar] [CrossRef] [PubMed]
- Tanner, G.J.; Howitt, C.A.; Forrester, R.I.; Campbell, P.M.; Tye-Din, J.A.; Anderson, R.P. Dissecting the T-cell response to hordeins in coeliac disease can develop barley with reduced immunotoxicity. Aliment. Pharmacol. Ther. 2010, 32, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Hileman, R.E.; Silvanovich, A.; Goodman, R.E.; Rice, E.A.; Holleschak, G.; Astwood, J.D.; Hefle, S.L. Bioinformatic methods for allergenicity assessment using a comprehensive allergen database. Int. Arch. Allergy Immunol. 2002, 128, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Molberg, O.; McAdam, S.; Lundin, K.E.; Kristiansen, C.; Arentz-Hansen, H.; Kett, K.; Sollid, L.M. T cells from celiac disease lesions recognize gliadin epitopes deamidated in situ by endogenous tissue transglutaminase. Eur. J. Immunol. 2001, 31, 1317–1323. [Google Scholar] [CrossRef]
- Keillor, J.W.; Apperley, K.Y.; Akbar, A. Inhibitors of tissue transglutaminase. Trends Pharmacol. Sci. 2015, 36, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Paterson, B.M.; Lammers, K.M.; Arrieta, M.C.; Fasano, A.; Meddings, J.B. The safety, tolerance, pharmacokinetic and pharmacodynamic effects of single doses of AT-1001 in coeliac disease subjects: A proof of concept study. Aliment. Pharmacol. Ther. 2007, 26, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Larche, M.; Wraith, D.C. Peptide-based therapeutic vaccines for allergic and autoimmune diseases. Nat. Med. 2005, 11, S69–S76. [Google Scholar] [CrossRef] [PubMed]
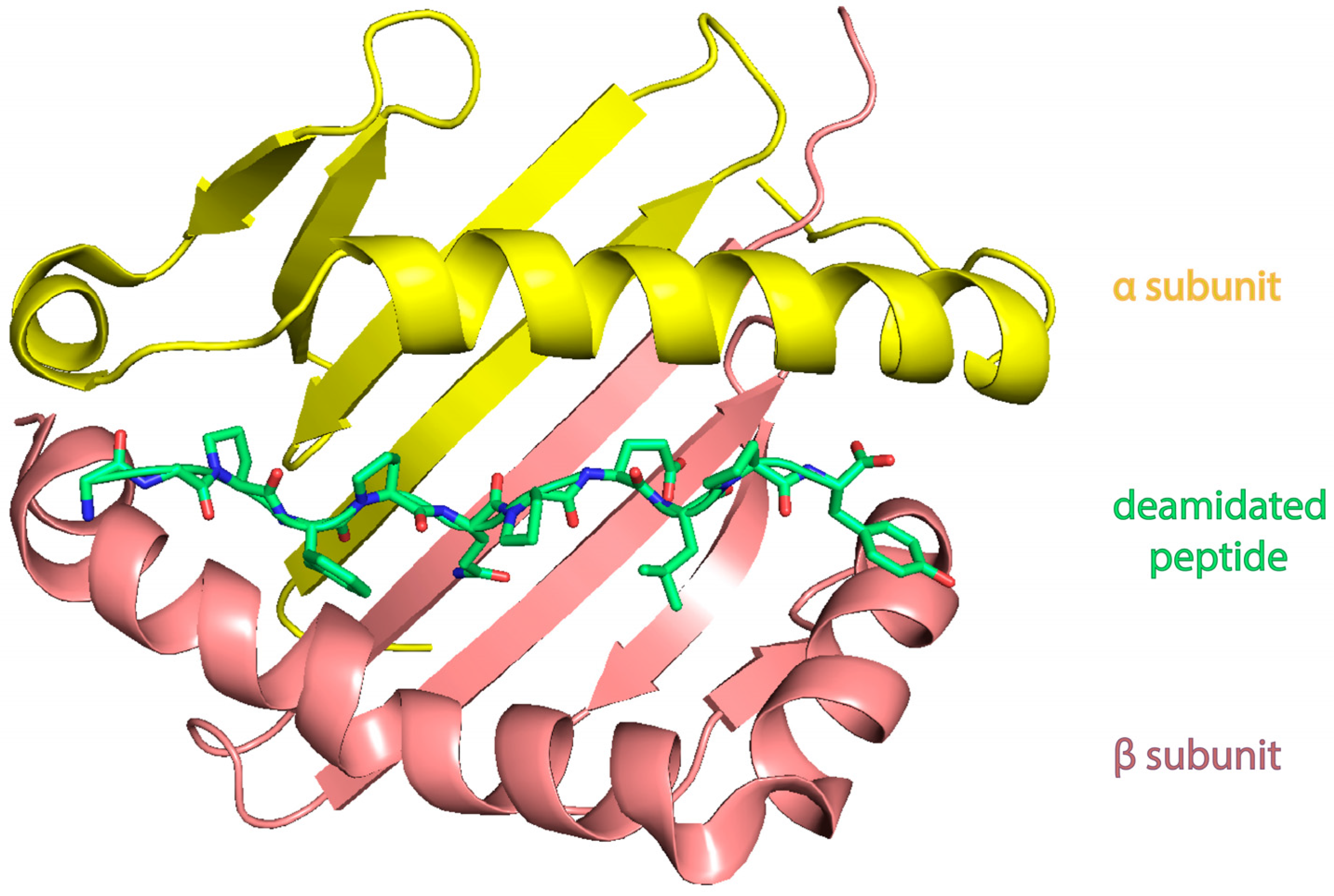
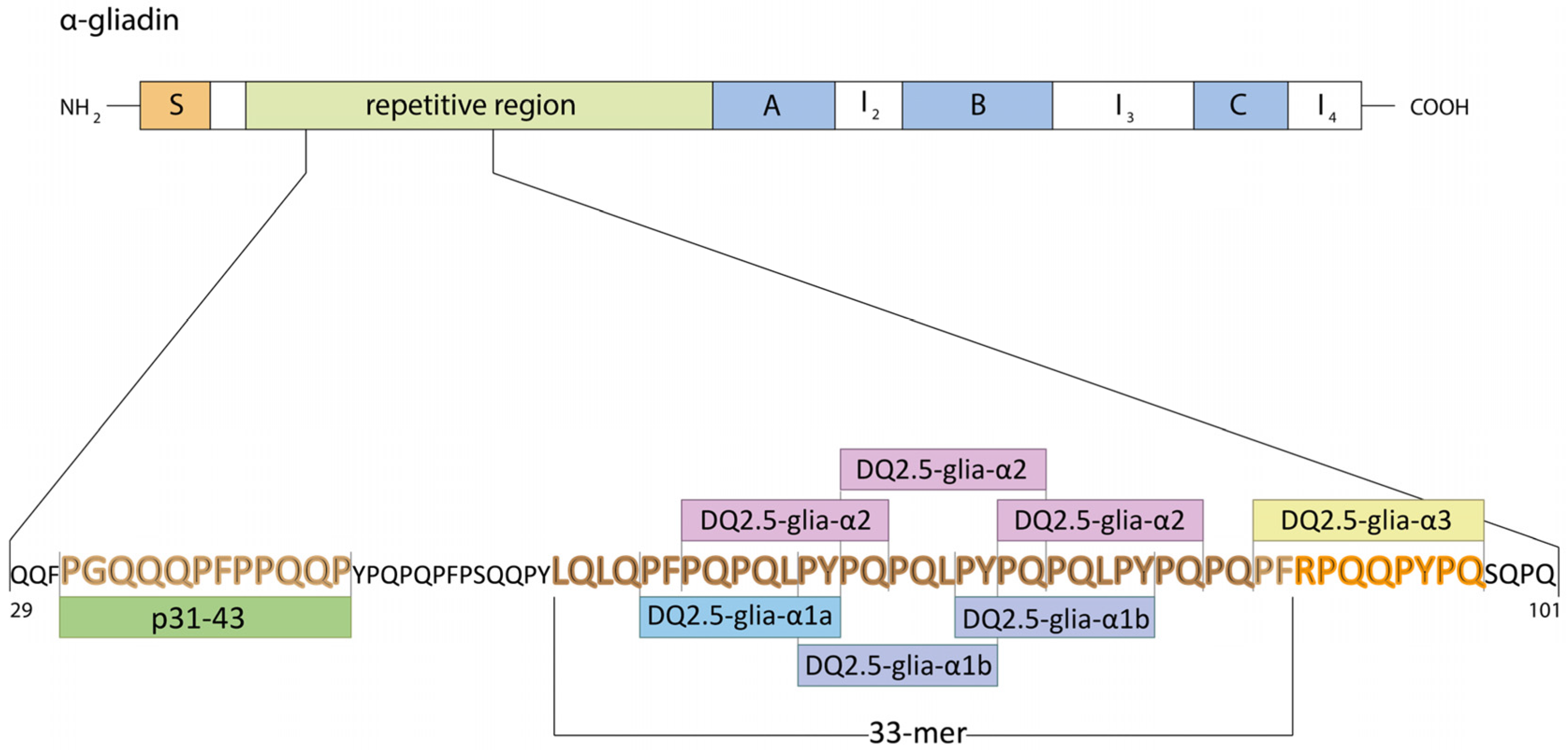
| Celiac Disease | Allergy | NCGS | |
|---|---|---|---|
| Underlying cause | Genetic: HLA-DQ2 or/and –DQ8 haplotype | Atopy (100%) | Probably, genetic: DQ2 and/or DQ8 (up to 50% of patients) |
| Laboratory markers | IgA (IgG) anti-tTG, IgA(IgG) anti-endomysium (anti-EMA), anti-deamidated gliadin peptides antibodies | Specific IgE for wheat, specific IgE for ω-5 gliadin, specific IgE for non-specific lipid transfer proteins | IgG antigliadin antibodies (in only a part of the patients) |
| Histopathological intestine symptoms | Atrophy of villi, crypt hyperplasia, increased infiltration by intraepithelial lymphocytes | Any mucosal damage or increased infiltration by intraepithelial lymphocytes or atrophy of villi and crypt hyperplasia | Any mucosal damage or increased infiltration by intraepithelial lymphocytes |
| Grain Species | Components | Molecular Weight (% Total) | Polymers or Monomers |
|---|---|---|---|
| HMW Prolamins | |||
| Wheat | HMW subunits of glutenin | 65–90 kDa (6%–10%) | Polymers |
| Barley | D-hordeins | >100 kDa (2%–4%) | Polymers |
| Rye | HMW secalins | >100 kDa (2%) | Polymers |
| S-rich prolamins | |||
| Wheat | γ-gliadins | 30–45 kDa (70%–80%) | Monomers |
| α-gliadins | Monomers | ||
| B- and C-type LMW subunits of glutenin | Polymers | ||
| Barley | B-hordeins and γ-hordeins | 32–45 kDa (80%) | Aggregated type, monomers or single chain polypeptide |
| Rye | γ-secalins | 40–75 kDa (80%) | Polymers |
| S-poor prolamins | |||
| Wheat | ω-gliadins | 30–75 kDa (10%–20%) | Monomers |
| D-type LMW subunits of glutenin | Aggregated type, polymers | ||
| Barley | C-hordeins | 40–72 kDa (10%–15%) | Monomers |
| Rye | ω-secalins | 48–55 kDa (10%–15%) | Monomers |
| Other gluten prolamins | |||
| Oat | avenins | 18.5–23.5 kDa (10%) | Monomers |
| Grain Species | Gluten Protein | Number of DQ2-Restricted Peptides Identified (Confirmed in Vitro on TCLs/TCCs or/and on PBMCs after in Vivo Challenge) | Number of DQ8-Restricted Peptides Identified (Confirmed in Vitro on TCLs/TCCs or/and on PBMCs after in Vivo Challenge) |
|---|---|---|---|
| Wheat | α-gliadins | 3 | 3 |
| γ-gliadins | 11 | 4 | |
| ω-gliadins | 3 | 4 | |
| Glutenins | 3 | 1 | |
| Barley | Hordeins | 8 | - |
| Rye | Secalins | 11 | - |
| Oat | Avenins | 6 | - |
| Protein | IgE-Binding Epitope Motifs |
|---|---|
| α/β-gliadin | QQQFPGQQ, LQQQ |
| γ-gliadin | QPQQPFPQ |
| ω5-gliadin | QQXPXQQ * |
| ω1,2-gliadin | QQPXPXQ |
| HMW-GS | QQPGQ(GQQ) |
| LMW-GS | QQPIQQQP |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balakireva, A.V.; Zamyatnin, A.A. Properties of Gluten Intolerance: Gluten Structure, Evolution, Pathogenicity and Detoxification Capabilities. Nutrients 2016, 8, 644. https://doi.org/10.3390/nu8100644
Balakireva AV, Zamyatnin AA. Properties of Gluten Intolerance: Gluten Structure, Evolution, Pathogenicity and Detoxification Capabilities. Nutrients. 2016; 8(10):644. https://doi.org/10.3390/nu8100644
Chicago/Turabian StyleBalakireva, Anastasia V., and Andrey A. Zamyatnin. 2016. "Properties of Gluten Intolerance: Gluten Structure, Evolution, Pathogenicity and Detoxification Capabilities" Nutrients 8, no. 10: 644. https://doi.org/10.3390/nu8100644
APA StyleBalakireva, A. V., & Zamyatnin, A. A. (2016). Properties of Gluten Intolerance: Gluten Structure, Evolution, Pathogenicity and Detoxification Capabilities. Nutrients, 8(10), 644. https://doi.org/10.3390/nu8100644






