Genome-Wide Association Study of Serum Selenium Concentrations
Abstract
:1. Introduction
2. Subjects and Methods
2.1. Study Population
2.2. Genotyping and Serum Selenium Measurement
2.3. Statistical Analysis
3. Results
| Variable | PLCO ( n = 582 men) | WHI ( n = 621 women) | PLCO + WHI ( n = 1203) |
|---|---|---|---|
| Age (years) | 64 (5) a | 67 (7) | 65 (6) |
| BMI (kg/m2) | 27.5 (3.9) | 27.8 (5.5) | 27.7 (4.8) |
| Ever smoker (Yes/No) | 363 (62) b | 343 (55) | 706 (59) |
| Cancer status (Yes/No) | 20 (3) | 584 (94) | 604 (50) |
| Selenium levels (mg/dL) | 142.4 (27.0) | 138.1 (21.3) | 140.2 (24.3) |
| SNP | Position a | Gene b | Coded allele | CAF c | PLCO | WHI | Combined analysis | |||
|---|---|---|---|---|---|---|---|---|---|---|
| beta | p value | beta | p value | Beta | p value | |||||
| rs1506807 | 4:178554686 | AGA | A | 0.73 | −0.059 | 8.39E−06 | −0.025 | 1.32E−0 | −0.043 | 2.63E−07 |
| rs1395479 | 4:178555185 | AGA | A | 0.27 | 0.059 | 8.31E−06 | 0.025 | 1.33E−02 | 0.043 | 2.62E−07 |
| rs891684 | 17:68225134 | SLC39A11 | A | 0.05 | −0.133 | 8.73E−06 | −0.057 | 9.99E−03 | −0.093 | 4.04E−07 |
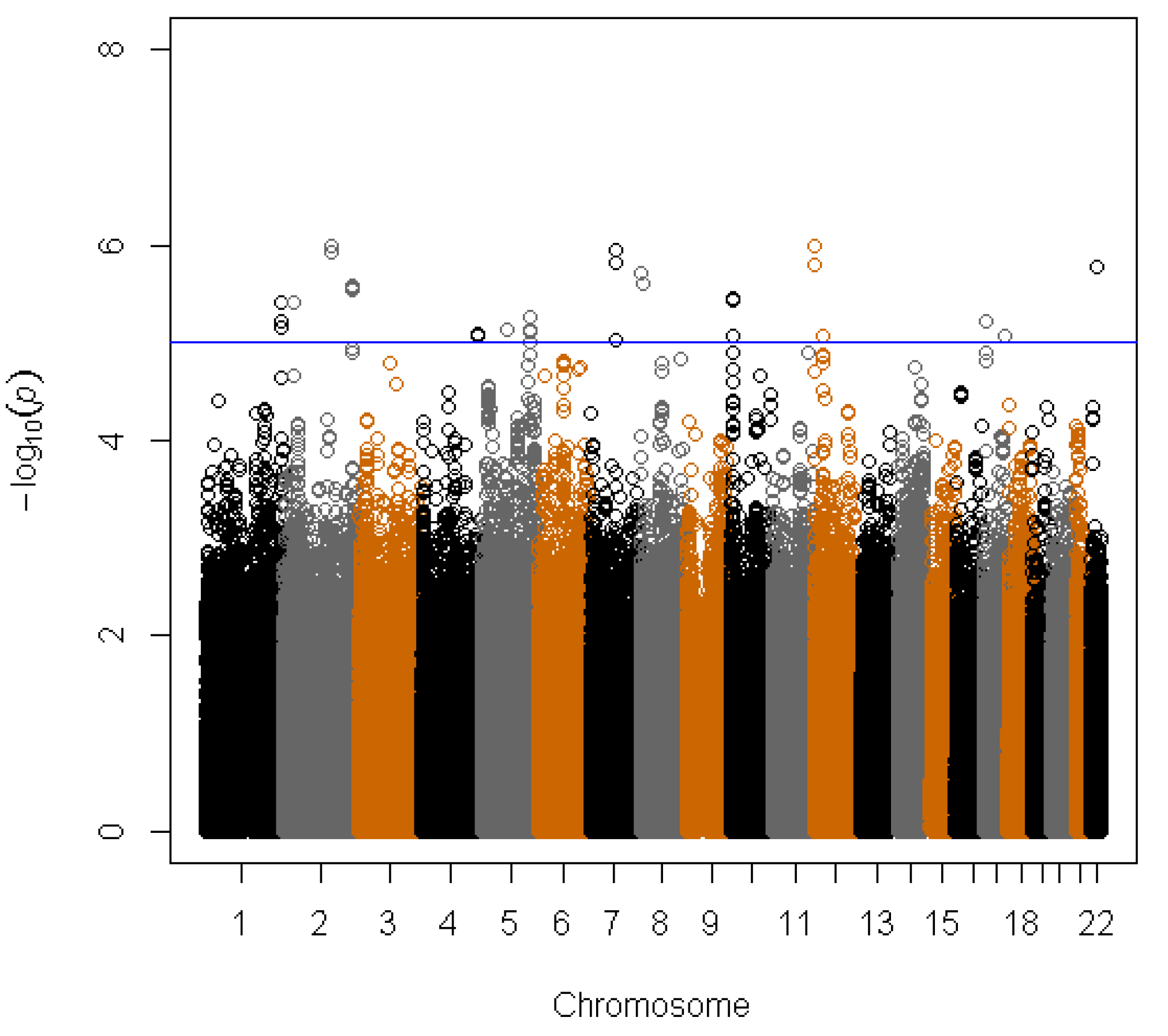
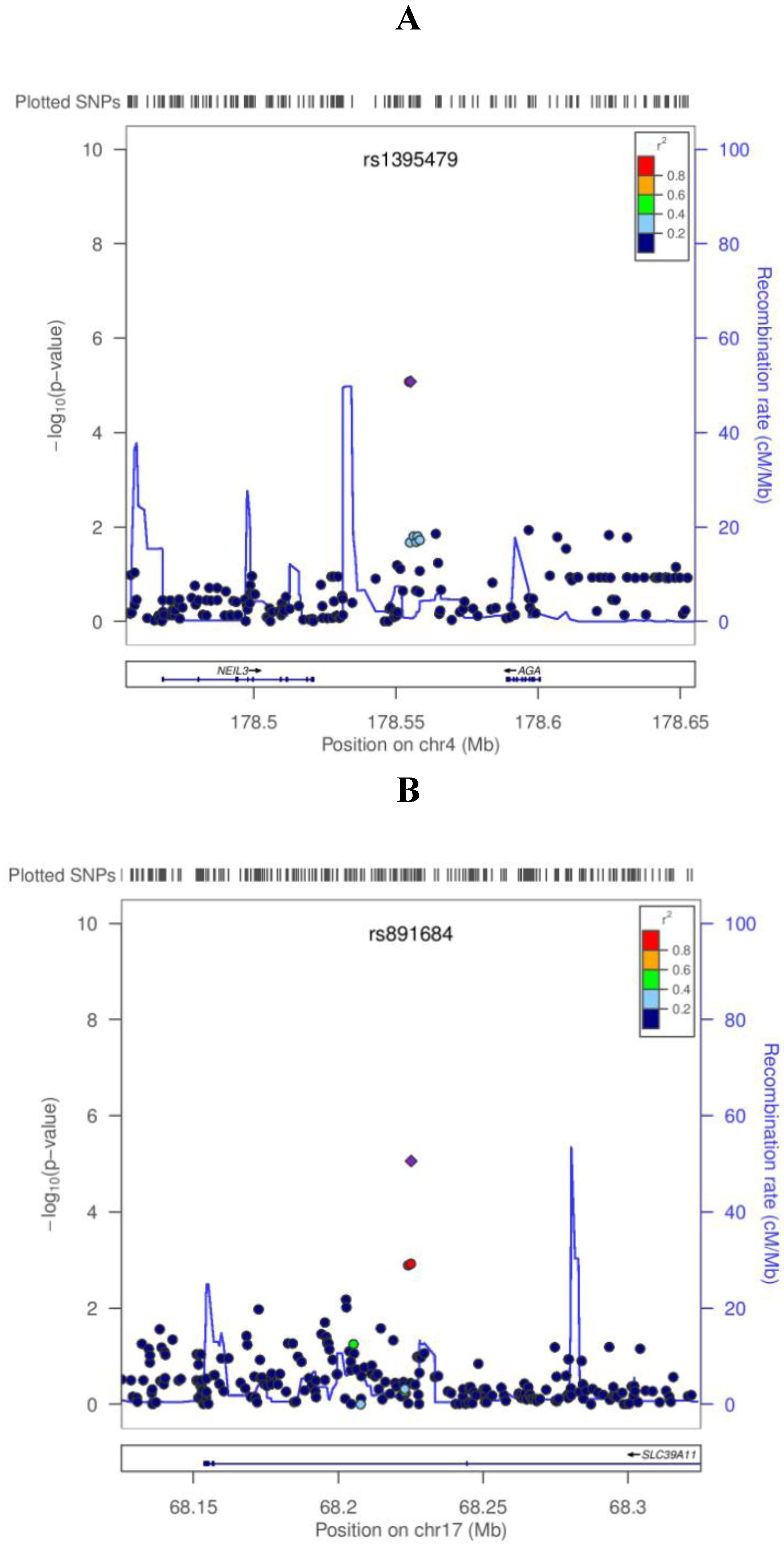
4. Discussion
5. Conclusions
Acknowledgements
Conflict of Interest
References
- Dennert, G.; Zwahlen, M.; Brinkman, M.; Vinceti, M.; Zeegers, M.P.; Horneber, M. Selenium for preventing cancer. Cochrane Database Syst. Rev. 2012, 5, CD005195. [Google Scholar]
- Flores-Mateo, G.; Navas-Acien, A.; Pastor-Barriuso, R.; Guallar, E. Selenium and coronary heart disease: A meta-analysis. Am. J. Clin. Nutr. 2006, 84, 762–773. [Google Scholar]
- Hurst, R.; Hooper, L.; Norat, T.; Lau, R.; Aune, D.; Greenwood, D.C.; Vieira, R.; Collings, R.; Harvey, L.J.; Sterne, J.A.; et al. Selenium and prostate cancer: Systematic review and meta-analysis. Am. J. Clin. Nutr. 2012, 96, 111–122. [Google Scholar] [CrossRef]
- Combs, G.F., Jr. Chemopreventive mechanisms of selenium. Med. Klin. (Munich) 1999, 94, 18–24. [Google Scholar] [CrossRef]
- Ge, K.; Yang, G. The epidemiology of selenium deficiency in the etiological study of endemic diseases in China. Am. J. Clin. Nutr. 1993, 57, 259S–263S. [Google Scholar]
- Rayman, M.P. The importance of selenium to human health. Lancet 2000, 356, 233–241. [Google Scholar] [CrossRef]
- Thomson, C.D.; Robinson, M.F. Urinary and fecal excretions and absorption of a large supplement of selenium: Superiority of selenate over selenite. Am. J. Clin. Nutr. 1986, 44, 659–663. [Google Scholar]
- Yang, G.Q.; Ge, K.Y.; Chen, J.S.; Chen, X.S. Selenium-related endemic diseases and the daily selenium requirement of humans. World Rev. Nutr. Diet. 1988, 55, 98–152. [Google Scholar]
- Taylor, E.W.; Nadimpalli, R.G.; Ramanathan, C.S. Genomic structures of viral agents in relation to the biosynthesis of selenoproteins. Biol. Trace Elem. Res. 1997, 56, 63–91. [Google Scholar] [CrossRef]
- Beck, M.A.; Shi, Q.; Morris, V.C.; Levander, O.A. Rapid genomic evolution of a non-virulent coxsackievirus B3 in selenium-deficient mice results in selection of identical virulent isolates. Nat. Med. 1995, 1, 433–436. [Google Scholar] [CrossRef]
- Arthur, J.R.; McKenzie, R.C.; Beckett, G.J. Selenium in the immune system. J. Nutr. 2003, 133, 1457S–1459S. [Google Scholar]
- Beck, M.A.; Levander, O.A.; Handy, J. Selenium deficiency and viral infection. J. Nutr. 2003, 133, 1463S–1467S. [Google Scholar]
- Brown, K.M.; Arthur, J.R. Selenium, selenoproteins and human health: A review. Public Health Nutr. 2001, 4, 593–599. [Google Scholar]
- Thomson, C.D. Assessment of requirements for selenium and adequacy of selenium status: A review. Eur. J. Clin. Nutr. 2004, 58, 391–402. [Google Scholar] [CrossRef]
- Yoon, S.O.; Yun, C.H.; Chung, A.S. Dose effect of oxidative stress on signal transduction in aging. Mech. Ageing Dev. 2002, 123, 1597–1604. [Google Scholar] [CrossRef]
- Rayman, M.P. Food-chain selenium and human health: Emphasis on intake. Br. J. Nutr. 2008, 100, 254–268. [Google Scholar]
- Davis, C.D.; Tsuji, P.A.; Milner, J.A. Selenoproteins and cancer prevention. Annu. Rev. Nutr. 2012, 32, 73–95. [Google Scholar] [CrossRef]
- Gromadzinska, J.; Reszka, E.; Bruzelius, K.; Wasowicz, W.; Akesson, B. Selenium and cancer: Biomarkers of selenium status and molecular action of selenium supplements. Eur. J. Nutr. 2008, 47, 29–50. [Google Scholar] [CrossRef]
- Arthur, J.R. The glutathione peroxidases. Cell. Mol. Life Sci. 2000, 57, 1825–1835. [Google Scholar] [CrossRef]
- Forsberg, L.; de Faire, U.; Marklund, S.L.; Andersson, P.M.; Stegmayr, B.; Morgenstern, R. Phenotype determination of a common Pro-Leu polymorphism in human glutathione peroxidase 1. Blood Cells Mol. Dis. 2000, 26, 423–426. [Google Scholar] [CrossRef]
- Zhuo, P.; Goldberg, M.; Herman, L.; Lee, B.S.; Wang, H.; Brown, R.L.; Foster, C.B.; Peters, U.; Diamond, A.M. Molecular consequences of genetic variations in the glutathione peroxidase 1 selenoenzyme. Cancer Res. 2009, 69, 8183–8190. [Google Scholar] [CrossRef]
- Takata, Y.; King, I.B.; Lampe, J.W.; Burk, R.F.; Hill, K.E.; Santella, R.M.; Kristal, A.R.; Duggan, D.J.; Vaughan, T.L.; Peters, U. Genetic variation in GPX1 is associated with GPX1 activity in a comprehensive analysis of genetic variations in selenoenzyme genes and their activity and oxidative stress in humans. J. Nutr. 2012, 142, 419–426. [Google Scholar] [CrossRef]
- Jablonska, E.; Gromadzinska, J.; Reszka, E.; Wasowicz, W.; Sobala, W.; Szeszenia-Dabrowska, N.; Boffetta, P. Association between GPx1 Pro198Leu polymorphism, GPx1 activity and plasma selenium concentration in humans. Eur. J. Nutr. 2009, 48, 383–386. [Google Scholar] [CrossRef]
- Combs, G.F., Jr.; Jackson, M.I.; Watts, J.C.; Johnson, L.K.; Zeng, H.; Idso, J.; Schomburg, L.; Hoeg, A.; Hoefig, C.S.; Chiang, E.C.; et al. Differential responses to selenomethionine supplementation by sex and genotype in healthy adults. Br. J. Nutr. 2012, 107, 1514–1525. [Google Scholar] [CrossRef]
- Moschos, M.P. Selenoprotein P. Cell. Mol. Life Sci. 2000, 57, 1836–1845. [Google Scholar] [CrossRef]
- Al-Taie, O.H.; Seufert, J.; Mork, H.; Treis, H.; Mentrup, B.; Thalheimer, A.; Starostik, P.; Abel, J.; Scheurlen, M.; Kohrle, J.; Jakob, F. A complex DNA-repeat structure within the Selenoprotein P promoter contains a functionally relevant polymorphism and is genetically unstable under conditions of mismatch repair deficiency. Eur. J. Hum. Genet. 2002, 10, 499–504. [Google Scholar] [CrossRef]
- Cooper, M.L.; Adami, H.O.; Gronberg, H.; Wiklund, F.; Green, F.R.; Rayman, M.P. Interaction between single nucleotide polymorphisms in selenoprotein P and mitochondrial superoxide dismutase determines prostate cancer risk. Cancer Res. 2008, 68, 10171–10177. [Google Scholar] [CrossRef]
- Steinbrecher, A.; Meplan, C.; Hesketh, J.; Schomburg, L.; Endermann, T.; Jansen, E.; Akesson, B.; Rohrmann, S.; Linseisen, J. Effects of selenium status and polymorphisms in selenoprotein genes on prostate cancer risk in a prospective study of European men. Cancer Epidemiol. Biomakers Prev. 2010, 19, 2958–2968. [Google Scholar] [CrossRef]
- Prorok, P.C.; Andriole, G.L.; Bresalier, R.S.; Buys, S.S.; Chia, D.; Crawford, E.D.; Fogel, R.; Gelmann, E.P.; Gilbert, F.; Hasson, M.A.; et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control. Clin. Trials 2000, 21, 273S–309S. [Google Scholar] [CrossRef]
- The Women’s Health Initiative Study Group. Design of the Women’s Health Initiative clinical trial and observational study. Control. Clin. Trials 1998, 19, 61–109. [CrossRef]
- Peters, U.; Hutter, C.M.; Hsu, L.; Schumacher, F.R.; Conti, D.V.; Carlson, C.S.; Edlund, C.K.; Haile, R.W.; Gallinger, S.; Zanke, B.W.; et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum. Genet. 2012, 131, 217–234. [Google Scholar] [CrossRef]
- Li, Y.; Willer, C.J.; Ding, J.; Scheet, P.; Abecasis, G.R. MaCH: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010, 34, 816–834. [Google Scholar] [CrossRef]
- Sturup, S.; Hayes, R.B.; Peters, U. Development and application of a simple routine method for the determination of selenium in serum by octopole reaction system ICPMS. Anal. Bioanal. Chem. 2005, 381, 686–694. [Google Scholar] [CrossRef]
- Peters, U.; Foster, C.B.; Chatterjee, N.; Schatzkin, A.; Reding, D.; Andriole, G.L.; Crawford, E.D.; Sturup, S.; Chanock, S.J.; Hayes, R.B. Serum selenium and risk of prostate cancer—A nested case-control study. Am. J. Clin. Nutr. 2007, 85, 209–217. [Google Scholar]
- Rudolph, R.E.; Vaughan, T.L.; Kristal, A.R.; Blount, P.L.; Levine, D.S.; Galipeau, P.C.; Prevo, L.J.; Sanchez, C.A.; Rabinovitch, P.S.; Reid, B.J. Serum selenium levels in relation to markers of neoplastic progression among persons with Barrett’s esophagus. J. Natl. Cancer Inst. 2003, 95, 750–757. [Google Scholar] [CrossRef]
- Goodman, G.E.; Schaffer, S.; Bankson, D.D.; Hughes, M.P.; Omenn, G.S. Carotene and Retinol Efficacy Trial Co-Investigators. Predictors of serum selenium in cigarette smokers and the lack of association with lung and prostate cancer risk. Cancer Epidemiol. Biomakers Prev. 2001, 10, 1069–1076. [Google Scholar]
- Ericson, S.P.; McHalsky, M.L.; Rabinow, B.E.; Kronholm, K.G.; Arceo, C.S.; Weltzer, J.A.; Ayd, S.W. Sampling and analysis techniques for monitoring serum for trace elements. Clin. Chem. 1986, 32, 1350–1356. [Google Scholar]
- Takata, Y.; Kristal, A.R.; King, I.B.; Song, X.; Diamond, A.M.; Foster, C.B.; Hutter, C.M.; Hsu, L.; Duggan, D.J.; Langer, R.D.; et al. Serum selenium, genetic variation in selenoenzymes, and risk of colorectal cancer: Primary analysis from the Women’s Health Initiative Observational Study and meta-analysis. Cancer Epidemiol. Biomakers Prev. 2011, 20, 1822–1830. [Google Scholar] [CrossRef]
- Pruim, R.J.; Welch, R.P.; Sanna, S.; Teslovich, T.M.; Chines, P.S.; Gliedt, T.P.; Boehnke, M.; Abecasis, G.R.; Willer, C.J. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 2010, 26, 2336–2337. [Google Scholar] [CrossRef]
- Panagiotou, O.A.; Ioannidis, J.P.; Genome-Wide Significance, P. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int. J. Epidemiol. 2012, 41, 273–286. [Google Scholar] [CrossRef]
- Newton-Cheh, C.; Guo, C.Y.; Wang, T.J.; O’Donnell, C.J.; Levy, D.; Larson, M.G. Genome-wide association study of electrocardiographic and heart rate variability traits: The Framingham Heart Study. BMC Med. Genet. 2007, 8, S7. [Google Scholar] [CrossRef]
- Aronson, N.N., Jr. Aspartylglycosaminuria: Biochemistry and molecular biology. Biochim. Biophys. Acta 1999, 1455, 139–154. [Google Scholar] [CrossRef]
- Funakoshi, Y.; Suzuki, T. Glycobiology in the cytosol: The bitter side of a sweet world. Biochim. Biophys. Acta 2009, 1790, 81–94. [Google Scholar] [CrossRef]
- Gottschalk, A. Glycoproteins: Their Composition, Structure and Function; Elsevier Pub. Co.: New York, NY, USA, 1972; p. 2. [Google Scholar]
- Ivatt, R.J. The Biology of Glycoproteins; Plenum Press: New York, NY, USA, 1984; p. 449. [Google Scholar]
- Lennarz, W.J. The Biochemistry of Glycoproteins and Proteoglycans; Plenum Press: New York, NY, USA, 1980; p. 381. [Google Scholar]
- Mathews, C.K.; van Holde, K.E.; Ahern, K.G. Biochemistry; Addison Wesley Longman: San Francisco, CA, USA, 2000. [Google Scholar]
- Montreuil, J.; Vliegenthart, J.F.G.; Schachter, H. Glycoproteins; Elsevier: Amsterdam, The Netherlands, 1995; p. 641. [Google Scholar]
- Steinbrenner, H.; Alili, L.; Stuhlmann, D.; Sies, H.; Brenneisen, P. Post-translational processing of selenoprotein P: Implications of glycosylation for its utilisation by target cells. Biol. Chem. 2007, 388, 1043–1051. [Google Scholar]
- Bandaru, V.; Sunkara, S.; Wallace, S.S.; Bond, J.P. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst.) 2002, 1, 517–529. [Google Scholar] [CrossRef]
- Landers, J.E.; Melki, J.; Meininger, V.; Glass, J.D.; van den Berg, L.H.; van Es, M.A.; Sapp, P.C.; van Vught, P.W.; McKenna-Yasek, D.M.; Blauw, H.M.; et al. Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2009, 106, 9004–9009. [Google Scholar] [CrossRef]
- Fox, C.S.; Liu, Y.; White, C.C.; Feitosa, M.; Smith, A.V.; Heard-Costa, N.; Lohman, K.; Consortium, G.; Consortium, M.; Consortium, G.; et al. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012, 8, e1002695. [Google Scholar] [CrossRef]
- Sun, J.; Purcell, L.; Gao, Z.; Isaacs, S.D.; Wiley, K.E.; Hsu, F.C.; Liu, W.; Duggan, D.; Carpten, J.D.; Gronberg, H.; et al. Association between sequence variants at 17q12 and 17q24.3 and prostate cancer risk in European and African Americans. Prostate 2008, 68, 691–697. [Google Scholar] [CrossRef]
- Prigol, M.; Bruning, C.A.; Martini, F.; Nogueira, C.W. Comparative excretion and tissue distribution of selenium in mice and rats following treatment with diphenyl diselenide. Biol. Trace Elem. Res. 2012, 150, 272–277. [Google Scholar] [CrossRef]
- Hakkak, R. Obesity decreases serum selenium levels in a mammary Tumor Zucker Rat Model. Vitam. Trace Elem. 2012, 1, 1000106. [Google Scholar] [CrossRef]
- Takata, Y.; Kristal, A.R.; Santella, R.M.; King, I.B.; Duggan, D.J.; Lampe, J.W.; Rayman, M.P.; Blount, P.L.; Reid, B.J.; Vaughan, T.L.; Peters, U. Selenium, selenoenzymes, oxidative stress and risk of neoplastic progression from Barrett’s esophagus: results from biomarkers and genetic variants. PLoS One 2012, 7, e38612. [Google Scholar]
- Hindorff, L.A.; Sethupathy, P.; Junkins, H.A.; Ramos, E.M.; Mehta, J.P.; Collins, F.S.; Manolio, T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 2009, 106, 9362–9367. [Google Scholar] [CrossRef]
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gong, J.; Hsu, L.; Harrison, T.; King, I.B.; Stürup, S.; Song, X.; Duggan, D.; Liu, Y.; Hutter, C.; Chanock, S.J.; et al. Genome-Wide Association Study of Serum Selenium Concentrations. Nutrients 2013, 5, 1706-1718. https://doi.org/10.3390/nu5051706
Gong J, Hsu L, Harrison T, King IB, Stürup S, Song X, Duggan D, Liu Y, Hutter C, Chanock SJ, et al. Genome-Wide Association Study of Serum Selenium Concentrations. Nutrients. 2013; 5(5):1706-1718. https://doi.org/10.3390/nu5051706
Chicago/Turabian StyleGong, Jian, Li Hsu, Tabitha Harrison, Irena B. King, Stefan Stürup, Xiaoling Song, David Duggan, Yan Liu, Carolyn Hutter, Stephen J. Chanock, and et al. 2013. "Genome-Wide Association Study of Serum Selenium Concentrations" Nutrients 5, no. 5: 1706-1718. https://doi.org/10.3390/nu5051706
APA StyleGong, J., Hsu, L., Harrison, T., King, I. B., Stürup, S., Song, X., Duggan, D., Liu, Y., Hutter, C., Chanock, S. J., Eaton, C. B., Marshall, J. R., & Peters, U. (2013). Genome-Wide Association Study of Serum Selenium Concentrations. Nutrients, 5(5), 1706-1718. https://doi.org/10.3390/nu5051706




