Unraveling MASLD: The Role of Gut Microbiota, Dietary Modulation, and AI-Driven Lifestyle Interventions
Abstract
1. Introduction
2. Gut Dysbiosis and Its Association with MASLD
2.1. Human Gut Microbiota
2.2. Gut Microbiota in MASLD
2.3. Conflicting Evidence in Gut Microbiota and MASLD Research
3. Microbiota-Derived Metabolites in MASLD Pathogenesis
3.1. Gut Microbiota and SCFA Metabolism
3.2. Gut Microbiota and BA Metabolism
3.3. Gut Microbiota and Choline Metabolism
3.4. Gut Microbiota and Ammonia Metabolism
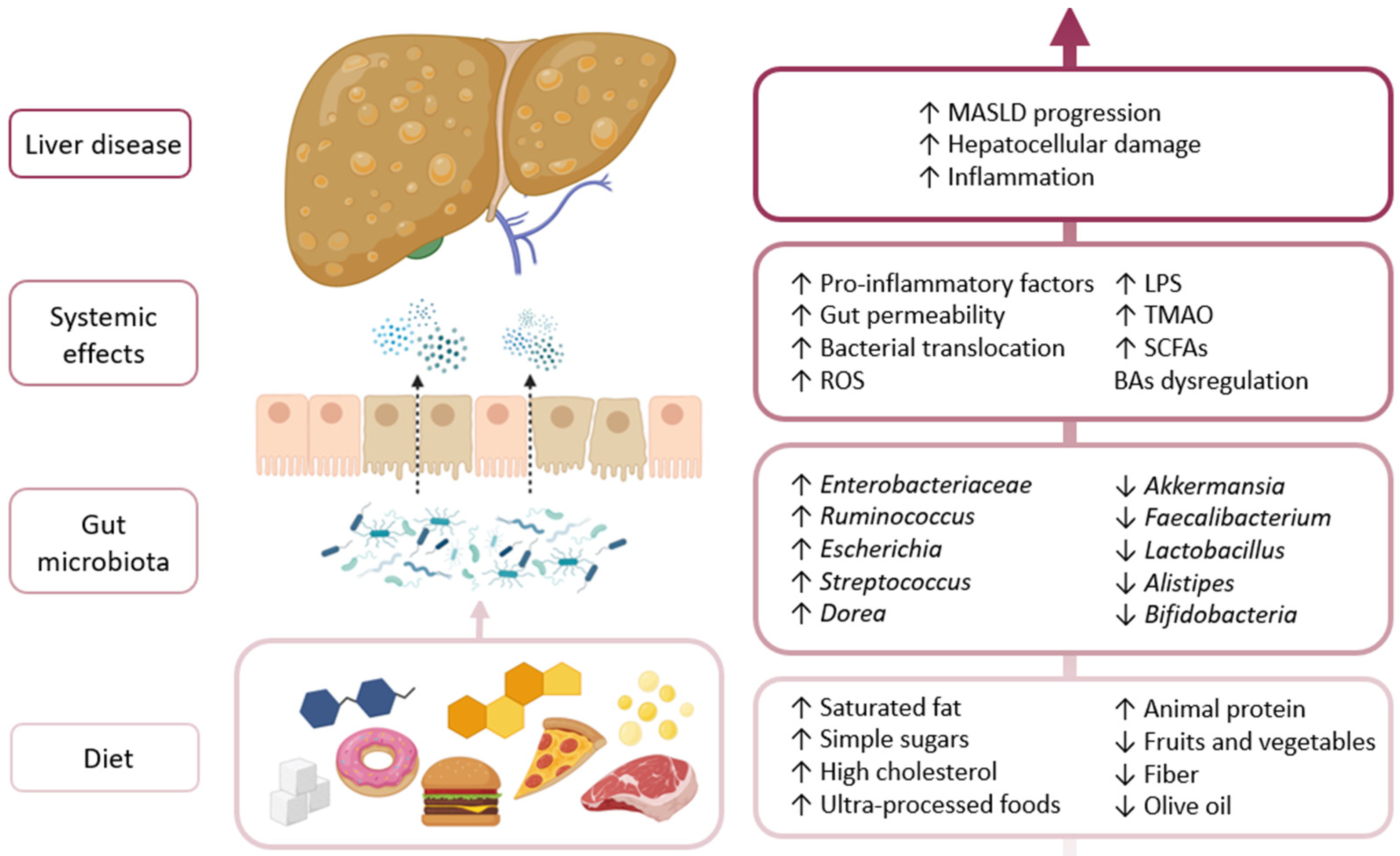
4. Influence of Dietary Patterns in MASLD
4.1. Western Diet and Dysbiosis
4.2. Mediterranean Diet and Microbiota Restoration
4.3. High-Fiber Diets and SCFA Production
4.4. Polyphenols, Flavonoids, and Gut–Liver Interactions
4.5. Other Dietary Components and Microbial Modulation
4.6. Diet–Microbiota Interactions in MASLD Progression
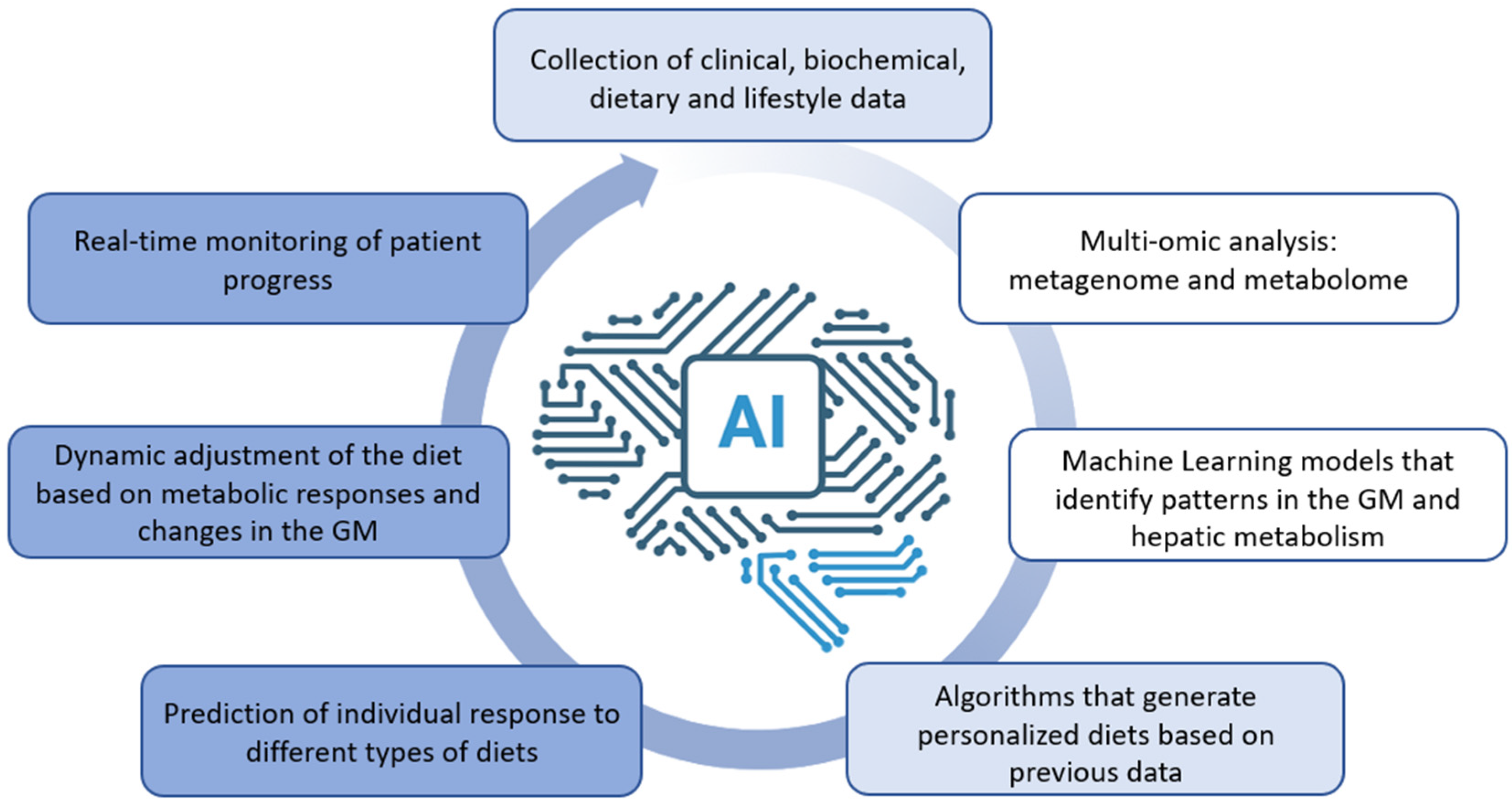
5. Artificial Intelligence in Lifestyle Interventions for MASLD
5.1. AI-Driven Dietary and Exercise Programs
5.2. Conversational Agents
5.3. Future Perspectives
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| BA | Bile acid |
| BSH | Bile salt hydrolase |
| EGCG | Epigallocatechin-3-gallate |
| FMT | Fecal microbiota transplantation |
| FXR | Farnesoid X receptor |
| GLP-1 | Glucagon-like peptide-1 |
| GM | Gut microbiota |
| GPCRs | G-protein-coupled receptors |
| HCC | Hepatocellular carcinoma |
| HSC | Hepatic stellate cells |
| HSDH | Hydroxysteroid dehydrogenases |
| LPS | Lipopolysaccharide |
| LPSN | List of prokaryotic names with standing in nomenclature |
| MASH | Metabolic dysfunction-associated steatohepatitis |
| MASLD | Metabolic-associated steatotic liver disease |
| ML | Machine learning |
| NLP | Natural language processing |
| PAMPs | Pathogen-associated molecular patterns |
| PRR | Pattern recognition receptors |
| PYY | Peptide YY |
| SCFAs | Short-chain fatty acids |
| T2DM | Type 2 diabetes mellitus |
| TLR | Toll-like receptor |
| TMA | Trimethylamine |
| TMAO | Trimethylamine N-oxide |
| UDCA | Ursodeoxycholic acid |
| VFA | Volatile fatty acids |
| VLDL | Very-low-density lipoproteins |
References
- Younossi, Z.M.; Golabi, P.; Paik, J.M.; Henry, A.; Van Dongen, C.; Henry, L. The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): A systematic review. Hepatology 2023, 77, 1335–1347. [Google Scholar] [CrossRef]
- Ekstedt, M.; Nasr, P.; Kechagias, S. Natural History of NAFLD/NASH. Curr. Hepatol. Rep. 2017, 16, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Estes, C.; Razavi, H.; Loomba, R.; Younossi, Z.; Sanyal, A.J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018, 67, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, F.; Kramer, J.R.; Mapakshi, S.; Natarajan, Y.; Chayanupatkul, M.; Richardson, P.A.; Li, L.; Desiderio, R.; Thrift, A.P.; Asch, S.M.; et al. Risk of Hepatocellular Cancer in Patients with Non-Alcoholic Fatty Liver Disease. Gastroenterology 2018, 155, 1828–1837.e2. [Google Scholar] [CrossRef]
- Harrison, S.A.; Bedossa, P.; Guy, C.D.; Schattenberg, J.M.; Loomba, R.; Taub, R.; Labriola, D.; Moussa, S.E.; Neff, G.W.; Rinella, M.E.; et al. A Phase 3, Randomized, Controlled Trial of Resmetirom in NASH with Liver Fibrosis. N. Engl. J. Med. 2024, 390, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Tacke, F.; Horn, P.; Wong, V.W.-S.; Ratziu, V.; Bugianesi, E.; Francque, S.; Zelber-Sagi, S.; Valenti, L.; Roden, M.; Schick, F.; et al. EASL–EASD–EASO Clinical Practice Guidelines on the management of metabolic dysfunction-associated steatotic liver disease (MASLD). J. Hepatol. 2024, 81, 492–542. [Google Scholar] [CrossRef]
- Promrat, K.; Kleiner, D.E.; Niemeier, H.M.; Jackvony, E.; Kearns, M.; Wands, J.R.; Fava, J.L.; Wing, R.R. Randomized controlled trial testing the effects of weight loss on nonalcoholic steatohepatitis. Hepatology 2010, 51, 121–129. [Google Scholar] [CrossRef]
- Vilar-Gomez, E.; Martinez-Perez, Y.; Calzadilla-Bertot, L.; Torres-Gonzalez, A.; Gra-Oramas, B.; Gonzalez-Fabian, L.; Friedman, S.L.; Diago, M.; Romero-Gomez, M. Weight loss through lifestyle modification significantly reduces features of nonalcoholic steatohepatitis. Gastroenterology 2015, 149, 367–378.e5. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Samuel, B.S.; Shaito, A.; Motoike, T.; Rey, F.E.; Backhed, F.; Manchester, J.K.; Hammer, R.E.; Williams, S.C.; Crowley, J.; Yanagisawa, M.; et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. USA 2008, 105, 16767–16772. [Google Scholar] [CrossRef]
- Dumas, M.-E.; Barton, R.H.; Toye, A.; Cloarec, O.; Blancher, C.; Rothwell, A.; Fearnside, J.; Tatoud, R.; Blanc, V.; Lindon, J.C.; et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. USA 2006, 103, 12511–12516. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Vallianou, N.; Christodoulatos, G.S.; Karampela, I.; Tsilingiris, D.; Magkos, F.; Stratigou, T.; Kounatidis, D.; Dalamaga, M. Understanding the role of the gut microbiome and microbial metabolites in non-alcoholic fatty liver disease: Current evidence and perspectives. Biomolecules 2022, 12, 56. [Google Scholar] [CrossRef] [PubMed]
- Bourganou, M.V.; Chondrogianni, M.E.; Kyrou, I.; Flessa, C.-M.; Chatzigeorgiou, A.; Oikonomou, E.; Lambadiari, V.; Randeva, H.S.; Kassi, E. Unraveling Metabolic Dysfunction-Associated Steatotic Liver Disease Through the Use of Omics Technologies. Int. J. Mol. Sci. 2025, 26, 1589. [Google Scholar] [CrossRef] [PubMed]
- Hamamah, S.; Iatcu, O.C.; Covasa, M. Dietary Influences on Gut Microbiota and Their Role in Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD). Nutrients 2024, 17, 143. [Google Scholar] [CrossRef]
- Singh, B.; Olds, T.; Brinsley, J.; Dumuid, D.; Virgara, R.; Matricciani, L.; Watson, A.; Szeto, K.; Eglitis, E.; Miatke, A.; et al. Systematic review and meta-analysis of the effectiveness of chatbots on lifestyle behaviours. npj Digit. Med. 2023, 6, 118. [Google Scholar] [CrossRef]
- Aggarwal, A.; Tam, C.C.; Wu, D.; Li, X.; Qiao, S. Artificial Intelligence–Based Chatbots for Promoting Health Behavioral Changes: Systematic Review. J. Med. Internet Res. 2023, 25, e40789. [Google Scholar] [CrossRef]
- Ramos, M.J.; Kendall, T.J.; Drozdov, I.; Fallowfield, J.A. A data-driven approach to decode metabolic dysfunction-associated steatotic liver disease. Ann. Hepatol. 2024, 29, 101278. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Parte, A.C. LPSN—List of prokaryotic names with standing in nomenclature (Bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef] [PubMed]
- Vallianou, N.; Liu, J.; Dalamaga, M. What are the key points in the association between the gut microbiome and nonalcoholic fatty liver disease? Metab. Open 2019, 1, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy Gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Reynoso-García, J.; Miranda-Santiago, A.E.; Meléndez-Vázquez, N.M.; Acosta-Pagán, K.; Sánchez-Rosado, M.; Díaz-Rivera, J.; Rosado-Quiñones, A.M.; Acevedo-Márquez, L.; Cruz-Roldán, L.; Tosado-Rodríguez, E.L.; et al. A complete guide to human microbiomes: Body niches, transmission, development, dysbiosis, and restoration. Front. Syst. Biol. 2022, 2, 951403. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005056. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.; DeCoffe, D.; Molcan, E.; Gibson, D.L. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients 2012, 4, 1095–1119. [Google Scholar] [CrossRef]
- Adak, A.; Khan, M.R. An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. 2019, 76, 473–493. [Google Scholar] [CrossRef]
- Vallianou, N.; Stratigou, T.; Christodoulatos, G.S.; Dalamaga, M. Understanding the Role of the Gut Microbiome and Microbial Metabolites in Obesity and Obesity-Associated Metabolic Disorders: Current Evidence and Perspectives. Curr. Obes. Rep. 2019, 8, 317–332. [Google Scholar] [CrossRef]
- Hernández-Ceballos, W.; Cordova-Gallardo, J.; Mendez-Sanchez, N. Gut microbiota in metabolic-associated fatty liver disease and in other chronic metabolic diseases. J. Clin. Transl. Hepatol. 2021, 9, 227. [Google Scholar] [CrossRef]
- Spencer, M.D.; Hamp, T.J.; Reid, R.W.; Fischer, L.M.; Zeisel, S.H.; Fodor, A.A. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 2011, 140, 976–986. [Google Scholar] [CrossRef]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.-S.; Tse, C.-H.; Lam, T.-T.Y.; Wong, G.L.-H.; Chim, A.M.-L.; Chu, W.C.-W.; Yeung, D.K.-W.; Law, P.T.-W.; Kwan, H.S.; Yu, J.; et al. Molecular Characterization of the Fecal Microbiota in Patients with Nonalcoholic Steatohepatitis–A Longitudinal Study. PLoS ONE 2013, 8, e62885. [Google Scholar] [CrossRef]
- Raman, M.; Ahmed, I.; Gillevet, P.M.; Probert, C.S.; Ratcliffe, N.M.; Smith, S.; Greenwood, R.; Sikaroodi, M.; Lam, V.; Crotty, P.; et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2013, 11, 868–875.e3. [Google Scholar] [CrossRef] [PubMed]
- Hrncir, T.; Hrncirova, L.; Kverka, M.; Hromadka, R.; Machova, V.; Trckova, E.; Kostovcikova, K.; Kralickova, P.; Krejsek, J.; Tlaskalova-Hogenova, H. Gut Microbiota and NAFLD: Pathogenetic Mechanisms, Microbiota Signatures, and Therapeutic Interventions. Microorganisms 2021, 9, 957. [Google Scholar] [CrossRef]
- Di Vincenzo, F.; Del Gaudio, A.; Petito, V.; Lopetuso, L.R.; Scaldaferri, F. Gut microbiota, intestinal permeability, and systemic inflammation: A narrative review. Intern. Emerg. Med. 2024, 19, 275–293. [Google Scholar] [CrossRef]
- Aron-Wisnewsky, J.; Vigliotti, C.; Witjes, J.; Le, P.; Holleboom, A.G.; Verheij, J.; Nieuwdorp, M.; Clément, K. Gut microbiota and human NAFLD: Disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 279–297. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Jasirwan, C.O.M.; Muradi, A.; Hasan, I.; Simadibrata, M.; Rinaldi, I. Correlation of gut Firmicutes/Bacteroidetes ratio with fibrosis and steatosis stratified by body mass index in patients with non-alcoholic fatty liver disease. Biosci. Microbiota Food Health 2020, 40, 50–58. [Google Scholar] [CrossRef]
- Boursier, J.; Mueller, O.; Barret, M.; Machado, M.; Fizanne, L.; Araujo-Perez, F.; Guy, C.D.; Seed, P.C.; Rawls, J.F.; David, L.A.; et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016, 63, 764–775. [Google Scholar] [CrossRef]
- Loomba, R.; Seguritan, V.; Li, W.; Long, T.; Klitgord, N.; Bhatt, A.; Dulai, P.S.; Caussy, C.; Bettencourt, R.; Highlander, S.K.; et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017, 25, 1054–1062.e5. [Google Scholar] [CrossRef] [PubMed]
- Nistal, E.; Saenz de Miera, L.E.; Ballesteros Pomar, M.; Sánchez-Campos, S.; García-Mediavilla, M.V.; Álvarez-Cuenllas, B.; Linares, P.; Olcoz, J.L.; Arias-Loste, M.T.; García-Lobo, J.M.; et al. An altered fecal microbiota profile in patients with non-alcoholic fatty liver disease (NAFLD) associated with obesity. Rev. Española Enferm. Dig. 2019, 111, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Mogna-Peláez, P.; Riezu-Boj, J.I.; Milagro, F.I.; Clemente-Larramendi, I.; Echeverría, S.E.; Herrero, J.I.; Elorz, M.; Benito-Boillos, A.; Tobaruela-Resola, A.L.; González-Muniesa, P.; et al. Sex-Dependent Gut Microbiota Features and Functional Signatures in Metabolic Disfunction-Associated Steatotic Liver Disease. Nutrients 2024, 16, 4198. [Google Scholar] [CrossRef] [PubMed]
- Satthawiwat, N.; Jinato, T.; Sutheeworapong, S.; Tanpowpong, N.; Chuaypen, N.; Tangkijvanich, P. Distinct Gut Microbial Signature and Host Genetic Variants in Association with Liver Fibrosis Severity in Patients with MASLD. Nutrients 2024, 16, 1800. [Google Scholar] [CrossRef]
- Hjorth, M.F.; Blædel, T.; Bendtsen, L.Q.; Lorenzen, J.K.; Holm, J.B.; Kiilerich, P.; Roager, H.M.; Kristiansen, K.; Larsen, L.H.; Astrup, A. Prevotella-to-Bacteroides ratio predicts body weight and fat loss success on 24-week diets varying in macronutrient composition and dietary fiber: Results from a post-hoc analysis. Int. J. Obes. 2019, 43, 149–157. [Google Scholar] [CrossRef]
- Oh, T.G.; Kim, S.M.; Caussy, C.; Fu, T.; Guo, J.; Bassirian, S.; Singh, S.; Madamba, E.V.; Bettencourt, R.; Richards, L.; et al. A Universal Gut-Microbiome-Derived Signature Predicts Cirrhosis. Cell Metab. 2020, 32, 878–888.e6. [Google Scholar] [CrossRef]
- Shen, F.; Zheng, R.-D.; Sun, X.-Q.; Ding, W.-J.; Wang, X.-Y.; Fan, J.-G. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat. Dis. Int. 2017, 16, 375–381. [Google Scholar] [CrossRef]
- Caussy, C.; Tripathi, A.; Humphrey, G.; Bassirian, S.; Singh, S.; Faulkner, C.; Bettencourt, R.; Rizo, E.; Richards, L.; Xu, Z.Z.; et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat. Commun. 2019, 10, 1406. [Google Scholar] [CrossRef]
- Medina, J.M.; Iruzubieta, P.; Fernández-López, R.; Crespo, J.; Cruz, F. Bacterial metabolic signatures in MASLD predicted through gene-centric studies in stool metagenomes. Marqués de Valdecilla University Hospital, Santander, Spain. 2025; submitted for publication. [Google Scholar]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Björck, I.; Bäckhed, F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.-Z.; et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 2018, 174, 1388–1405.e21. [Google Scholar] [CrossRef] [PubMed]
- Iruzubieta, P.; Medina, J.M.; Fernández-López, R.; Crespo, J.; de la Cruz, F. A role for gut microbiome fermentative pathways in fatty liver disease progression. J. Clin. Med. 2020, 9, 1369. [Google Scholar] [CrossRef]
- Parlesak, A.; Schäfer, C.; Schütz, T.; Bode, J.C.; Bode, C. Increased intestinal permeability to macromolecules and endotoxemia in patients with chronic alcohol abuse in different stages of alcohol-induced liver disease. J. Hepatol. 2000, 32, 742–747. [Google Scholar] [CrossRef]
- Son, G.; Kremer, M.; Hines, I.N. Contribution of Gut Bacteria to Liver Pathobiology. Gastroenterol. Res. Pract. 2010, 2010, 453563. [Google Scholar] [CrossRef]
- Quesada-Vázquez, S.; Bone, C.; Saha, S.; Triguero, I.; Colom-Pellicer, M.; Aragonès, G.; Hildebrand, F.; del Bas, J.M.; Caimari, A.; Beraza, N.; et al. Microbiota Dysbiosis and Gut Barrier Dysfunction Associated with Non-Alcoholic Fatty Liver Disease Are Modulated by a Specific Metabolic Cofactors’ Combination. Int. J. Mol. Sci. 2022, 23, 13675. [Google Scholar] [CrossRef]
- Compare, D.; Coccoli, P.; Rocco, A.; Nardone, O.M.; De Maria, S.; Cartenì, M.; Nardone, G. Gut–liver axis: The impact of gut microbiota on non alcoholic fatty liver disease. Nutr. Metab. Cardiovasc. Dis. 2012, 22, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Seki, E.; Schnabl, B. Role of innate immunity and the microbiota in liver fibrosis: Crosstalk between the liver and gut. J. Physiol. 2012, 590, 447–458. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Gaudier, E.; Jarry, A.; Blottière, H.M.; de Coppet, P.; Buisine, M.P.; Aubert, J.P.; Laboisse, C.; Cherbut, C.; Hoebler, C. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am. J. Physiol. Liver Physiol. 2004, 287, G1168–G1174. [Google Scholar] [CrossRef]
- Zhou, D.; Pan, Q.; Xin, F.-Z.; Zhang, R.-N.; He, C.-X.; Chen, G.-Y.; Liu, C.; Chen, Y.-W.; Fan, J.-G. Sodium butyrate attenuates high-fat diet-induced steatohepatitis in mice by improving gut microbiota and gastrointestinal barrier. World J. Gastroenterol. 2017, 23, 60–75. [Google Scholar] [CrossRef]
- Tolhurst, G.; Heffron, H.; Lam, Y.S.; Parker, H.E.; Habib, A.M.; Diakogiannaki, E.; Cameron, J.; Grosse, J.; Reimann, F.; Gribble, F.M. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 2012, 61, 364–371. [Google Scholar] [CrossRef]
- Lee, J.; Hong, S.-W.; Rhee, E.-J.; Lee, W.-Y. GLP-1 Receptor Agonist and Non-Alcoholic Fatty Liver Disease. Diabetes Metab. J. 2012, 36, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Svegliati-Baroni, G.; Saccomanno, S.; Rychlicki, C.; Agostinelli, L.; De Minicis, S.; Candelaresi, C.; Faraci, G.; Pacetti, D.; Vivarelli, M.; Nicolini, D.; et al. Glucagon-like peptide-1 receptor activation stimulates hepatic lipid oxidation and restores hepatic signalling alteration induced by a high-fat diet in nonalcoholic steatohepatitis. Liver Int. 2011, 31, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Niioka, M.; Kobayashi, N.; Tanaka, M.; Watanabe, T. Butyrate-Producing Probiotics Reduce Nonalcoholic Fatty Liver Disease Progression in Rats: New Insight into the Probiotics for the Gut-Liver Axis. PLoS ONE 2013, 8, e63388. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Loomba, R.; Hwang, S.; O’donnell, C.J.; Ellison, R.C.; Vasan, R.S.; D’agostino, R.B.; Liang, T.J.; Fox, C.S. Parental Obesity and Offspring Serum Alanine and Aspartate Aminotransferase Levels: The Framingham Heart Study. Gastroenterology 2008, 134, 953–959.e1. [Google Scholar] [CrossRef]
- Juanola, O.; Ferrusquía-Acosta, J.; García-Villalba, R.; Zapater, P.; Magaz, M.; Marín, A.; Olivas, P.; Baiges, A.; Bellot, P.; Turon, F.; et al. Circulating levels of butyrate are inversely related to portal hypertension, endotoxemia, and systemic inflammation in patients with cirrhosis. FASEB J. 2019, 33, 11595–11605. [Google Scholar] [CrossRef]
- Zhai, S.; Qin, S.; Li, L.; Zhu, L.; Zou, Z.; Wang, L. Dietary butyrate suppresses inflammation through modulating gut microbiota in high-fat diet-fed mice. FEMS Microbiol. Lett. 2019, 366, fnz153. [Google Scholar] [CrossRef]
- Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The Potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, H.; Xu, J.; Guo, X.; Zhao, H.; Chen, Y.; Zhou, Y.; Nie, Y. F. prausnitzii and its supernatant increase SCFAs-producing bacteria to restore gut dysbiosis in TNBS-induced colitis. AMB Express 2021, 11, 33. [Google Scholar] [CrossRef]
- Segers, A.; de Vos, W.M. Mode of action of Akkermansia muciniphila in the intestinal dialogue: Role of extracellular proteins, metabolites and cell envelope components. Microbiome Res. Rep. 2023, 2, 6. [Google Scholar] [CrossRef]
- Rodríguez-Daza, M.C.; de Vos, W.M. Polyphenols as Drivers of a Homeostatic Gut Microecology and Immuno-Metabolic Traits of Akkermansia muciniphila: From Mouse to Man. Int. J. Mol. Sci. 2023, 24, 45. [Google Scholar] [CrossRef] [PubMed]
- Anhê, F.F.; Pilon, G.; Roy, D.; Desjardins, Y.; Levy, E.; Marette, A. Triggering Akkermansia with dietary polyphenols: A new weapon to combat the metabolic syndrome? Gut Microbes 2016, 7, 146–153. [Google Scholar] [CrossRef]
- Roopchand, D.E.; Carmody, R.N.; Kuhn, P.; Moskal, K.; Rojas-Silva, P.; Turnbaugh, P.J.; Raskin, I. Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high-fat diet–induced metabolic syndrome. Diabetes 2015, 64, 2847–2858. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, S.; Li, T.; Li, N.; Han, D.; Zhang, B.; Xu, Z.Z.; Zhang, S.; Pang, J.; Wang, S.; et al. Gut microbiota from green tea polyphenol-dosed mice improves intestinal epithelial homeostasis and ameliorates experimental colitis. Microbiome 2021, 9, 184. [Google Scholar] [CrossRef] [PubMed]
- Mao, S. Effects of six flavonoid compounds addition on short-chain fatty acids production and human fecal microbial community change during in vitro fermentation. Afr. J. Microbiol. Res. 2011, 5, 4484–4491. [Google Scholar] [CrossRef][Green Version]
- Slijepcevic, D.; van de Graaf, S.F. Bile Acid Uptake Transporters as Targets for Therapy. Dig. Dis. 2017, 35, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Wahlström, A.; Sayin, S.I.; Marschall, H.-U.; Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.-J.; Hylemon, P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016, 7, 22–39. [Google Scholar] [CrossRef]
- Ferslew, B.C.; Xie, G.; Johnston, C.K.; Su, M.; Stewart, P.W.; Jia, W.; Brouwer, K.L.; Barritt, A.S. Altered Bile Acid Metabolome in Patients with Nonalcoholic Steatohepatitis. Dig. Dis. Sci. 2015, 60, 3318–3328. [Google Scholar] [CrossRef]
- Dawson, P.A.; Karpen, S.J. Intestinal transport and metabolism of bile acids. J. Lipid Res. 2015, 56, 1085–1099. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, S.; Biagioli, M.; Zampella, A.; Distrutti, E. Bile acids activated receptors regulate innate immunity. Front. Immunol. 2018, 9, 1853. [Google Scholar] [CrossRef] [PubMed]
- Keitel, V.; Donner, M.; Winandy, S.; Kubitz, R.; Häussinger, D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem. Biophys. Res. Commun. 2008, 372, 78–84. [Google Scholar] [CrossRef]
- Feng, S.; Xie, X.; Li, J.; Xu, X.; Chen, C.; Zou, G.; Lin, G.; Huang, T.; Hu, R.; Ran, T.; et al. Bile acids induce liver fibrosis through the NLRP3 inflammasome pathway and the mechanism of FXR inhibition of NLRP3 activation. Hepatol. Int. 2024, 18, 1040–1052. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, M.; Luo, Y.; Yang, W.; Yang, J.; Liu, J.; Zhou, J.; Xu, C.; Zhao, F.; Su, M.; et al. Ratio of Conjugated Chenodeoxycholic to Muricholic Acids is Associated with Severity of Nonalcoholic Steatohepatitis. Obesity 2019, 27, 2055–2066. [Google Scholar] [CrossRef]
- Pols, T.W.; Noriega, L.G.; Nomura, M.; Auwerx, J.; Schoonjans, K. The bile acid membrane receptor TGR5: A valuable metabolic target. Dig. Dis. 2011, 29, 37–44. [Google Scholar] [CrossRef]
- Larabi, A.B.; Masson, H.L.P.; Bäumler, A.J. Bile acids as modulators of gut microbiota composition and function. Gut Microbes 2023, 15, 2172671. [Google Scholar] [CrossRef]
- Huang, W.; Wang, G.; Xia, Y.; Xiong, Z.; Ai, L. Bile salt hydrolase-overexpressing Lactobacillus strains can improve hepatic lipid accumulation in vitro in an NAFLD cell model. Food Nutr. Res. 2020, 64. [Google Scholar] [CrossRef] [PubMed]
- Paik, D.; Yao, L.; Zhang, Y.; Bae, S.; D’agostino, G.D.; Zhang, M.; Kim, E.; Franzosa, E.A.; Avila-Pacheco, J.; Bisanz, J.E.; et al. Human gut bacteria produce ΤH17-modulating bile acid metabolites. Nature 2022, 603, 907–912. [Google Scholar] [CrossRef]
- Song, P.; Zhang, X.; Feng, W.; Xu, W.; Wu, C.; Xie, S.; Yu, S.; Fu, R. Biological synthesis of ursodeoxycholic acid. Front. Microbiol. 2023, 14, 1140662. [Google Scholar] [CrossRef]
- Li, H.; Wang, M.; Chen, P.; Zhu, M.; Chen, L. A high-dose of ursodeoxycholic acid treatment alleviates liver inflammation by remodeling gut microbiota and bile acid profile in a mouse model of non-alcoholic steatohepatitis. Biomed. Pharmacother. 2024, 174, 116617. [Google Scholar] [CrossRef] [PubMed]
- Nian, F.; Wu, L.; Xia, Q.; Tian, P.; Ding, C.; Lu, X. Akkermansia muciniphila and Bifidobacterium bifidum Prevent NAFLD by Regulating FXR Expression and Gut Microbiota. J. Clin. Transl. Hepatol. 2023, 11, 763–776. [Google Scholar] [CrossRef]
- Stephenson, K.; Kennedy, L.; Hargrove, L.; Demieville, J.; Thomson, J.; Alpini, G.; Francis, H. Updates on dietary models of nonalcoholic fatty liver disease: Current studies and insights. Gene Expr. 2018, 18, 5–17. [Google Scholar] [CrossRef]
- Velasquez, M.T.; Ramezani, A.; Manal, A.; Raj, D.S. Trimethylamine N-oxide: The good, the bad and the unknown. Toxins 2016, 8, 326. [Google Scholar] [CrossRef] [PubMed]
- Jomard, A.; Liberale, L.; Doytcheva, P.; Reiner, M.F.; Müller, D.; Visentin, M.; Bueter, M.; Lüscher, T.F.; Vettor, R.; Lutz, T.A.; et al. Effects of acute administration of trimethylamine N-oxide on endothelial function: A translational study. Sci. Rep. 2022, 12, 8664. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.W.; Hazen, S.L. Microbiome, trimethylamine N-oxide, and cardiometabolic disease. Transl. Res. 2017, 179, 108–115. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Liu, Y.; Zhou, R.-F.; Chen, X.-L.; Wang, C.; Tan, X.-Y.; Wang, L.-J.; Zheng, R.-D.; Zhang, H.-W.; Ling, W.-H.; et al. Associations of gut-flora-dependent metabolite trimethylamine-N-oxide, betaine and choline with non-alcoholic fatty liver disease in adults. Sci. Rep. 2016, 6, 19076. [Google Scholar] [CrossRef]
- León-Mimila, P.; Villamil-Ramírez, H.; Li, X.; Shih, D.; Hui, S.; Ocampo-Medina, E.; López-Contreras, B.; Morán-Ramos, S.; Olivares-Arevalo, M.; Grandini-Rosales, P.; et al. Trimethylamine N-oxide levels are associated with NASH in obese subjects with type 2 diabetes. Diabetes Metab. 2021, 47, 101183. [Google Scholar] [CrossRef]
- Zhu, W.; Gregory, J.C.; Org, E.; Buffa, J.A.; Gupta, N.; Wang, Z.; Li, L.; Fu, X.; Wu, Y.; Mehrabian, M.; et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell 2016, 165, 111–124. [Google Scholar] [CrossRef]
- Thomsen, K.L.; De Chiara, F.; Rombouts, K.; Vilstrup, H.; Andreola, F.; Mookerjee, R.P.; Jalan, R. Ammonia: A novel target for the treatment of non-alcoholic steatohepatitis. Med. Hypotheses 2018, 113, 91–97. [Google Scholar] [CrossRef]
- Richardson, A.J.; McKain, N.; Wallace, R.J. Ammonia production by human faecal bacteria, and the enumeration, isolation and characterization of bacteria capable of growth on peptides and amino acids. BMC Microbiol. 2013, 13, 6. [Google Scholar] [CrossRef] [PubMed]
- De Chiara, F.; Thomsen, K.L.; Habtesion, A.; Jones, H.; Davies, N.; Gracia-Sancho, J.; Manicardi, N.; Hall, A.; Andreola, F.; Paish, H.L.; et al. Ammonia Scavenging Prevents Progression of Fibrosis in Experimental Nonalcoholic Fatty Liver Disease. Hepatology 2020, 71, 874–892. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.; Jalan, R. Ammonia and inflammation in the pathogenesis of hepatic encephalopathy: Pandora’s box? Hepatology 2007, 46, 291–294. [Google Scholar] [CrossRef]
- Szudzik, M.; Hutsch, T.; Chabowski, D.; Zajdel, M.; Ufnal, M. Normal caloric intake with high-fat diet induces metabolic dysfunction-associated steatotic liver disease and dyslipidemia without obesity in rats. Sci. Rep. 2024, 14, 22796. [Google Scholar] [CrossRef]
- Savard, C.; Tartaglione, E.V.; Kuver, R.; Haigh, G.W.; Farrell, G.C.; Subramanian, S.; Chait, A.; Yeh, M.M.; Quinn, L.S.; Ioannou, G.N. Synergistic interaction of dietary cholesterol and dietary fat in inducing experimental steatohepatitis. Hepatology 2013, 57, 81–92. [Google Scholar] [CrossRef]
- Zhou, X.; Qiao, K.; Wu, H.; Zhang, Y. The Impact of Food Additives on the Abundance and Composition of Gut Microbiota. Molecules 2023, 28, 631. [Google Scholar] [CrossRef]
- Ross, F.C.; Patangia, D.; Grimaud, G.; Lavelle, A.; Dempsey, E.M.; Ross, R.P.; Stanton, C. The interplay between diet and the gut microbiome: Implications for health and disease. Nat. Rev. Microbiol. 2024, 22, 671–686. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Cao, H. Role of diet and nutritional management in non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 2013, 28, 81–87. [Google Scholar] [CrossRef]
- Vancells Lujan, P.; Viñas Esmel, E.; Sacanella Meseguer, E. Overview of non-alcoholic fatty liver disease (Nafld) and the role of sugary food consumption and other dietary components in its development. Nutrients 2021, 13, 1442. [Google Scholar] [CrossRef]
- Tan, J.K.; Macia, L.; Mackay, C.R. Dietary fiber and SCFAs in the regulation of mucosal immunity. J. Allergy Clin. Immunol. 2023, 151, 361–370. [Google Scholar] [CrossRef]
- Sonnenburg, E.D.; Sonnenburg, J.L. Starving our microbial self: The deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 2014, 20, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Jegatheesan, P.; De Bandt, J.P. Fructose and NAFLD: The multifaceted aspects of fructose metabolism. Nutrients 2017, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.F.; Cotter, P.D.; Healy, S.; Marques, T.M.; O’Sullivan, O.; Fouhy, F.; Clarke, S.F.; O’Toole, P.W.; Quigley, E.M.; Stanton, C.; et al. Composition and energy harvesting capacity of the gut microbiota: Relationship to diet, obesity and time in mouse models. Gut 2010, 59, 1635–1642. [Google Scholar] [CrossRef]
- Yu, J.; Sun, H.; Yang, Y.; Yan, Y. Sesamolin Alleviates Nonalcoholic Fatty Liver Disease through Modulating Gut Microbiota and Metabolites in High-Fat and High-Fructose Diet-Fed Mice. Int. J. Mol. Sci. 2022, 23, 13853. [Google Scholar] [CrossRef]
- Crawford, M.; Whisner, C.; Al-Nakkash, L.; Sweazea, K.L. Six-Week High-Fat Diet Alters the Gut Microbiome and Promotes Cecal Inflammation, Endotoxin Production, and Simple Steatosis without Obesity in Male Rats. Lipids 2019, 54, 119–131. [Google Scholar] [CrossRef]
- Le Roy, T.; Llopis, M.; Lepage, P.; Bruneau, A.; Rabot, S.; Bevilacqua, C.; Martin, P.; Philippe, C.; Walker, F.; Bado, A.; et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut 2012, 62, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, T.S.; Rampelli, S.; Jeffery, I.B.; Santoro, A.; Neto, M.; Capri, M.; Giampieri, E.; Jennings, A.; Candela, M.; Turroni, S.; et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: The NU-AGE 1-year dietary intervention across five European countries. Gut 2020, 69, 1218–1228. [Google Scholar] [CrossRef]
- Meslier, V.; Laiola, M.; Roager, H.M.; De Filippis, F.; Roume, H.; Quinquis, B.; Giacco, R.; Mennella, I.; Ferracane, R.; Pons, N.; et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 2020, 69, 1258–1268. [Google Scholar] [CrossRef]
- Bartimoccia, S.; Cammisotto, V.; Nocella, C.; Del Ben, M.; D’amico, A.; Castellani, V.; Baratta, F.; Pignatelli, P.; Loffredo, L.; Violi, F.; et al. Extra Virgin Olive Oil Reduces Gut Permeability and Metabolic Endotoxemia in Diabetic Patients. Nutrients 2022, 14, 2153. [Google Scholar] [CrossRef]
- Ma, G.; Chen, Y. Polyphenol supplementation benefits human health via gut microbiota: A systematic review via meta-analysis. J. Funct. Foods 2020, 66, 103829. [Google Scholar] [CrossRef]
- Etxeberria, U.; Fernandez-Quintela, A.; Milagro, F.I.; Aguirre, L.; Martínez, J.A.; Portillo, M.P. Impact of polyphenols and polyphenol-rich dietary sources on gut microbiota composition. J. Agric. Food Chem. 2013, 61, 9517–9533. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Nguyen, L.H.; Li, Y.; Yan, Y.; Ma, W.; Rinott, E.; Ivey, K.L.; Shai, I.; Willett, W.C.; Hu, F.B.; et al. The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk. Nat. Med. 2021, 27, 333–343. [Google Scholar] [CrossRef]
- Oliver, A.; Chase, A.B.; Weihe, C.; Orchanian, S.B.; Riedel, S.F.; Hendrickson, C.L.; Lay, M.; Sewall, J.M.; Martiny, J.B.H.; Whiteson, K. High-Fiber, Whole-Food Dietary Intervention Alters the Human Gut Microbiome but Not Fecal Short-Chain Fatty Acids. mSystems 2021, 6. [Google Scholar] [CrossRef]
- Siddiqui, M.T.; Cresci, G.A.M. The Immunomodulatory Functions of Butyrate. J. Inflamm. Res. 2021, 14, 6025–6041. [Google Scholar] [CrossRef]
- Singh, V.; Lee, G.; Son, H.; Koh, H.; Kim, E.S.; Unno, T.; Shin, J.H. Butyrate producers, “The Sentinel of Gut”: Their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics. Front. Microbiol. 2023, 13, 1103836. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, B.; Ren, L.; Du, H.; Fei, C.; Qian, C.; Li, B.; Zhang, R.; Liu, H.; Li, Z.; et al. High-fiber diet ameliorates gut microbiota, serum metabolism and emotional mood in type 2 diabetes patients. Front. Cell. Infect. Microbiol. 2023, 13, 1069954. [Google Scholar] [CrossRef]
- Chandalia, M.; Garg, A.; Lutjohann, D.; Von Bergmann, K.; Grundy, S.M.; Brinkley, L.J. Beneficial Effects of High Dietary Fiber Intake in Patients with Type 2 Diabetes Mellitus. N. Engl. J. Med. 2000, 342, 1392–1398. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.-X.; Chen, G.-R.; Xu, H.; Ge, R.-S.; Lin, J. Activation of the AMP activated protein kinase by short-chain fatty acids is the main mechanism underlying the beneficial effect of a high fiber diet on the metabolic syndrome. Med. Hypotheses 2010, 74, 123–126. [Google Scholar] [CrossRef]
- Ottman, N.; Reunanen, J.; Meijerink, M.; Pietilä, T.E.; Kainulainen, V.; Klievink, J.; Huuskonen, L.; Aalvink, S.; Skurnik, M.; Boeren, S.; et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS ONE 2017, 12, e0173004. [Google Scholar] [CrossRef]
- Cao, Y.; Liu, H.; Qin, N.; Ren, X.; Zhu, B.; Xia, X. Impact of food additives on the composition and function of gut microbiota: A review. Trends Food Sci. Technol. 2020, 99, 295–310. [Google Scholar] [CrossRef]
- Naimi, S.; Viennois, E.; Gewirtz, A.T.; Chassaing, B. Direct impact of commonly used dietary emulsifiers on human gut microbiota. Microbiome 2021, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Koren, O.; Goodrich, J.K.; Poole, A.C.; Srinivasan, S.; Ley, R.E.; Gewirtz, A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015, 519, 92–96. [Google Scholar] [CrossRef]
- Mitsou, E.K.; Kakali, A.; Antonopoulou, S.; Mountzouris, K.C.; Yannakoulia, M.; Panagiotakos, D.B.; Kyriacou, A. Adherence to the Mediterranean diet is associated with the gut microbiota pattern and gastrointestinal characteristics in an adult population. Br. J. Nutr. 2017, 117, 1645–1655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, J.; Tan, H.; Zhong, Y.; Nie, S. Akkermansia muciniphila, an important link between dietary fiber and host health. Curr. Opin. Food Sci. 2022, 47, 100905. [Google Scholar] [CrossRef]
- Volynets, V.; Louis, S.; Pretz, D.; Lang, L.; Ostaff, M.J.; Wehkamp, J.; Bischoff, S.C. Intestinal barrier function and the gut microbiome are differentially affected in mice fed a western-style diet or drinking water supplemented with fructose. J. Nutr. 2017, 147, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Coker, O.O.; Chu, E.S.; Fu, K.; Lau, H.C.H.; Wang, Y.-X.; Chan, A.W.H.; Wei, H.; Yang, X.; Sung, J.J.Y.; et al. Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut 2020, 70, 761–774. [Google Scholar] [CrossRef]
- Romualdo, G.R.; Valente, L.C.; Sprocatti, A.C.; Bacil, G.P.; de Souza, I.P.; Rodrigues, J.; Rodrigues, M.A.M.; Vinken, M.; Cogliati, B.; Barbisan, L.F. Western diet–induced mouse model of non-alcoholic fatty liver disease associated with metabolic outcomes: Features of gut microbiome-liver-adipose tissue axis. Nutrition 2022, 103–104, 111836. [Google Scholar] [CrossRef]
- Nagpal, R.; Shively, C.A.; Register, T.C.; Craft, S.; Yadav, H. Gut microbiome-Mediterranean diet interactions in improving host health. F1000Research 2019, 8, 699. [Google Scholar] [CrossRef]
- Chiu, C.-C.; Ching, Y.-H.; Li, Y.-P.; Liu, J.-Y.; Huang, Y.-T.; Huang, Y.-W.; Yang, S.-S.; Huang, W.-C.; Chuang, H.-L. Nonalcoholic fatty liver disease is exacerbated in high-fat diet-fed gnotobiotic mice by colonization with the gut microbiota from patients with nonalcoholic steatohepatitis. Nutrients 2017, 9, 1220. [Google Scholar] [CrossRef]
- Matchado, M.S.; Rühlemann, M.; Reitmeier, S.; Kacprowski, T.; Frost, F.; Haller, D.; Baumbach, J.; List, M. On the limits of 16S rRNA gene-based metagenome prediction and functional profiling. Microb. Genom. 2024, 10, 001203. [Google Scholar] [CrossRef]
- Naska, A.; Lagiou, A.; Lagiou, P. Dietary assessment methods in epidemiological research: Current state of the art and future prospects. F1000Research 2017, 6, 926. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, X.; Xu, J.; Yuan, J.; Cai, W.; Chen, T.; Wang, K.; Gao, Y.; Nie, S.; Xu, X.; et al. Deep-learning models for the detection and incidence prediction of chronic kidney disease and type 2 diabetes from retinal fundus images. Nat. Biomed. Eng. 2021, 5, 533–545. [Google Scholar] [CrossRef]
- Colmenarejo, G. Machine learning models to predict childhood and adolescent obesity: A review. Nutrients 2020, 12, 2466. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lim, D.H.; Kim, Y. Classification and prediction on the effects of nutritional intake on overweight/obesity, dyslipidemia, hypertension and type 2 diabetes mellitus using deep learning model: 4–7th Korea national health and nutrition examination survey. Int. J. Environ. Res. Public Health 2021, 18, 5597. [Google Scholar] [CrossRef]
- Moshawrab, M.; Adda, M.; Bouzouane, A.; Ibrahim, H.; Raad, A. Cardiovascular Events Prediction using Artificial Intelligence Models and Heart Rate Variability. Procedia Comput. Sci. 2022, 203, 231–238. [Google Scholar] [CrossRef]
- Park, I.-G.; Yoon, S.J.; Won, S.-M.; Oh, K.-K.; Hyun, J.Y.; Suk, K.T.; Lee, U. Gut microbiota-based machine-learning signature for the diagnosis of alcohol-associated and metabolic dysfunction-associated steatotic liver disease. Sci. Rep. 2024, 14, 16122. [Google Scholar] [CrossRef] [PubMed]
- McTeer, M.; Applegate, D.; Mesenbrink, P.; Ratziu, V.; Schattenberg, J.M.; Bugianesi, E.; Geier, A.; Gomez, M.R.; Dufour, J.-F.; Ekstedt, M.; et al. Machine learning approaches to enhance diagnosis and staging of patients with MASLD using routinely available clinical information. PLoS ONE 2024, 19, e0299487. [Google Scholar] [CrossRef] [PubMed]
- Côté, M.; Lamarche, B. Artificial intelligence in nutrition research: Perspectives on current and future applications. Appl. Physiol. Nutr. Metab. 2022, 47, 1–8. [Google Scholar] [CrossRef]
- Kassem, H.; Beevi, A.A.; Basheer, S.; Lutfi, G.; Ismail, L.C.; Papandreou, D. Investigation and Assessment of AI’s Role in Nutrition—An Updated Narrative Review of the Evidence. Nutrients 2025, 17, 190. [Google Scholar] [CrossRef]
- Ahn, J.C.; Connell, A.; Simonetto, D.A.; Hughes, C.; Shah, V.H. Application of Artificial Intelligence for the Diagnosis and Treatment of Liver Diseases. Hepatology 2021, 73, 2546–2563. [Google Scholar] [CrossRef]
- Shea, B.; Bakre, S.; Carano, K.; Scharen, J.; Langheier, J.; A Hu, E. Changes in glycemic control among individuals with diabetes who used a personalized digital nutrition platform: Longitudinal study. JMIR Diabetes 2021, 6, e32298. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, B.J.; Gutierrez-Osuna, R. A Review of Digital Innovations for Diet Monitoring and Precision Nutrition. J. Diabetes Sci. Technol. 2023, 17, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Kim, G.; Hwang, Y.-C.; Lee, W.J.; Park, H.; Kim, J.H. Validation of the effectiveness of a digital integrated healthcare platform utilizing an AI-based dietary management solution and a real-time continuous glucose monitoring system for diabetes management: A randomized controlled trial. BMC Med. Inform. Decis. Mak. 2020, 20, 156. [Google Scholar] [CrossRef]
- A Hu, E.; Scharen, J.; Nguyen, V.; Langheier, J. Evaluating the impact of a digital nutrition platform on cholesterol levels in users with dyslipidemia: Longitudinal study. JMIR Cardio 2021, 5, e28392. [Google Scholar] [CrossRef]
- Knights, V.; Kolak, M.; Markovikj, G.; Kljusurić, J.G. Modeling and Optimization with Artificial Intelligence in Nutrition. Appl. Sci. 2023, 13, 7835. [Google Scholar] [CrossRef]
- Saeed, H.; Díaz, L.A.; Gil-Gómez, A.; Burton, J.; Bajaj, J.S.; Romero-Gomez, M.; Arrese, M.; Arab, J.P.; Khan, M.Q. Microbiome-centered therapies for the management of metabolic dysfunction-associated steatotic liver disease. Clin. Mol. Hepatol. 2025, 31, S94–S111. [Google Scholar] [CrossRef]
- Oh, Y.J.; Zhang, J.; Fang, M.-L.; Fukuoka, Y. A systematic review of artificial intelligence chatbots for promoting physical activity, healthy diet, and weight loss. Int. J. Behav. Nutr. Phys. Act. 2021, 18, 160. [Google Scholar] [CrossRef]
- Armand, T.P.T.; Nfor, K.A.; Kim, J.-I.; Kim, H.-C. Applications of Artificial Intelligence, Machine Learning, and Deep Learning in Nutrition: A Systematic Review. Nutrients 2024, 16, 1073. [Google Scholar] [CrossRef]
- Maher, C.A.; Davis, C.R.; Curtis, R.G.; Short, C.E.; Murphy, K.J. A physical activity and diet program delivered by artificially intelligent virtual health coach: Proof-of-concept study. JMIR mHealth uHealth 2020, 8, e17558. [Google Scholar] [CrossRef]
- Vasiloglou, M.F.; Lu, Y.; Stathopoulou, T.; Papathanail, I.; Faeh, D.; Ghosh, A.; Baumann, M.; Mougiakakou, S. Assessing mediterranean diet adherence with the smartphone: The medipiatto project. Nutrients 2020, 12, 3763. [Google Scholar] [CrossRef]
- Zhang, X.; Chan, F.T.; Yan, C.; Bose, I. Towards risk-aware artificial intelligence and machine learning systems: An overview. Decis. Support Syst. 2022, 159, 113800. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-González, C.; Alonso-Peña, M.; Argos Vélez, P.; Crespo, J.; Iruzubieta, P. Unraveling MASLD: The Role of Gut Microbiota, Dietary Modulation, and AI-Driven Lifestyle Interventions. Nutrients 2025, 17, 1580. https://doi.org/10.3390/nu17091580
Jiménez-González C, Alonso-Peña M, Argos Vélez P, Crespo J, Iruzubieta P. Unraveling MASLD: The Role of Gut Microbiota, Dietary Modulation, and AI-Driven Lifestyle Interventions. Nutrients. 2025; 17(9):1580. https://doi.org/10.3390/nu17091580
Chicago/Turabian StyleJiménez-González, Carolina, Marta Alonso-Peña, Paula Argos Vélez, Javier Crespo, and Paula Iruzubieta. 2025. "Unraveling MASLD: The Role of Gut Microbiota, Dietary Modulation, and AI-Driven Lifestyle Interventions" Nutrients 17, no. 9: 1580. https://doi.org/10.3390/nu17091580
APA StyleJiménez-González, C., Alonso-Peña, M., Argos Vélez, P., Crespo, J., & Iruzubieta, P. (2025). Unraveling MASLD: The Role of Gut Microbiota, Dietary Modulation, and AI-Driven Lifestyle Interventions. Nutrients, 17(9), 1580. https://doi.org/10.3390/nu17091580




