Abstract
Obesity (OB) has become a serious health issue owing to its ever-increasing prevalence over the past few decades due to its contribution to severe metabolic and inflammatory disorders such as cardiovascular disease, type 2 diabetes, and cancer. The unbalanced energy metabolism in OB is associated with substantial epigenetic changes mediated by the gut microbiome (GM) structure and composition alterations. Remarkably, experimental evidence also indicates that OB-induced epigenetic modifications in adipocytes can lead to cellular “memory” alterations, predisposing individuals to weight regain after caloric restriction and subsequently inducing inflammatory pathways in the liver. Various environmental factors, especially diet, play key roles in the progression or prevention of OB and OB-related disorders by modulating the GM structure and composition and affecting epigenetic mechanisms. Here, we will first focus on the key role of epigenetic aberrations in the development of OB. Then, we discuss the association between abnormal alterations in the composition of the microbiome and OB and the interplays between the microbiome and the epigenome in the development of OB. Finally, we review promising strategies, including prebiotics, probiotics, a methyl-rich diet, polyphenols, and herbal foods for the prevention and/or treatment of OB via modulating the GM and their metabolites influencing the epigenome.
1. Introduction
Obesity (OB) is known as a multifactorial trait characterized by an abnormal accumulation of fat mass and a high body mass index (BMI) [1,2]. One of the main public health problems around the world is the ever-increasing prevalence of OB due to altered lifestyles, regular use of high-calorie foods, and insufficient physical activities, the core causative factors of metabolic disorders [3,4]. Excessive weight also increases the risk for the development of some serious disorders like cancer, nonalcoholic fatty liver disease (NAFLD), type 2 diabetes (T2DM), and cardiovascular diseases [5,6,7]. Additionally, the adipose tissues in obese subjects comprises an elevated amount of classically activated M1 macrophages and reduced amounts of activated M2 macrophages, which produce great amounts of pro-inflammatory cytokines, like tumor necrosis factor alpha (TNF-α), interleukin-1 beta (IL-1β), and interleukin 6 (IL-6), which in turn give rise to the initiation of systemic inflammation and insulin resistance [8,9]. OB-induced inflammation is linked to epigenetic aberrations in adipose tissue. For example, DNA hypermethylation at the peroxisome proliferator-activated receptor γ (PPARγ) 1 promoter induced by OB-dependent factors is a key regulator in the activation of adipose tissue macrophages, leading to pro-inflammatory phenotype, triggering inflammation, and insulin resistance in OB [10]. Furthermore, it has been shown that suppressing M2 macrophage polarization in OB-induced inflammation occurred by adipocyte-derived exosomal microRNA (miRNA)-1224 through Musashi RNA binding protein 2 (MSI2)-mediated Wnt/β-Catenin axis [11]. Remarkably, experimental evidence also indicates that OB-induced epigenetic modifications in adipocytes can be registered in the cellular “memory”, predisposing individuals to weight regain after caloric restriction and subsequently inducing the liver inflammatory pathways [12].
In addition to epigenetic abnormalities, aberrations in the composition and diversity of body microbial communities due to environmental factors and diet can contribute to the development of OB and related metabolic diseases [13]. For example, obese children exhibited an elevated abundance of Proteobacteria at the phylum level and decreased abundance of Oscillibacter and Alistipes at the genus level with corresponding reduction in the concentrations of butyrate and isobutyrate in their fecal samples [14]. Moreover, subjects with T2DM and OB exhibit alterations in the structure of microbial community of the saliva, like reduction in the bacterial richness and diversity that is linked to the consumption of specific foods [15].
The human gastrointestinal (GI) tract is one of the biggest organs in the body, with a total surface area of about 250–400 m2, which is the habitat of an intricate and dynamic population of microorganisms involved in the homeostasis or pathogenesis of many illnesses [16,17]. The total number of microorganisms in the human GI tract is about 1014 and their collective genomic content is almost 100 times more than the human genome [18]. High-throughput sequencing technologies have demonstrated remarkable reductions in the alpha diversity and changes in the composition at the phylum or genus level of the gut microbiome (GM) in OB [19]. NAFLD, one of the most common consequences of OB is also associated with significant GM and epigenetic alterations [20,21]. In this line, recent studies have elucidated the complex interactions between OB, epigenetic alterations, and GM in NAFLD development [22]. Moreover, gut dysbiosis can enhance intestinal permeability, allowing the translocation of harmful substances to the liver, thus exacerbating liver inflammation and fibrosis [23]. These interconnected mechanisms provide valuable insights into potential therapeutic strategies, including targeting epigenetic modifications, modulating GM, and developing personalized dietary interventions to mitigate OB and the progression of NAFLD and its associated metabolic complications.
In this review, we briefly discuss the potential role of epigenetic aberrations in the onset and development of OB. Then, we review the correlation between abnormal changes in the composition of GM and OB and the interactions between the GM and the epigenome in the development of OB. At the end, we discuss how promising strategies, such as dietary modifications and administration of probiotics, prebiotics, and polyphenols can contribute to the prevention and/or treatment of OB through modulating the microbiota and their epigenetic metabolites. Figure 1 provides an overview of the interconnected topics to be explored in this work.
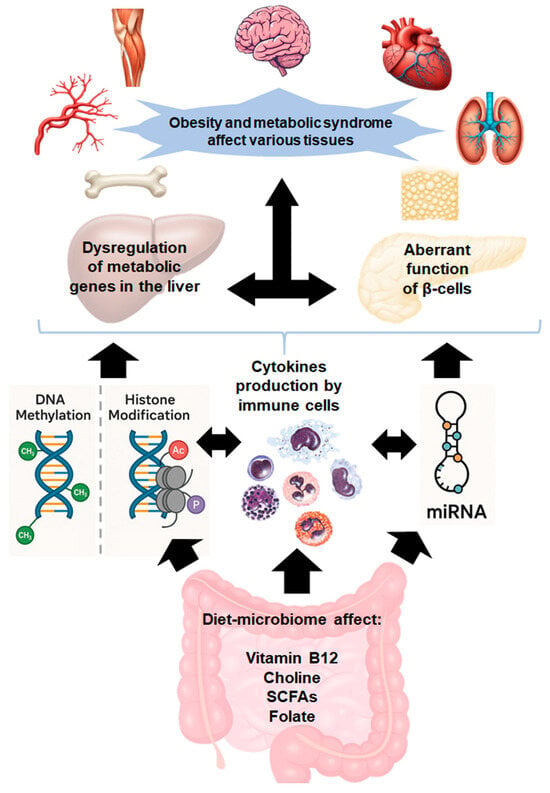
Figure 1.
Mechanistic cascade linking gut dysbiosis to obesity and metabolic syndrome.
The gut microbiota metabolizes dietary components, producing bioactive compounds such as folic acid, vitamin B12, and short-chain fatty acids (SCFAs), which influence the epigenome and gene expression. Gut dysbiosis not only disrupts the synthesis of these bioactive compounds but also triggers inflammation, compromising intestinal barrier integrity. This allows harmful metabolites to translocate into the bloodstream, activating immune cells and initiating a cytokine surge. The resulting inflammatory response disrupts metabolic gene regulation in the liver, pancreas, and other tissues, contributing to obesity and metabolic syndrome.
2. Epigenetics and Development of Obesity
Epigenetic mechanisms are involved in genes expression fine-tuning without changing the nucleotide sequences [24]. Such mechanisms are capable of regulating the genes activities at the transcriptional and post-transcriptional levels and/or at the translation and post-translational levels, which in turn lead to more diversity in morphogenesis, cell differentiation, stress responses, and adaptability of an organism to the everchanging environmental conditions [25,26,27]. Similarly to genetic codes, epigenetic marks are heritable but flexible and can guarantee the precise transmission of tissue-specific chromatin states and gene expression profiles across numerous cell generations [28]. The main epigenetic mechanisms discussed in this work, include DNA methylation, histone modifications, and RNA interference which is mediated by non-coding RNAs [29]. DNA methylation (DNAmet) involves the transfer of a methyl group onto the C5 position of the cytosine by the mediation of DNMTs (DNA methylal transferase enzymes) to form 5-methylcytosine (5-mC), which further can be modified to 5-hydroxymethylcytosine (5-hmC) by TET enzymes (Ten-Eleven Translocation methylcytosine dioxygeneases) during demethylation reaction. DNAmet contributes to modulation of gene expression by turning genes “on” or “off” in an age- and tissue-related fashion largely through recruitment of DNA binding proteins and suppressing transcription factor binding to the corresponding DNA. As a consequence of a dynamic process involved in both de novo DNAmet and demethylation, methylation patterns are changeable during development, while differentiated cells possess a unique DNAmet pattern capable of modulating tissue-specific gene transcription [30]. Likewise, acetylation/deacetylation or methylation/demethylation of the amino acids of diverse histone proteins tails are versatile post-translational modifications that affect genes expression levels. In this process, the addition or removal of acetyl groups is performed by lysine acetyltransferases and lysine deacetylases, respectively, where acetylation induces but deacetylation suppresses gene expression. Histone methylation can induce or suppress gene expression depend on the amino acid position [31,32].
Non-coding RNAs (ncRNAs) are a heterogeneous class of transcripts, which are not translated into proteins. However, ncRNAs, in particular miRNAs, are involved in a myriad of cellular processes such as RNA maturation and processing, metabolism, signaling, cell proliferation and differentiation, gene expression, and protein synthesis [33,34]. In the following section, we briefly discussed the relationship between epigenetics and the development of OB.
2.1. Altered DNAmet in Obesity
OB is connected to derangements in transmethylation and one-carbon metabolism in various body organs. Multiple DNAmet sites play key roles in increasing the risk of OB. Hypomethylation at the cg21178254 site upstream of CCNL1 elevates the risk of OB through escalating the expression of this gene [35]. Hypermethylation at cg02814054 contributes to OB via reducing the MAST3 expression, while hypomethylation at cg06028605 elevates the risk of OB by reducing the SLC5A11 expression [35]. Likewise, rare variants within the 2p23.3 genome site influence OB by increasing susceptibility of the cg01884057 site to methylation, which in turn decreases the expression of POMC, ADCY3, and DNAJC27 [35].
Consumption of an obesogenic diet can contribute to the onset and development of OB and other related disorders by causing abnormalities in DNAmet [36,37]. For example, Vander Velden et al. found that consumption of an obesogenic diet could cause differentially methylated regions (DMRs) in the hippocampus of male mice, which in turn altered the expression of some genes, including BIN1 and histone deacetylase 5 (HDAC5) [38]. Wu et al. reported that consuming a high-fat diet (HFD) by male mice caused disturbance in the gluconeogenesis of offspring by changing Igf2/H19 DNAmet in the liver [39]. HFD-induced OB in mice led to the induction of the expression of DNA methyltransferase-1 (Dnmt1) and Dnmt3a enzymes, elevating DNAmet in the teste, but reducing global DNAmet in the ovaries [40]. Another study by the same group also showed increase in the total expression of Dnmt1, Dnmt3a, and Dnmt3b and a decrease in Dnmt3L and global DNAmet level in the uterus of mice with HFD-induced OB [41]. Additional studies on the association between the consumption of obesogenic diet and abnormalities in DNAmet, and OB in humans are summarized in Table 1.

Table 1.
DNA methylation alterations associated with obesity (OB).
2.2. Histone Modifications in Obesity
In addition to DNAmet, the development of OB is linked to changes in histone modifications. For example, Guo et al. found a total of 281 differentially expressed proteins and 147 differentially modified proteomic and lysine acetylation sites in subjects with metabolically unhealthy OB [57]. Elevated acetylation in visceral adipose tissue of subjects with metabolically unhealthy OB was characterized by specific differentially acetylated sites, such as H3.7K80, H1.2K63, and H1XK90 on histone proteins [57]. As another example, Wu et al. reported that the monocytes of individuals with OB showed elevated levels of mitochondrial acetyl-CoA acetyltransferase 1 and histone acetylation [58]. Alawathugoda et al. recently examined the impact of maternal OB on the genes expression levels and cell fate during embryonic cortical neurogenesis and found that HFD could change EZH2 expression/phosphorylation and reduce H3K27me3 level and associated transcriptional derepression of EZH2 target genes, which in turn disrupted embryonic neurogenesis in rats [59]. More studies on histone modifications in OB are summarized in Table 2.

Table 2.
Histone modifications alterations related to obesity (OB).
2.3. Altered miRNAs in Obesity
The adipose tissue is one of the core sources of circulating miRNAs, which are involved in adipogenesis, lipid metabolism, and homeostasis [67]. Therefore, the adipose-derived miRNAs levels are considered worthy biomarkers for forecasting the development of OB in adulthood, and for identifying the degree of adipose tissue inflammation [68,69]. In this line, Ahmad Mir et al. identified differences in the expression of a total of 64 miRNAs between subjects with metabolically healthy OB in comparison with metabolically unhealthy OB in the blood samples. In their study, miR-130b-5p, miR-363-3p, and miR-636 were connected to cholesterol level, while miR-130a-3p was linked to low-density lipoprotein (LDL) level [70]. Other examples of altered expression of miRNAs in OB are summarized in Table 3.

Table 3.
miRNA alterations related to obesity (OB).
While various epigenetic alterations are associated with OB, the epigenome itself is modulated by multiple internal and external factors, like endocrine disruptors, nutritional factors, pharmaceuticals, and inorganic chemicals [77]. The relationship between GM and OB will be addressed after first elucidating the links between GM and OB.
3. Gut Microbiota and Obesity
In addition to genetic factors and lifestyle, changes in the GM and oral microbiome are also involved in the development of OB [78]. For example, the prevalence of OB among 647 obese individuals (compared to 969 non-obese subjects) from African American populations was associated with five bacterial taxa in Firmicutes and two bacterial taxa in Actinobacteria and Proteobacteria [79]. Gut dysbiosis, characterized by an increased abundance of Firmicutes and an altered Firmicutes/Bacteroidetes ratio, has been implicated in metabolic dysfunctions affecting overall systemic homeostasis.
OB can also influence cellular turnover of the intestine by elevating cell death and changing the expression of genes involved in survival/proliferation, which in turn lead to heightened intestinal permeability, alterations in villi/crypt length, and reductions in the expression of genes responsible for mucus synthesis and tight junctions [80]. More investigations into the association between changes in the oral and GM and the development of OB are shown in Table 4.

Table 4.
Microbiome alterations associated with obesity (OB).
4. Mutual Connections Between Gut Microbial Changes and Epigenetic Alterations
Alterations in the composition of GM involved in the pathogenesis of OB are linked to epigenetic modifications [95]. In this regard, it has been shown that gut bacterial communities by modulating genes expression through epigenetic mechanisms can influence key regulators of glucose and energy homeostasis, such as HDAC7 and IGF2BP2 in the adipose tissue [96]. For example, obese subjects with low Bacteroidetes-to-Firmicutes ratio exhibited DNA hypomethylation and up-regulation of HDAC7 and IGF2BP2 genes in the adipose tissue versus individuals with high Bacteroidetes-to-Firmicutes ratio [96]. As another example, Salas-Perez et al. reported decreased abundance of Ruminococcus in obese individuals that could influence BMI via altering DNAmet, particularly in a DMR located at the MACROD2 gene [97]. Moreover, accumulation of the visceral fat induces the degradation of short-chain fatty acids (SCFAs) and produces lipopolysaccharide (LPS), which in turn increases systemic inflammation [98]. The obesogenic diet is also capable of re-shaping GM, changing bacterial metabolite production, altering histone methylation and acetylation, and subsequently activating pathways involved in the development of colon cancer [99]. Furthermore, a significant difference in colonic acetate production, a known HDAC inhibitor, was observed in lean versus obese youth, which was attributed to variations in colonic lactulose fermentation [100].
Other epigenetic metabolites of the GM are strongly linked to BMI. For example, butyrate was negatively connected to BMI, visceral fat area, and the percentage of body fat in obese individuals [101]. In addition to altering histone methylation and acetylation, SCFAs may exert their beneficial or detrimental effects in OB via changing DNAmet levels. For example, propionate may induce DNA hypermethylation at the cg26345888 locus, leading to the suppression of DAB1 expression in obese individuals. This epigenetic modification has been associated with vitamin D deficiency, which in turn may contribute to the development of diabetes [102].
5. Therapeutic Strategies for Prevention or Treatment of Obesity by Microbiome Mediated Epigenetic Modulations
A fundamental therapeutic approach for OB and its associated metabolic disorders is lifestyle modification. In this regard the first and most important remedy is caloric restriction (CR) and physical activity [103], which appears to be difficult for many individuals, in particular in industrialized countries. Besides changing lifestyle, emerging evidence has shown that microbiome-based therapeutics such as the use of probiotics, prebiotics, and some specific diets, may help prevent or manage OB by modulating the oral and fecal microbiome via epigenetic mechanisms. This section will review these findings.
5.1. Caloric Restriction (CR) and Physical Activity and Their Influence on Gut Microbiome
CR is the most common option for the prevention or treatment of OB through promoting metabolic health [104,105]. In addition to fat burning, CR can prevent OB by modulating the composition of gut microbiota via increasing probiotic genera like Bifidobacterium and Lactobacillus and reducing the abundance of Helicobacter [106]. Ott et al. reported that a 4-week CR could improve weight loss, enhance gut barrier integrity and alleviate systemic inflammation in women with OB, possibly via epigenetic mechanisms [107]. Chen et al. reported that the anti-OB effects of CR are associated with the establishment of a gut microbiome profile opposing OB. Specifically, they identified an increased abundance of Christensenellaceae, a bacterial family involved in acetate and butyrate production. This microbial shift may serve as a potential biomarker for OB reduction [108]. Dong et al. found that high protein CR diet could contribute to weight loss in obese subjects by elevating microbial diversity, increasing the abundance of Akkermansia spp. and Bifidobacterium spp. and depleting of Prevotella spp. [109]. It is important to note that, while CR exhibits beneficial effects in metabolic disorders and OB, excessive CR can lead to liver injury and exacerbate hepatic inflammation by increasing the expression of pro-inflammatory cytokines [110].
Physical activity or exercise also contribute to improving gut dysbiosis in OB and reducing fat accumulation by modulating the composition of GM and their epigenetic metabolites [111]. Exercise has been shown to enhance the abundance of butyrate-producing fecal bacteria and stimulates lipid metabolism through the butyrate-SESN2/CRTC2 signaling pathway [112]. Additionally, Nagano et al. demonstrated that exercise mitigates HFD-induced OB in mice by increasing the relative abundance of butyrate-producing bacterial taxa, such as Ruminococcaceae and Eubacteriaceae, while reducing the abundance of Erysipelotrichaceae and Rikenellaceae [113]. In humans, a 12-week combined strength and endurance training program in obese children led to an increased abundance of butyrate-producing gut microbiota, including Blautia, Dialister, and Roseburia, while concurrently down-regulating the OB-associated NLR family pyrin domain containing 3 (NLRP3) signaling pathway [114]. Eight weeks of exercise training in obese individuals could also improve insulin sensitivity and reduce visceral adiposity by increasing the abundance of the genera Ruminococcus gauvreauii, Lachnospiraceae FCS020 group, and Anaerostipes [115]. Furthermore, Qian et al. found that exercise training in obese subjects could reduce the levels of Akkermansia, Intestinimonas microbiome and elevate the levels of Ruminococcaceae UCG011, and Holdemania microbiome [116].
Different intensities of exercise may have various impacts on the composition of GM and butyrate-producing bacteria. For example, lower exercise intensity elevated relative abundance of Bifidobacterium, A. municiphila, and butyrate-producers Lachnospira eligens, Enterococcus spp., and Clostridium Cluster IV, while higher exercise intensity escalated the abundance of methane producer Methanobrevibacter smithii, and butyrate-producers, including Eryspelothrichales and Oscillospirales [117].
5.2. Dietary Methyl Donors and GM
Dietary methyl donors play a key role in the modulation of glucose and lipid metabolism. Individuals with OB and greater dietary intakes of methyl donors, including betaine, methionine, choline, and vitamins B6, B9, and B12, are less susceptible to being metabolically unhealthy [118]. These ingredients may be considered as good resources to prevent OB via regulation of the GM and epigenetic mechanisms. As an example, Du et al. found that betaine supplementation in mice could improve gut dysbiosis caused by HFD and it is able to increase the abundance of anti-OB strains, including Bifidobacterium, Lactobacillus, and Akkermansia muciniphila. Akkermansia muciniphila is one of the master modulators of betaine effects in promoting microbiome ecology and elevating the levels of strains involved in SCFAs production, specially acetate and butyrate [119]. They demonstrated that acetate and butyrate attenuate the development of OB and glucose intolerance by modulating DNA methylation at the promoter of the host miR-378a [119]. As another example [120] demonstrated that a 4-week dietary supplementation with betaine reduced OB in HFD mice by restoring the balance of glucose and lipid metabolisms. This effect was accompanied by the reversal of elevated levels of miR-27a, miR-27b, miR-34a, miR-200b, and miR-223 in the liver of obese mice [120].
5.3. GM-Derived Metabolites for Treatment of Obesity
5.3.1. Short Chain Fatty Acids (SCFAs) for Treatment of Obesity
GM-derived metabolites such as butyrate and acetate not only act as inhibitors of HDAC activity but also play key roles in reducing OB-induced inflammation and maintaining gut barrier integrity via regulating the expression of tight junction proteins. These metabolites are also involved in promoting fat thermogenesis, improving mitochondrial function, reducing endoplasmic reticulum stress, hampering autophagy dysfunction, and elevation of the anti-inflammatory M2 macrophages [121,122,123,124].
Additionally, butyrate, as one of the most abundant GM metabolites, plays a key role in decreasing appetite and activation of brown adipose tissue through the gut–brain neural circuit by inhibiting the action of orexigenic neurons involved in the expression of neuropeptide Y in the hypothalamus and reducing the activity of neurons in the nucleus tractus solitarius and dorsal vagal complex in the brainstem [125]. Butyrate and propionate have demonstrated ability for hampering diet-induced OB in mice by stimulating gut hormones and suppressing food intake as well [126]. In this context, sodium butyrate (SB)-treated obese mice exhibited reduction in diet-induced OB, which was associated with increased level of H3K9Ac on the promoter of Adipor1/2, Ucp2 and Ucp3 genes, reduced level of HDAC1, and elevated expression of AMP kinase (AMPK) and adiponectin receptors (adipoR1/2) in muscles [127]. As another example, a two-months butyrate intervention in mid-adult obese mice alleviated neuroinflammation, liver fibrosis, and other adverse impacts of OB [128]. Butyrate also increases glucagon-like peptide-1 (GLP-1) production in colonic cells that is known for its anti-OB effects [129].
In other studies, Mollica et al. found that butyrate could promote mitochondrial cell energy metabolism, respiratory capacity, fatty acid oxidation, and glucose homeostasis in insulin-resistant obese mice by changing the mitochondrial dynamic toward fusion [130]. Furthermore, Wanjun et al. reported that SB administration could inhibit body weight gain, restore the GM composition altered by a HFD, and strengthen intestinal barrier integrity [131]. These effects led to a reduction in serum LPS concentrations (53% lower compared to the HFD group) and a decrease in inflammation [131]. The protective effect of butyrate against OB-induced intestinal barrier disruption is associated with the induction of Paneth cell α-defensin expression, which occurs through histone deacetylation and signal transducer and activator of transcription 3 (STAT3)-dependent mechanism [132]. SB intervention for 12 weeks could also decrease body weight in OB-prone and OB-resistant rats through overexpression of antioxidant enzymes, elevating glutathione (GSH)/glutathione disulphide (GSSG) ratio, reducing reactive oxygen species (ROS) and malondialdehyde (MDA) levels, increasing the expression of Pi3k, Nrf2, Nqo-1, and Ho-1, and reductions in the Gsk-3β mRNA expression [133]. Furthermore, Fu et al. reported that SB supplementation could decrease the body fat content and blood lipids in ovariectomized mice by inducing estrogen receptor alpha (ERα) expression, promoting mitochondrial aerobic respiration, and enhancing the phosphorylated form of AMPK and PGC1α [134].
Butyrate can also suppress quinolinic acid-induced BDNF reduction and OB-induced cognitive dysfunction through epigenetic enhancement of H3K18ac at BDNF promoter region [135]. In another interesting study, Shon et al. found that anti-OB effect of butyrate was associated with reshaping gut microbiome and epigenetic modulation of muscular circadian clock [136]. In their study, butyrate was found to reduce weight gain in HFD rats by increasing the abundance of Firmicutes and upregulating the expression of muscle circadian clock genes (Clock, Arntl, and Per2). This effect was linked to increased transcription of genes involved in fatty acid oxidation, which was associated with elevated pan-histone acetylation at the promoter regions of circadian clock genes, likely due to the inhibition of histone deacetylases [136].
Notably, engineered butyrate-producing bacteria may be considered, amongst other promising remedies to hamper OB. For example, engineered Bacillus subtilis SCK6 could reduce food intake and body weight gain in HFD mice by improving butyric acid production [137]. The engineered butyrate-producing strain BsS-RS06551 could also hamper OB in mice by improving insulin resistance and glucose tolerance as well as modulating the composition and structure of the GM [138].
In addition to butyrate, human acetate produced by GM and diets containing acetate, like vinegar, have demonstrated the ability to improve insulin sensitivity and glucose homeostasis. From a mechanistic perspective, acetate is capable of promoting the performance of adipose tissue by elevating oxidative capacity and regulating glucose-stimulated insulin secretion in the pancreas [139].
Dietary supplementation of sodium acetate (SA) could reduce appetite and fat deposition in HFD mice by regulating genes and hormones relevant to mitochondrial function, adipogenesis, lipolysis, and beige adipogenesis [140]. In a 12-week intervention in HFD-fed mice, Den Besten et al. reported that oral SA supplementation present in the diet (at 5%, weight/weight proportion) could hamper HFD-induced weight gain (~30%) versus control HFD-fed mice [141]. Likewise, in a study by Lu et al., 16 weeks of oral SA supplementation (5%, wt./wt.) could prevent HFD-induced weight gain by 72% versus control HFD-fed mice [142]. Another study demonstrated that a six-week intragastric administration of acetic acid (50, 250 mmol/L) to HFD-fed mice could contribute to the reduction in weight gain (7% and 8%, respectively) and body fat accumulation than HFD alone [143]. In a human study by Petersen et al., rates of endogenous acetate turnover were ∼30% higher in the lean individuals than obese individuals [144].
SA is capable of decreasing serum triacylglycerol, inflammatory markers like IL-6, free fatty acids, and glucose, as well as elevating serum GLP-1, and leptin levels [140]. Furthermore, Wang et al. showed that dietary acetic acid could attenuate HFD-induced OB in mice by changing taurine conjugated bile acids metabolism [145]. Another study demonstrated that both acetate and propionate, as epigenetic modifying metabolites of the GM, could alert T cell polarization and reduce inflammatory markers, including IL-1β and IL-6 in HFD-fed mice [146]. The protective impact of acetate and propionate against systemic inflammation in OB may be linked to the reduction in TNF-α and IL-6 and changes in free fatty acid receptor (FFAR) and HDAC mRNA expression in monocytes and macrophages from obese individuals as well [147]. Additionally, propionate or a mixture of SCFAs could induce the expression of adiponectin and resistin by altering their promoter DNAmet via reducing the expressions of DNMT1, 3a, 3b and MBD2 and suppressing the binding of DNMT enzymes to adiponectin and resistin promoter regions in the adipose tissue of the HFD-fed mice [148]. Weitkunat et al. reported that propionate could attenuate hepatic gene and protein expression of lipogenic enzymes and subsequently hepatic triglyceride concentration and HFD-induced insulin resistance after 22 weeks in mice [149]. In a study, propionate could reduce interferon-γ and interleukin-17 and blunt histone deacetylase activity in CD4+ T cells and nuclear factor κB activity which further could decrease IL-6 release in primary cells from obese individuals [150]. In 2025, Miyamoto et al. found that elevating propionate production by Acidipropionibacterium acidipropionici, a propionate-producing bacterium, could promote metabolism in HFD-fed mice, modulate glucose tolerance, and suppress hepatic inflammation through free fatty acid receptor 3 (FFAR3, also termed GPR41) signaling [151].
Figure 2 depicts a summary of events that through which GM-derived metabolites may affect OB.
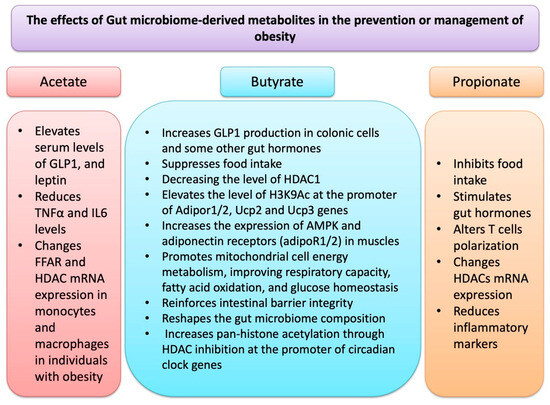
Figure 2.
GM-derived metabolites, in particular SCFAs such as acetate, butyrate and propionate affect epigenetic regulation and other molecular events related to obesity.
5.3.2. Indole and Its Derivatives for Treatment of Obesity and Obesity-Related Disorders
Dietary tryptophan can be metabolized into indole and its derivatives by gut microorganisms. The indole derivatives include indole-3-butyric acid (IBA), indole-3-acetic acid (IAA), indole-3-propionic acid (IPA), indoleacrylic acid (IA), and indole-3-aldehyde (I3A). Indole and its derivatives participate in numerous physiological processes such as maintaining intestinal mucosal homeostasis, improving glucose metabolism, modulation of GI barrier function, and preventing gut dysbiosis and endotoxin leakage [152,153,154,155]. HFD accelerates the release of antimicrobial peptides by Paneth cells, which in turn gives rise to suppressing the Clostridia growth in the gut and subsequently reducing the generation of the beneficial metabolite IPA. Therefore, indole and its derivatives may possess potential in the management and treatment of diet-induced OB and fatty liver disease by stimulating GLP-1 secretion and targeting some cellular pathways [156,157]. As an example, Wang et al. found that the GM-derived metabolite IPA could increase leptin sensitivity by targeting STAT3 to prevent diet-induced OB [158]. In another study, Wang et al. reported that elevated I3A from the intestine after sleeve gastrostomy could attenuate the M1/M2 macrophage ratio in the liver and hence improve NAFLD in obese subjects [159].
5.4. Probiotics
Probiotics are live microorganisms that possess health benefits at proper concentrations by improving gut barrier integrity to prevent the penetration of bacterial antigens and toxic metabolites to blood circulation [160]. Probiotics also contribute to remodeling of the GM and restoring normal performance in subject with OB and gut dysbiosis by introducing beneficial microbes, reducing fecal abundance of pathogenic bacteria, and attenuating the adipose tissue inflammation through restoring the adipose invariant natural killer T cells [161,162,163,164]. Beneficial impact of probiotics against OB and OB-related disorders is through mitigating epigenetic aberrations. For example, probiotic supplementation during pregnancy can reduce the risk of OB in mothers and their infants by influencing DNAmet status of some genes’ promoter regions relevant to OB and weight gain [165]. In this context, Vähämiko et al. found that probiotic supplementation during pregnancy reduced DNAmet levels of 37 gene promoters and increased DNAmet level of one gene promoter in mothers while decreased DNAmet levels of 68 gene promoters in their infants [165]. Moreover, probiotics have demonstrated the ability to reduce body weight and fat accumulation not only through modulation of the GM but also increasing the concentrations of methyl donors such as folic acid. For instance, the administration of Bacteroides thetaiotaomicron to HFD mice for 12 weeks suppressed hyperlipidemia and insulin resistance and decreased body weight and fat accumulation by reducing Firmicutes/Bacteroidetes ratio and elevating gut–liver folate and unsaturated fatty acid metabolism [166].
In addition to DNAmet alterations, beneficial effects of probiotics against OB are connected to targeting histone modifications due to their key roles in the production of butyrate and acetate, as well known HDAC inhibitors. For example, neuroprotective impact of clostridium butyricum in managing cognitive impairment in obese mice is mediated by butyrate production [167]. Choi et al. examined the impact of clostridium butyricum on HFD-induced intestinal inflammation and reported its ability for ameliorating inflammatory markers (e.g., TNF-α) and restoring butyric acid in stools in obese males, but not in obese female rats [168]. Vu et al. reported that probiotic treatment could improve OB-associated colitis in mice by enhancing intestinal tight junctions, possibly via epigenetic mechanisms [169].
Cai et al. found that Lactobacillus plantarum FRT4, a probiotic present in a type of local yogurt, could attenuate the HFD-induced body weight gain, serum cholesterol, triglyceride, liver and fat weight, and the level of alanine aminotransferase (ALT) in the liver of HFD-induced obese mice by elevating microbial diversity and escalating the abundance of species involved in producing SCFAs, including Butyicicoccus, Butyricimonas, Alistipes, Bacteroides, Parabateroides, Intestinimonas, and Anaerotruncus [170]. Additionally, Won et al. examined the anti-OB effect of Lactobacillus sakei (L. sakei) ADM14 administration in a HFD-induced obese mouse model and found that it was capable of reducing weight gain and blood cholesterol levels by improving the Firmicutes to Bacteroidetes ratio, elevating the levels of Bacteroides faecichinchillae and Alistipes, and enhancing butyrate production [171]. Joung et al. reported that treatment of HFD-induced obese mice with Lactobacillus plantarum K50 for 12 weeks hampered fat accumulation and low-grade inflammation by promoting the GM composition, attenuating the Firmicutes/Bacteroidetes ratio, and elevating the concentrations of butyrate and isovalerate [172].
Dietary supplementation with probiotic SF68 has also been shown to reduce intestinal inflammation and improve the integrity and functionality of the intestinal epithelial barrier in obese mice. This effect is mediated by an increase in the expression of tight junction proteins and the intestinal butyrate transporter [173]. Furthermore, Pediococcus acidilactici (pA1c®) supplementation could decrease body weight, reduce OB-related dyslipidemia, and normalize cholesterol profile in Wistar rats by improving the GM richness, reducing inflammation, and elevating the abundance of butyrate-producing bacteria (Lachnospiraceae and Ruminococcaceae), Bacillaceae, and Dehalobacteriaceae families and reducing the abundance of Aerococcaceae [174]. Hosomi et al. also found that oral administration of Blautia wexlerae to mice could mitigate both HFD–induced OB and diabetes by increasing S-adenosylmethionine, acetylcholine, L-ornithine, succinate, lactate, and acetate via reshaping the composition of the GM [175].
In a double-blind, parallel, randomized, controlled clinical trial involving 32 adult women with OB, probiotic treatment (Bifidobacterium lactis UBBLa-70) and symbiotic treatment (Bifidobacterium lactis UBBLa-70 combined with fructooligosaccharide) were found to reduce inflammation and have beneficial effects on body weight. These interventions were associated with increased levels of pyruvate and alanine, along with reduced citrate level and decreased ^1H NMR lipid signals [176]. In another human study, a 12 weeks treatment of obese children with probiotics containing L. rhamnosus bv-77, Lactobacillus salivarius AP-32, and Bifidobacterium animalis CP-9 improved OB-related gut dysbiosis by reducing LDL, serum total cholesterol and TNF-α levels and elevating the abundance of Lactobacillus spp. and B. animalis, species involved in the production of SCFAs [177]. Figure 3 shows a summary of events that mediate probiotics effects on the epigenome.
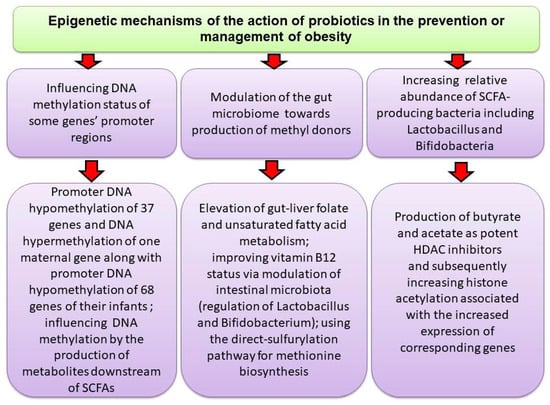
Figure 3.
Probiotics through different mechanisms may affect DNAmet or produce SCFAs that promote gene expression by inducing histone acetylation.
5.5. Engineered Probiotics by Synthetic Biology Approaches for Management of Obesity
Although conventional probiotics have demonstrated efficiency for the treatment of OB and OB-related disorders, they face several limitations, including instability in harsh environments, limited targeting precision, and lack of control over the release of therapeutic agents. Microbes can be reprogrammed via synthetic design to create a variety of therapeutic molecules such as enzymes, cytokines, and signaling molecules from the host and bacterial proteins during pathological condition in the host [178].
Synthetic biology and its related tools like synthetic gene circuits, plasmid-based mechanisms, and CRISPR-Cas systems can improve the functionality, stability, and specificity of engineered probiotics in biomedicine for targeted therapeutic delivery [179,180]. The probiotic survival can be ensured by technologies like encapsulation. The gene circuits facilitate programmable, stimulus-responsive therapeutic release of the engineered probiotics. Therefore, engineered probiotics may be considered promising strategies for managing metabolic and endocrine disorders like OB and diabetes [181,182]. The engineered probiotics are considered non-invasive alternatives to insulin infusion and have demonstrated the ability for precise sensing alterations in glucose concentration and contribution to insulin release [183]. Likewise, the engineered probiotics are capable of managing OB through regulation of appetite-regulating hormones, increasing lipid metabolism, reducing cholesterol levels, elevating satiety, and decreasing food intake [184,185]. Genetically modified probiotics may also contribute to managing OB, diabetes mellitus, and metabolic syndromes by targeting the PI3K/AKT/mTOR signaling pathway [186]. Developing synthetic biological lactic acid bacteria (LAB) probiotics is a promising strategy to manage or treat OB and OB-related disorders, possibly through epigenetic mechanisms, since lactic acid can be employed as substrates in the biochemical synthesis of butyric acid [187,188]. As an example, in a study by Duan et al., the human commensal Lactobacillus gasseri ATCC 33323 (L), a GLP-1(1-37)–secreting bacterium was engineered to release GLP-1(1-37), which further contributes to reprogramming intestinal epithelial cells into glucose-responsive insulin-secreting cells [189]. They found that daily oral administration effectively reduced hyperglycemia in a rat model of diabetes.
Additionally, researchers have developed bacteria engineered to produce N-acylphosphatidylethanolamines (NAPEs), bioactive lipids with known anti-obesity properties. These engineered bacteria have shown effectiveness without requiring antibiotic pretreatment, and with administration periods as short as two weeks [190]. This approach has demonstrated success using human NAPE-producing enzymes, supporting the potential for clinical translation. Researchers also identified a strain of Bifidobacterium pseudocatenulatum JJ3 that interacts with an engineered bacterium (BsS-RS06551) through five dipeptides. This consortium showed more pronounced effects on alleviating obesity than either strain administered individually [191]. The mechanism involves perturbation of the vitamin B6 metabolism pathway in the gut and enrichment of beneficial Bifidobacterium longum populations [191]. Modern synthetic probiotics target specific metabolic pathways. For instance, these probiotics can produce SCFAs that interact with gut epithelial cell receptors to increase GLP-1 and PYY levels, enhancing satiety or regulate gene expression by upregulating peroxisome proliferator-activated receptor (PPAR)γ and PPARα while downregulating sterol regulatory element-binding protein-1c (SREBP-1c) and fatty acid synthase expression [192]. Synthetic probiotics may also target the gut–liver axis to preferentially regulate visceral and subcutaneous fat deposits [193]. Furthermore, droplet-based microfluidic approaches have enabled the isolation of bacteria that can utilize metabolites from engineered bacteria, facilitating the construction of synthetic microbial consortia with enhanced anti-obesity potential [191]. Considering the novelty of these approaches in the field, and despite their promise, further studies are warranted to assess their long-term and potential evolutionary impacts.
5.6. Prebiotics/Postbiotics
Prebiotics are ingredients in diets that boost the proliferation or function of the gut microorganisms [194]. Specific prebiotics are capable of managing OB by promoting intestinal microbial structure and function [195]. In this line, Wei et al. have shown that a dietary polysaccharide from Enteromorpha clathrata, a marine-derived green alga, could reduce the body weight and the serum concentrations of triacylglycerol and cholesterol in HFD-fed mice through increasing the abundance of butyrate-producing bacterium, Eubacterium xylanophilum in the gut, reshaping the GM structure, and improving the gut dysbiosis [196]. Raspberry polysaccharides, food-derived compounds, have shown abilities for reducing OB and related conditions such as hyperlipemia, hyperglycemia, endotoxemia, inflammation, and oxidative stress in the liver of obese mice by elevating the abundance of Dubosiella and its metabolite (butyrate) and enhancing intestinal barrier integrity [197]. Formononetin, an isoflavone, has been shown to normalize body weight and hyperglycemia while improving the high-density lipoprotein (HDL)-to-LDL ratio. This effect is attributed to the increased abundance of specific gut bacteria, including Clostridium aldenense, Clostridiaceae (unclassified), and Eubacterium plexicaum, as well as acetate and butyrate-producing species. Additionally, formononetin modulates liver miRNA expression associated with OB and reduces the levels of inflammatory markers, such as IL-6, IL-22, and TNF-α [198]. Using certain bioactive dietary fibers, such as apple pectin, inulin, β-glucan, xanthan gum, guar gum, xylan, and carrageenan is also a promising strategy to alleviate OB through the enrichment of commensal bacteria, including Ruminococcus, Butyricimonas, Akkermansia, Oscillospira, and Prevotella and possibly their epigenetic metabolites [199]. As another example, Zheng et al. found that Dendrobium officinale (D. officinale) dietary fiber could diminish OB, liver steatosis, inflammatory responses, and oxidative stress in HFD-induced obese mice by reducing the level of Bilophila, elevating the levels of Bifidobacterium, Akkermansia, and Muribaculum as well as enriching the concentrations of acetate and taurine [200].
Administration of soybean insoluble dietary fiber in HFD mice for 20 weeks could also normalize body weight, total cholesterol, triglyceride, and diminish hepatic lipid content by heightening the levels of commensal bacteria, including Lactobacillales, Lactobacillus [genus], Lachnospirace_Nk4A136_group [genus]), and decreasing levels of pathogenic bacteria, including Lachnospiraceae [family] and Bacteroides_acidifaciens [species] as well as increasing SCFAs concentrations [201]. Functional fiber-rich diets also strongly affect GM composition in various intestinal segments of obese mice. For example, Xu et al. reported that functional fiber supplementation could contribute to the transfer and colonization of microbes from the anterior to the posterior intestinal segments and elevate the abundance of bacteria critically involved in the production of acetate and butyrate [202]. Moreover, insoluble dietary fibers derived from brown seaweed Laminaria japonica can reduce serum cholesterol and glucose concentrations in obese mice by increasing the levels of Akkermansia muciniphila and restoring the cecal propionate and acetate concentrations [203]. Inulin, a natural soluble fiber, is also capable of improving imbalanced glucose and lipid metabolism in obese mice by enriching SCFAs-producing Ruminococcaceae and Lachnospiraceae bacteria at the family level and elevation of the fecal SCFAs concentrations [204].
Additionally, anti-OB capabilities of Tibetan highland barley fiber in obese mice was attributed to heightening the abundance of Lachnospiraceae_NK4A136_group, Akkermansiaceae, and Muribaculaceae, decreasing the abundance of Alloprevotella, Prevotellaceae, Rikenellaceae, and Bacteroidaceae, and increasing the concentrations of acetate, propionate and butyrate [205].
In humans, a randomized controlled trial among overweight or adults with OB showed that avocado (containing dietary fiber and monounsaturated fatty acids) consumption increased bacterial alfa diversity, the abundance of Faecalibacterium, Lachnospira, and Alistipes about 26–65% and increase the fecal concentrations of acetate, stearic acid, and palmitic acid versus controls [206]. Furthermore, Iversen et al. demonstrated that a dietary intervention involving high-fiber rye products could increase the abundance of the butyrate-producing bacterium Agathobacter, while decreasing the abundance of the Ruminococcus torques group in humans, a shift associated with weight loss. However, the anti-OB potential of corn peptides in combination with swimming has been linked to the elevation of the abundance of beneficial bacteria like Oscillospiraceae, reductions in the levels of Alloprevotella, and increasing folate synthesis, a methyl donor in one carbon metabolism [207].
Polyphenols are among other nutritional compounds that may function as prebiotics. Among them, resveratrol butyrate esters, derivatives of resveratrol and butyric acid, have demonstrated capabilities for inhibiting bisphenol A (BPA)-induced OB in rats female offspring by decreasing the Firmicutes/Bacteroidetes ratio and the fecal concentration of acetate [208]. Qiao et al. investigated the protective impact of apigenin (Api) against OB-associated metabolic syndrome and found that Api supplementation could ameliorate intestinal dysbiosis induced by HFD via reducing gut barrier damage, increasing the abundance of Akkermansia and Incertae_Sedis, involved in acetate and butyrate production, and decreasing the abundance of Faecalibaculum and Dubosiella at the genus level [209]. Li et al. also found that the anti-OB features of a chlorophyll-rich spinach extract supplementation for 13 weeks in HFD mice was due to elevating levels of Akkermansia, a bacterium involved in acetate production [210]. Furthermore, the preventative impact of alkaloids and polysaccharides present in black tea on HFD-induced OB in mice have been associated with improvements in gut dysbiosis and modulation of DNAmet in imprinted genes such as Magel2, Ctnna3, Gab1, and L3mbtl1 in the spermatozoa of the animals [211].
Postbiotics, which are bioactive compounds produced when gut bacteria break down prebiotics, also have significant anti-OB effects. For example, the postbiotics generated through the bioconversion of citrus pomace and whey by kefir LAB, such as Lactobacillus spp., may help reduce hypertriglyceridemia and body weight gain in HFD mice. This effect is likely mediated by an increase in the relative abundance of Akkermansia odorimutans, which is involved in butyrate production, and a decrease in the abundance of Odoribacter profusus, a bacterial marker of OB [212]. Nowadays, postbiotics are increasingly recognized as promising alternatives to probiotics and prebiotics in the realm of gut or body health and microbiome modulation. While postbiotics show great promise, they are not intended to completely replace probiotics and prebiotics. Rather, they offer an additional tool in the growing field of microbiome modulation and gut health optimization. The ideal approach may involve a combination of these three elements, tailored to individual needs and health goals.
5.7. Antibiotics and Gut Microbiota Changes
Although antibiotics are known for their disturbing impacts on GM composition, in some conditions, antibiotics have the capacity for changing GM and influencing the expression of some proteins in adipose tissue in HFD mice by altering genes’ promoter DNAmet, which in turn could elevate fatty acid oxidation and reduce body weight gain [213]. For example, Yao et al. found that antibiotic use suppressed the body weight gain after 16 weeks of HFD feeding in mice by altering the composition of GM, improving the beta oxidation and thermogenesis (associated with the elevations in the expression of the related genes, including PPAR-α, Pgc-1α, and Atgl), decreasing DNAmet of the adiponectin and resistin promoters, and reducing the expression of DNMT1 and DNMT3a [213]. Nevertheless, this line of research requires further investigation to draw definitive conclusions.
6. Existing Challenges and Research Directions
Although mounting evidence has shown that probiotics and prebiotics are capable of preventing or managing OB, the lack of knowledge of the long-term impact of these microbiome-based therapeutics may hamper their clinical applications. Therefore, future investigations should focus on developing effective long-term preventive strategies with the capacity for inclusion into one’s lifestyle. Additionally, most of the current findings related to the effects of microbiome-based therapeutics in preventing and managing OB have been obtained using animal models, especially rodents. More investigations in humans are needed before microbiome-based therapeutics can be extensively prescribed for the prevention or treatment of OB. Although numerous studies have shown that GM-derived metabolites such as butyrate are capable of preventing OB via epigenetic regulation, some current studies have demonstrated the potential obesogenic role of a large amount of butyrate in early life [214]. For example, as maternal smoking during pregnancy, which increases the risk of OB and overweight in offspring, is connected to an elevated abundance of Firmicutes and, subsequently, increased microbial butyrate production [215]. Therefore, numerous factors such as age and other environmental factors should be considered to establish successful GM-targeted therapeutic approaches for the prevention or management of OB. Another major challenge is that findings on the association between the microbiome and OB development remain relatively weak, often confounded by small sample sizes and a high degree of inter-individual variability. Therefore, large-scale investigations with larger sample sizes are essential to precisely clarify the host metabolic-bacterial cross-talk and the interplays between GM and host genetics in the development of OB and OB-related disorders.
Microbiome research also faces several methodological limitations that can impact the interpretation of results. These include sequencing approach differences, standardization issues, and challenges in determining causation. 16S rRNA sequencing involves amplifying and sequencing the 16S rRNA gene, a marker specific to bacteria and archaea. This cost-effective method provides a broad overview of microbial community composition. However, it offers limited taxonomic resolution (often only to the genus level) and does not provide functional information. It can also suffer from PCR bias, where certain sequences are preferentially amplified. Metagenomic sequencing (whole-genome shotgun sequencing) sequences the entire DNA content of a sample, providing species-level resolution and revealing the functional potential of the microbial community by identifying genes and metabolic pathways. However, metagenomic sequencing is more expensive and requires greater computational resources. It can also be biased toward organisms with larger genomes or those that are more easily lysed [216]. Therefore, the choice of sequencing method affects the level of detail and type of information obtained. 16S rRNA sequencing is suitable for broad community surveys, while metagenomics is necessary for in-depth functional analysis. Critically, insufficient sequencing depth can lead to underestimation of microbial diversity and inaccurate relative abundance estimates, regardless of the method used.
In both methods, variability can be introduced by factors such as sampling site, time of day, and collection method. For example, stool samples may not fully represent the gut microbiome, and swabs can be affected by environmental contaminants. Standardizing these factors across studies is challenging due to logistical and practical constraints. Furthermore, DNA extraction methods can selectively recover DNA from certain microbial groups, leading to biased results, and improper storage (e.g., temperature fluctuations and long storage times) can alter microbial composition and DNA integrity [217]. The lack of standardization can lead to inconsistencies between studies, making it difficult to compare results and draw general conclusions. Furthermore, most microbiome studies are observational, which complicates determining whether changes in the microbiome cause a particular outcome or are simply correlated with it. Many factors (e.g., diet, genetics, and environment) can also influence both the microbiome and the outcome of interest, leading to spurious associations.
In respect to intervention studies, while various interventions (e.g., fecal microbiota transplantation and dietary interventions) can provide stronger evidence for causation, they are often complex and may have unintended effects. Even when a causal link is established, the underlying mechanisms may remain unclear. Therefore, without careful experimental design and mechanistic validation, determining whether the microbiome plays a causal role in a particular disease or condition remains difficult. Eventually, these methodological limitations can compromise the validity and reproducibility of microbiome research.
In summary, due to the necessity for diverse resources and interdisciplinary teams and their high costs, exploring the microbiome’s association with OB development can be challenging. Nevertheless, basic, translational, and human mechanistic investigations that particularly discover functional associations between the GM and OB and encompass cross-cutting interdisciplinary teams could help drive the field forward. The design of sophisticated bioinformatics tools and innovative and promising technologies capable of integrating the information from both host and microbiome would enhance a systems-level understanding.
7. Conclusions
The GM has a dynamic metabolic potential by affecting various metabolic pathways in the host. Recent findings have shown that the pathogenesis of OB is associated with alterations in the microbiota composition and subsequently epigenetic aberrations. The clarification of microbial features relevant to OB across various populations and their interplays with diet, epigenetics, and host genetics paves the way to explore promising strategies with more efficiency and minimal side effects for managing OB and OB-related disorders. Generally, OB and OB-related disorders are associated with low diversity in microbial composition and gene count.
Reshaping the composition of the GM and normalizing epigenetic abnormalities using microbiome-based therapeutics may be considered a promising strategy to obtain stable weight reduction. Moreover, targeting the epigenetic metabolites produced by these microorganisms and the host using dietary intervention and drugs may be considered a promising approach for managing, and preventing OB and OB-related disorders. Promising strategies for the prevention and/or treatment of OB via modifying the GM composition via epigenetic shifts are consuming probiotics, prebiotics, post-biotics, and some specific diets, such as dietary methyl donors and polyphenols. These interventions may be considered the most efficient approaches with minimal side effects to mitigate OB and OB-related disorders in the clinic, as they can selectively promote the number or the growth and activity of health-promoting bacteria and improve the intestinal microecological environment of intestinal bacteria under obese conditions.
However, more detailed investigations are needed to explore the precise mechanisms of action of epigenetic metabolites/alterations and the roles played by the microbial communities in the development and prevention of OB.
Author Contributions
Conceptualization, S.T. and H.M.A.; writing—original draft preparation S.N. and H.M.A.; editing and reorganization of the article H.M.A., S.T., A.P. and G.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We acknowledge Faria Ashrafi for her assistance with the illustrations in this manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Nomenclature
| OB | obesity |
| GM | gut microbiome |
| T2DM | type 2 diabetes |
| BMI | body mass index |
| NAFLD | nonalcoholic fatty liver disease |
| TNF-α | tumor necrosis factor alpha |
| IL-6 | interleukin 6 |
| IL-1β | interleukin-1 beta |
| PPARγ | peroxisome proliferator-activated receptor γ |
| GI | gastrointestinal |
| ncRNAs | non-coding RNAs |
| miRNAs | microRNAs |
| DNAmet | DNA methylation |
| DMRs | differentially methylated regions |
| HFD | high-fat diet |
| HDACs | histone deacetylases |
| DNMTs | DNA methyltransferases |
| 5mC | 5-methylcytosine |
| VAT | visceral adipose tissue |
| SAT | subcutaneous adipose tissue |
| WAT | white adipose tissue |
| SCFAs | short-chain fatty acids |
| CR | caloric restriction |
| LDL | low-density lipoprotein |
| SB | sodium butyrate |
| GLP-1 | glucagon-like peptide-1 |
| STAT3 | signal transducer and activator of transcription 3 |
| GSH/GSSG ratio | glutathione (GSH)/glutathione disulfide (GSSG) ratio |
| ROS | reactive oxygen species |
| MDA | malondialdehyde |
| FFAR | free fatty acid receptor |
| IBA | indole-3-butyric acid |
| IAA | indole-3-acetic acid |
| IPA | indole-3-propionic acid |
| IA | indoleacrylic acid |
| I3A | indole-3-aldehyde |
| ALT | alanine aminotransferase |
| Microbiota | A microbial community including commensal, symbiotic and pathogenic microorganisms; typically defined based on the habitat that it occupies (e.g., the oral or gut microbiota) |
| Microbiome | The aggregate genomes and genes present in the members of microbial community in the body |
| Dysbiosis | A perturbation in microbial balance in the gastrointestinal tract that causes host maladaptation and disease |
| Alpha diversity | Demonstration of the number of different species within a specific community or individual sample |
| Beta diversity | Demonstration of the similarity of one community or individual sample to another |
| Short chain fatty acids | Metabolites created by bacteria during the metabolization of cellulose or other polysaccharides |
| MicroRNA | A class of non-coding RNA in the form of tiny fragment with a crucial regulatory function |
| Histone modification | The occurrence of acetylation, methylation, and phosphorylation on the N-terminal tail of histones H3 and H4 for controlling gene transcription |
| Histone deacetylases (HDAC) | A class of enzymes in charge of the removal of an acetyl group from lysine residues on the histone tail and subsequently affecting the interplay between histones and DNA |
| Polyphenols | Naturally occurring plant ingredients including phenol groups |
| Probiotics | Live microorganisms capable of creating a health benefit on the host when administered in adequate concentrations |
| Prebiotics | Substrates, fermented nondigestible food ingredients, or substances that are selectively consumed by health-promoting bacteria for creating a health benefit via increasing their growth and/or activity |
| Postbiotics | Bacterial fragments with or without bioactive compounds as a product of microbial growth with a health benefit for the host |
| Caloric restriction (CR) | The decrease in caloric intake without the induction of malnutrition |
| Glucagon-like peptide-1 (GLP-1) | An incretin released by the intestines that promotes insulin secretion and sensitivity, improves satiety, and suppresses secretion of glucagon |
| Lipopolysaccharide (LPS) | An endotoxin and the main component of the outer membrane of Gram-negative bacteria involved in the release of cytokines and activation of the innate immune system |
References
- Lin, X.; Li, H. Obesity: Epidemiology, pathophysiology, and therapeutics. Front. Endocrinol. 2021, 12, 706978. [Google Scholar] [CrossRef] [PubMed]
- Lingvay, I.; Cohen, R.V.; le Roux, C.W.; Sumithran, P. Obesity in adults. Lancet 2024, 404, 972–987. [Google Scholar] [CrossRef] [PubMed]
- Engin, A. The definition and prevalence of obesity and metabolic syndrome. In Obesity and Lipotoxicity; Springer: Cham, Switzerland, 2017; pp. 1–17. [Google Scholar]
- Magkos, F.; Sørensen, T.I.; Raubenheimer, D.; Dhurandhar, N.V.; Loos, R.J.; Bosy-Westphal, A.; Clemmensen, C.; Hjorth, M.F.; Allison, D.B.; Taubes, G. On the pathogenesis of obesity: Causal models and missing pieces of the puzzle. Nat. Metab. 2024, 6, 1856–1865. [Google Scholar] [CrossRef] [PubMed]
- Devericks, E.N.; Carson, M.S.; McCullough, L.E.; Coleman, M.F.; Hursting, S.D. The obesity-breast cancer link: A multidisciplinary perspective. Cancer Metastasis Rev. 2022, 41, 607–625. [Google Scholar] [CrossRef]
- Battineni, G.; Sagaro, G.G.; Chintalapudi, N.; Amenta, F.; Tomassoni, D.; Tayebati, S.K. Impact of obesity-induced inflammation on cardiovascular diseases (CVD). Int. J. Mol. Sci. 2021, 22, 4798. [Google Scholar] [CrossRef]
- Chandrasekaran, P.; Weiskirchen, R. The role of obesity in type 2 diabetes mellitus—An overview. Int. J. Mol. Sci. 2024, 25, 1882. [Google Scholar] [CrossRef]
- Savulescu-Fiedler, I.; Mihalcea, R.; Dragosloveanu, S.; Scheau, C.; Baz, R.O.; Caruntu, A.; Scheau, A.-E.; Caruntu, C.; Benea, S.N. The interplay between obesity and inflammation. Life 2024, 14, 856. [Google Scholar] [CrossRef]
- Varra, F.-N.; Varras, M.; Varra, V.-K.; Theodosis-Nobelos, P. Molecular and pathophysiological relationship between obesity and chronic inflammation in the manifestation of metabolic dysfunctions and their inflammation-mediating treatment options. Mol. Med. Rep. 2024, 29, 95. [Google Scholar] [CrossRef]
- Wang, X.; Cao, Q.; Yu, L.; Shi, H.; Xue, B.; Shi, H. Epigenetic regulation of macrophage polarization and inflammation by DNA methylation in obesity. JCI Insight 2016, 1, e87748. [Google Scholar] [CrossRef]
- Zhang, D.; Yao, X.; Teng, Y.; Zhao, T.; Lin, L.; Li, Y.; Shang, H.; Jin, Y.; Jin, Q. Adipocytes-derived Exosomal microRNA-1224 inhibits M2 macrophage polarization in obesity-induced adipose tissue inflammation via MSI2-mediated Wnt/β-catenin Axis. Mol. Nutr. Food Res. 2022, 66, 2100889. [Google Scholar] [CrossRef]
- Fischer, I.; Irmler, M.; Meyer, C.; Sachs, S.; Neff, F.; Hrabě de Angelis, M.; Beckers, J.; Tschöp, M.; Hofmann, S.; Ussar, S. A history of obesity leaves an inflammatory fingerprint in liver and adipose tissue. Int. J. Obes. 2018, 42, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, D.; Venkatesan, M.; Sabarathinam, S. Efficacy of Microbiome-Targeted Interventions in Obesity Management—A Comprehensive Systematic Review. Diabetes Metab. Syndr. Clin. Res. Rev. 2025, 19, 103208. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ma, X.; Mei, H.; Chang, X.; He, P.; Sun, L.; Xiao, H.; Wang, S.; Li, R. Association between gut microbiota and short-chain fatty acids in children with obesity. Sci. Rep. 2025, 15, 483. [Google Scholar] [CrossRef] [PubMed]
- Shaalan, A.; Lee, S.; Feart, C.; Garcia-Esquinas, E.; Gomez-Cabrero, D.; Lopez-Garcia, E.; Morzel, M.; Neyraud, E.; Rodriguez-Artalejo, F.; Streich, R. Alterations in the oral microbiome associated with diabetes, overweight, and dietary components. Front. Nutr. 2022, 9, 914715. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Yuan, X.; Chang, C.; Chen, X.; Li, K. Emerging trends and focus of human gastrointestinal microbiome research from 2010–2021: A visualized study. J. Transl. Med. 2021, 19, 327. [Google Scholar] [CrossRef]
- Parker, A.; Lawson, M.A.; Vaux, L.; Pin, C. Host-microbe interaction in the gastrointestinal tract. Environ. Microbiol. 2018, 20, 2337–2353. [Google Scholar] [CrossRef]
- Pinart, M.; Dötsch, A.; Schlicht, K.; Laudes, M.; Bouwman, J.; Forslund, S.K.; Pischon, T.; Nimptsch, K. Gut microbiome composition in obese and non-obese persons: A systematic review and meta-analysis. Nutrients 2021, 14, 12. [Google Scholar] [CrossRef]
- Tang, R.; Liu, R.; Zha, H.; Cheng, Y.; Ling, Z.; Li, L. Gut microbiota induced epigenetic modifications in the non-alcoholic fatty liver disease pathogenesis. Eng. Life Sci. 2024, 24, 2300016. [Google Scholar] [CrossRef]
- Leung, C.; Rivera, L.; Furness, J.B.; Angus, P.W. The role of the gut microbiota in NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 412–425. [Google Scholar] [CrossRef]
- Tzeng, H.-T.; Lee, W.-C. Impact of transgenerational nutrition on nonalcoholic fatty liver disease development: Interplay between gut microbiota, epigenetics and immunity. Nutrients 2024, 16, 1388. [Google Scholar] [CrossRef] [PubMed]
- Saez, V.M.; Perdicaro, D.; Cremonini, E.; Costantino, V.; Fontana, A.; Oteiza, P.; Prieto, M.V. Grape pomace extract attenuates high fat diet-induced endotoxemia and liver steatosis in mice. Food Funct. 2025, 16, 2515–2529. [Google Scholar] [CrossRef] [PubMed]
- Al Aboud, N.M.; Tupper, C.; Jialal, I. Genetics, epigenetic mechanism. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2018. [Google Scholar]
- Selleri, L.; Rijli, F.M. Shaping faces: Genetic and epigenetic control of craniofacial morphogenesis. Nat. Rev. Genet. 2023, 24, 610–626. [Google Scholar] [CrossRef] [PubMed]
- Gökbuget, D.; Blelloch, R. Epigenetic control of transcriptional regulation in pluripotency and early differentiation. Development 2019, 146, dev164772. [Google Scholar] [CrossRef]
- Weiner, A.K.; Katz, L.A. Epigenetics as driver of adaptation and diversification in microbial eukaryotes. Front. Genet. 2021, 12, 642220. [Google Scholar] [CrossRef]
- Vaissière, T.; Sawan, C.; Herceg, Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat. Res. Rev. Mutat. Res. 2008, 659, 40–48. [Google Scholar] [CrossRef]
- Radford, E.J. An introduction to epigenetic mechanisms. Prog. Mol. Biol. Transl. Sci. 2018, 158, 29–48. [Google Scholar]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Shvedunova, M.; Akhtar, A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2022, 23, 329–349. [Google Scholar] [CrossRef]
- Calestagne-Morelli, A.; Ausio, J. Long-range histone acetylation: Biological significance, structural implications, and mechanisms. Biochem. Cell Biol. 2006, 84, 518–527. [Google Scholar] [CrossRef]
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2024, 25, 211–232. [Google Scholar] [CrossRef] [PubMed]
- Loganathan, T.; Doss C, G.P. Non-coding RNAs in human health and disease: Potential function as biomarkers and therapeutic targets. Funct. Integr. Genom. 2023, 23, 33. [Google Scholar] [CrossRef] [PubMed]
- Nikpay, M.; Ravati, S.; Dent, R.; McPherson, R. Epigenome-wide study identified methylation sites associated with the risk of obesity. Nutrients 2021, 13, 1984. [Google Scholar] [CrossRef] [PubMed]
- Haberman, M.; Menashe, T.; Cohen, N.; Kisliouk, T.; Yadid, T.; Marco, A.; Meiri, N.; Weller, A. Paternal high-fat diet affects weight and DNA methylation of their offspring. Sci. Rep. 2024, 14, 19874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zou, W.; Zhang, S.; Wu, H.; Gao, Y.; Zhang, J.; Zheng, J. Maternal high-fat diet orchestrates offspring hepatic cholesterol metabolism via MEF2A hypermethylation-mediated CYP7A1 suppression. Cell. Mol. Biol. Lett. 2024, 29, 154. [Google Scholar] [CrossRef]
- Vander Velden, J.W.; Osborne, D.M. Prolonged diet-induced obesity modifies DNA methylation and gene expression in the hippocampus. Neurosci. Lett. 2022, 780, 136656. [Google Scholar] [CrossRef]
- Wu, H.-Y.; Cheng, Y.; Jin, L.-Y.; Zhou, Y.; Pang, H.-Y.; Zhu, H.; Yan, C.-C.; Yan, Y.-S.; Yu, J.-E.; Sheng, J.-Z. Paternal obesity impairs hepatic gluconeogenesis of offspring by altering Igf2/H19 DNA methylation. Mol. Cell. Endocrinol. 2021, 529, 111264. [Google Scholar] [CrossRef]
- Sukur, G.; Uysal, F.; Cinar, O. High-fat diet induced obesity alters Dnmt1 and Dnmt3a levels and global DNA methylation in mouse ovary and testis. Histochem. Cell Biol. 2023, 159, 339–352. [Google Scholar] [CrossRef]
- Bozdemir, N.; Kablan, T.; Altintas, M.O.; Sukur, G.; Cinar, O.; Uysal, F. Altered DNA methylation and Dnmt expression in obese uterus may cause implantation failure. J. Mol. Histol. 2024, 55, 427–436. [Google Scholar] [CrossRef]
- Keyhan, S.; Burke, E.; Schrott, R.; Huang, Z.; Grenier, C.; Price, T.; Raburn, D.; Corcoran, D.L.; Soubry, A.; Hoyo, C. Male obesity impacts DNA methylation reprogramming in sperm. Clin. Epigenetics 2021, 13, 17. [Google Scholar] [CrossRef]
- Chen, N.; Miao, L.; Lin, W.; Zou, D.; Huang, L.; Huang, J.; Shi, W.; Li, L.; Luo, Y.; Liang, H. Integrated DNA methylation and gene expression analysis identified S100A8 and S100A9 in the pathogenesis of obesity. Front. Cardiovasc. Med. 2021, 8, 631650. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, A.G.; Carreira, M.C.; Boughanem, H.; Moreno-Navarrete, J.M.; Nicoletti, C.F.; Oliver, P.; de Luis, D.; Nonino, C.B.; Portillo, M.P.; Martinez-Olmos, M.A. Adipose tissue and blood leukocytes ACE2 DNA methylation in obesity and after weight loss. Eur. J. Clin. Investig. 2022, 52, e13685. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.-B.; Ni, J.; Yao, R.; Goetzinger, K.R.; Harman, C.; Reece, E.A.; Wang, B.; Yang, P. Maternal obesity increases DNA methylation and decreases RNA methylation in the human placenta. Reprod. Toxicol. 2022, 107, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Murphy, K.E.; Briollais, L.; McGowan, P.O.; Matthews, S.G. DNA methylation profiles in the blood of newborn term infants born to mothers with obesity. PLoS ONE 2022, 17, e0267946. [Google Scholar] [CrossRef]
- Baca, P.; Barajas-Olmos, F.; Mirzaeicheshmeh, E.; Zerrweck, C.; Guilbert, L.; Sánchez, E.C.; Flores-Huacuja, M.; Villafán, R.; Martínez-Hernández, A.; García-Ortiz, H. DNA methylation and gene expression analysis in adipose tissue to identify new loci associated with T2D development in obesity. Nutr. Diabetes 2022, 12, 50. [Google Scholar] [CrossRef]
- Sehgal, R.; Perfilyev, A.; Männistö, V.; Ågren, J.; Nilsson, E.; Käkelä, P.; Ling, C.; de Mello, V.D.; Pihlajamäki, J. Liver saturated fat content associates with hepatic DNA methylation in obese individuals. Clin. Epigenetics 2023, 15, 21. [Google Scholar] [CrossRef]
- Lecorguillé, M.; McAuliffe, F.M.; Twomey, P.J.; Viljoen, K.; Mehegan, J.; Kelleher, C.C.; Suderman, M.; Phillips, C.M. Maternal glycaemic and insulinemic status and newborn DNA methylation: Findings in women with overweight and obesity. J. Clin. Endocrinol. Metab. 2023, 108, 85–98. [Google Scholar] [CrossRef]
- Han, F.; Zhu, S.; Kong, X.; Wang, W.; Wu, Y. Integrated genetic and epigenetic analyses uncovered GLP1R association with metabolically healthy obesity. Int. J. Obes. 2024, 48, 324–329. [Google Scholar] [CrossRef]
- Ren, Y.; Huang, P.; Huang, X.; Zhang, L.; Liu, L.; Xiang, W.; Liu, L.; He, X. Alterations of DNA methylation profile in peripheral blood of children with simple obesity. Health Inf. Sci. Syst. 2024, 12, 26. [Google Scholar] [CrossRef]
- Jesse, T.G.; Becer, E.; Kalkan, R. Identification of the Relationship Between DNA Methylation of Circadian Rhythm Genes and Obesity. Biochem. Genet. 2024, 62, 281–293. [Google Scholar] [CrossRef]
- Noronha, N.Y.; Diani, L.M.; Rodrigues, G.d.S.; Noma, I.H.Y.; Pereira, V.A.B.; Pinhel, M.A.d.S.; Watanabe, L.M.; Morais, D.A.; Barbosa Jr, F.; Nonino, C.B. Serum Cobalt Concentration and DNA Methylation Signatures in Women with Obesity. Obesities 2024, 4, 85–92. [Google Scholar] [CrossRef]
- Boonrong, C.; Roytrakul, S.; Shantavasinkul, P.C.; Sritara, P.; Sirivarasai, J. Role of Dietary Factors on DNA Methylation Levels of TNF-Alpha Gene and Proteome Profiles in Obese Men. Nutrients 2024, 16, 877. [Google Scholar] [CrossRef] [PubMed]
- Lariviere, D.; Craig, S.J.; Paul, I.M.; Hohman, E.E.; Savage, J.S.; Wright, R.O.; Chiaromonte, F.; Makova, K.D.; Reimherr, M.L. Methylation profiles at birth linked to early childhood obesity. J. Dev. Orig. Health Dis. 2024, 15, e7. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, L.M.; Pereira, V.A.B.; Noronha, N.Y.; de Souza Pinhel, M.A.; Wolf, L.S.; de Oliveira, C.C.; Plaça, J.R.; Noma, I.H.Y.; da Silva Rodrigues, G.; de Souza, V.C.O. The influence of serum selenium in differential epigenetic and transcriptional regulation of CPT1B gene in women with obesity. J. Trace Elem. Med. Biol. 2024, 83, 127376. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, Z.; Yang, J.; Liu, J.; Lin, H.; Yin, N. Depot-specific acetylation profiles of adipose tissues—Therapeutic targets for metabolically unhealthy obesity. Diabetol. Metab. Syndr. 2025, 17, 36. [Google Scholar] [CrossRef]
- Wu, J.; Singh, K.; Shing, V.; Gupta, A.; Arenberg, B.C.; Huffstutler, R.D.; Lee, D.-Y.; Sack, M.N. Mitochondrial fatty acid oxidation regulates monocytic type I interferon signaling via histone acetylation. Sci. Adv. 2025, 11, eadq9301. [Google Scholar] [CrossRef]
- Alawathugoda, T.T.; Sheikh, M.A.; Challagandla, A.K.; Dheen, S.T.; Emerald, B.S.; Ansari, S.A. Maternal obesity alters histone modifications mediated by the interaction between EZH2 and AMPK, impairing neural differentiation in the developing embryonic brain cortex. J. Biol. Chem. 2025, 301, 108173. [Google Scholar] [CrossRef]
- Glendining, K.A.; Jasoni, C.L. Maternal high fat diet-induced obesity modifies histone binding and expression of oxtr in offspring hippocampus in a sex-specific manner. Int. J. Mol. Sci. 2019, 20, 329. [Google Scholar] [CrossRef]
- Jannat Ali Pour, N.; Meshkani, R.; Toolabi, K.; Mohassel Azadi, S.; Zand, S.; Emamgholipour, S. Adipose tissue mRNA expression of HDAC1, HDAC3 and HDAC9 in obese women in relation to obesity indices and insulin resistance. Mol. Biol. Rep. 2020, 47, 3459–3468. [Google Scholar] [CrossRef]
- Wang, F.; Chen, H.; Chen, Y.; Cheng, Y.; Li, J.; Zheng, L.; Zeng, X.; Luo, T. Diet-induced obesity is associated with altered expression of sperm motility-related genes and testicular post-translational modifications in a mouse model. Theriogenology 2020, 158, 233–238. [Google Scholar] [CrossRef]
- Çakır, I.; Hadley, C.K.; Pan, P.L.; Bagchi, R.A.; Ghamari-Langroudi, M.; Porter, D.T.; Wang, Q.; Litt, M.J.; Jana, S.; Hagen, S. Histone deacetylase 6 inhibition restores leptin sensitivity and reduces obesity. Nat. Metab. 2022, 4, 44–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Lin, Y.; Gao, L.; Yang, Z.; Lin, J.; Ren, S.; Li, F.; Chen, J.; Wang, Z.; Dong, Z. PPAR-γ integrates obesity and adipocyte clock through epigenetic regulation of Bmal1. Theranostics 2022, 12, 1589. [Google Scholar] [CrossRef] [PubMed]
- Taghizadeh, N.; Mohammadi, S.; Yousefi, Z.; Golpour, P.; Taheri, A.; Maleki, M.H.; Nourbakhsh, M.; Nourbakhsh, M.; Azar, M.R. Assessment of global histone acetylation in pediatric and adolescent obesity: Correlations with SIRT1 expression and metabolic-inflammatory profiles. PLoS ONE 2023, 18, e0293217. [Google Scholar] [CrossRef] [PubMed]
- Fofana, M.; Li, Z.; Li, H.; Li, W.; Wu, L.; Lu, L.; Liu, Q. Decreased Ubiquitination and Acetylation of Histones 3 and 4 are Associated with Obesity-Induced Disorders of Spermatogenesis in Mice. Toxics 2024, 12, 296. [Google Scholar] [CrossRef]
- Elkhawaga, S.Y.; Ismail, A.; Elsakka, E.G.; Doghish, A.S.; Elkady, M.A.; El-Mahdy, H.A. miRNAs as cornerstones in adipogenesis and obesity. Life Sci. 2023, 315, 121382. [Google Scholar] [CrossRef]
- Liu, X.; Sun, H.; Zheng, L.; Zhang, J.; Su, H.; Li, B.; Wu, Q.; Liu, Y.; Xu, Y.; Song, X. Adipose-derived miRNAs as potential biomarkers for predicting adulthood obesity and its complications: A systematic review and bioinformatic analysis. Obes. Rev. 2024, 25, e13748. [Google Scholar] [CrossRef]
- Makarenkov, N.; Haim, Y.; Yoel, U.; Pincu, Y.; Tarnovscki, T.; Liberty, I.F.; Kukeev, I.; Baraf, L.; Dukhno, O.; Zilber, O. Circulating miRNAs Detect High vs. Low Visceral Adipose Tissue Inflammation in Patients Living with Obesity. J. Clin. Endocrinol. Metab. 2024, 109, 858–867. [Google Scholar] [CrossRef]
- Mir, F.A.; Mall, R.; Iskandarani, A.; Ullah, E.; Samra, T.A.; Cyprian, F.; Parray, A.; Alkasem, M.; Abdalhakam, I.; Farooq, F. Characteristic MicroRNAs linked to dysregulated metabolic pathways in Qatari adult subjects with obesity and metabolic syndrome. Front. Endocrinol. 2022, 13, 937089. [Google Scholar] [CrossRef]
- Simino, L.A.; Panzarin, C.; Fontana, M.F.; de Fante, T.; Geraldo, M.V.; Ignácio-Souza, L.M.; Milanski, M.; Torsoni, M.A.; Ross, M.G.; Desai, M. MicroRNA Let-7 targets AMPK and impairs hepatic lipid metabolism in offspring of maternal obese pregnancies. Sci. Rep. 2021, 11, 8980. [Google Scholar] [CrossRef]
- Lischka, J.; Schanzer, A.; Hojreh, A.; Ba-Ssalamah, A.; de Gier, C.; Valent, I.; Item, C.B.; Greber-Platzer, S.; Zeyda, M. Circulating microRNAs 34a, 122, and 192 are linked to obesity-associated inflammation and metabolic disease in pediatric patients. Int. J. Obes. 2021, 45, 1763–1772. [Google Scholar] [CrossRef]
- Mennitti, L.V.; Carpenter, A.A.; Loche, E.; Pantaleão, L.C.; Fernandez-Twinn, D.S.; Schoonejans, J.M.; Blackmore, H.L.; Ashmore, T.J.; Pisani, L.P.; Tadross, J.A. Effects of maternal diet-induced obesity on metabolic disorders and age-associated miRNA expression in the liver of male mouse offspring. Int. J. Obes. 2022, 46, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Eritja, À.; Caus, M.; Belmonte, T.; de Gonzalo-Calvo, D.; García-Carrasco, A.; Martinez, A.; Martínez, M.; Bozic, M. microRNA Expression Profile in Obesity-Induced Kidney Disease Driven by High-Fat Diet in Mice. Nutrients 2024, 16, 691. [Google Scholar] [CrossRef] [PubMed]
- Masoumi-Ardakani, Y.; Eghbalian, M.; Fallah, H.; Jafari, A.; Shahouzehi, B. Exploring serum miR-33b as a novel diagnostic marker for hypercholesterolemia and obesity: Insights from a pilot case-control study. BMC Endocr. Disord. 2025, 25, 27. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Hou, Y.; Yu, W.; Wang, J.; Chu, X.; Zhang, X.; Pang, H.; Ma, D.; Tang, Y.; Li, M. Adipose tissue-derived microRNA-450a-5p induces type 2 diabetes mellitus by downregulating DUSP10. Mol. Biomed. 2025, 6, 7. [Google Scholar] [CrossRef]
- Nalvarte, I.; Rüegg, J.; Guerrero-Bosagna, C. Intrinsic and extrinsic factors that influence epigenetics. In Epigenetics and Assisted Reproduction; CRC Press: Boca Raton, FL, USA, 2018; pp. 99–114. [Google Scholar]
- Bombin, A.; Yan, S.; Bombin, S.; Mosley, J.D.; Ferguson, J.F. Obesity influences composition of salivary and fecal microbiota and impacts the interactions between bacterial taxa. Physiol. Rep. 2022, 10, e15254. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, Q.; Zheng, W.; Steinwandel, M.; Blot, W.J.; Shu, X.-O.; Long, J. Oral microbiome and obesity in a large study of low-income and African-American populations. J. Oral Microbiol. 2019, 11, 1650597. [Google Scholar] [CrossRef]
- Nagpal, R.; Newman, T.M.; Wang, S.; Jain, S.; Lovato, J.F.; Yadav, H. Obesity-linked gut microbiome dysbiosis associated with derangements in gut permeability and intestinal cellular homeostasis independent of diet. J. Diabetes Res. 2018, 2018, 3462092. [Google Scholar] [CrossRef]
- Liu, R.; Hong, J.; Xu, X.; Feng, Q.; Zhang, D.; Gu, Y.; Shi, J.; Zhao, S.; Liu, W.; Wang, X. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 2017, 23, 859–868. [Google Scholar] [CrossRef]
- Wu, Y.; Chi, X.; Zhang, Q.; Chen, F.; Deng, X. Characterization of the salivary microbiome in people with obesity. PeerJ 2018, 6, e4458. [Google Scholar] [CrossRef]
- Sohail, M.U.; Elrayess, M.A.; Al Thani, A.A.; Al-Asmakh, M.; Yassine, H.M. Profiling the oral microbiome and plasma biochemistry of obese hyperglycemic subjects in Qatar. Microorganisms 2019, 7, 645. [Google Scholar] [CrossRef]
- Ke, X.; Walker, A.; Haange, S.-B.; Lagkouvardos, I.; Liu, Y.; Schmitt-Kopplin, P.; Von Bergen, M.; Jehmlich, N.; He, X.; Clavel, T. Synbiotic-driven improvement of metabolic disturbances is associated with changes in the gut microbiome in diet-induced obese mice. Mol. Metab. 2019, 22, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Maya-Lucas, O.; Murugesan, S.; Nirmalkar, K.; Alcaraz, L.D.; Hoyo-Vadillo, C.; Pizano-Zárate, M.L.; García-Mena, J. The gut microbiome of Mexican children affected by obesity. Anaerobe 2019, 55, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cuesta, M.C.; Del Campo, R.; Garriga-García, M.; Peláez, C.; Requena, T. Taxonomic characterization and short-chain fatty acids production of the obese microbiota. Front. Cell. Infect. Microbiol. 2021, 11, 598093. [Google Scholar] [CrossRef] [PubMed]
- Therdtatha, P.; Song, Y.; Tanaka, M.; Mariyatun, M.; Almunifah, M.; Manurung, N.E.P.; Indriarsih, S.; Lu, Y.; Nagata, K.; Fukami, K. Gut microbiome of Indonesian adults associated with obesity and type 2 diabetes: A cross-sectional study in an Asian city, Yogyakarta. Microorganisms 2021, 9, 897. [Google Scholar] [CrossRef]
- Duan, M.; Wang, Y.; Zhang, Q.; Zou, R.; Guo, M.; Zheng, H. Characteristics of gut microbiota in people with obesity. PLoS ONE 2021, 16, e0255446. [Google Scholar] [CrossRef]
- Thomas, C.; Minty, M.; Canceill, T.; Loubières, P.; Azalbert, V.; Tercé, F.; Champion, C.; Burcelin, R.; Barthet, P.; Laurencin-Dalicieux, S. Obesity drives an oral microbiota signature of female patients with periodontitis: A pilot study. Diagnostics 2021, 11, 745. [Google Scholar] [CrossRef]
- Dong, T.S.; Guan, M.; Mayer, E.A.; Stains, J.; Liu, C.; Vora, P.; Jacobs, J.P.; Lagishetty, V.; Chang, L.; Barry, R.L. Obesity is associated with a distinct brain-gut microbiome signature that connects Prevotella and Bacteroides to the brain’s reward center. Gut Microbes 2022, 14, 2051999. [Google Scholar] [CrossRef]
- Ma, X.; Brinker, E.; Graff, E.C.; Cao, W.; Gross, A.L.; Johnson, A.K.; Zhang, C.; Martin, D.R.; Wang, X. Whole-genome shotgun metagenomic sequencing reveals distinct gut microbiome signatures of obese cats. Microbiol. Spectr. 2022, 10, e0083722. [Google Scholar] [CrossRef]
- Gilley, S.P.; Ruebel, M.L.; Sims, C.; Zhong, Y.; Turner, D.; Lan, R.S.; Pack, L.M.; Piccolo, B.D.; Chintapalli, S.V.; Abraham, A. Associations between maternal obesity and offspring gut microbiome in the first year of life. Pediatr. Obes. 2022, 17, e12921. [Google Scholar] [CrossRef]
- Ma, T.; Wu, Z.; Lin, J.; Shan, C.; Abasijiang, A.; Zhao, J. Characterization of the oral and gut microbiome in children with obesity aged 3 to 5 years. Front. Cell. Infect. Microbiol. 2023, 13, 1102650. [Google Scholar] [CrossRef]
- Vallès, Y.; Arshad, M.; Abdalbaqi, M.; Inman, C.K.; Ahmad, A.; Drou, N.; Gunsalus, K.C.; Ali, R.; Tahlak, M.; Abdulle, A. The infants’ gut microbiome: Setting the stage for the early onset of obesity. Front. Microbiol. 2024, 15, 1371292. [Google Scholar] [CrossRef] [PubMed]
- Stols-Gonçalves, D.; Tristão, L.S.; Henneman, P.; Nieuwdorp, M. Epigenetic markers and microbiota/metabolite-induced epigenetic modifications in the pathogenesis of obesity, metabolic syndrome, type 2 diabetes, and non-alcoholic fatty liver disease. Curr. Diabetes Rep. 2019, 19, 31. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Molina, B.; Sánchez-Alcoholado, L.; Cabrera-Mulero, A.; Lopez-Dominguez, R.; Carmona-Saez, P.; Garcia-Fuentes, E.; Moreno-Indias, I.; Tinahones, F.J. Gut microbiota composition is associated with the global DNA methylation pattern in obesity. Front. Genet. 2019, 10, 613. [Google Scholar] [CrossRef] [PubMed]
- Salas-Perez, F.; Assmann, T.S.; Ramos-Lopez, O.; Martínez, J.A.; Riezu-Boj, J.I.; Milagro, F.I. Crosstalk between gut microbiota and epigenetic markers in obesity development: Relationship between Ruminococcus, BMI, and MACROD2/SEL1L2 methylation. Nutrients 2023, 15, 1550. [Google Scholar] [CrossRef]
- Yan, H.; Qin, Q.; Chen, J.; Yan, S.; Li, T.; Gao, X.; Yang, Y.; Li, A.; Ding, S. Gut microbiome alterations in patients with visceral obesity based on quantitative computed tomography. Front. Cell. Infect. Microbiol. 2022, 11, 823262. [Google Scholar] [CrossRef]
- Qin, Y.; Roberts, J.D.; Grimm, S.A.; Lih, F.B.; Deterding, L.J.; Li, R.; Chrysovergis, K.; Wade, P.A. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 2018, 19, 7. [Google Scholar] [CrossRef]
- Galuppo, B.; Cline, G.; Van Name, M.; Shabanova, V.; Wagner, D.; Kien, C.L.; Santoro, N. Colonic fermentation and acetate production in youth with and without obesity. J. Nutr. 2021, 151, 3292–3298. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Y.; Li, J.; Wang, X.; Zhang, M.; Du, M.; Jiang, W.; Li, C. Butyrate and propionate are negatively correlated with obesity and glucose levels in patients with type 2 diabetes and obesity. Diabetes Metab. Syndr. Obes. 2024, 17, 1533–1541. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, Z.; Li, L.; Liang, X.; Wu, Y.; Wang, X.; Ma, H.; Cheng, J.; Zhang, A.; Tang, P. Gut microbiota induces DNA methylation via SCFAs predisposing obesity-prone individuals to diabetes. Pharmacol. Res. 2022, 182, 106355. [Google Scholar] [CrossRef]
- Kern, L.; Kviatcovsky, D.; He, Y.; Elinav, E. Impact of caloric restriction on the gut microbiota. Curr. Opin. Microbiol. 2023, 73, 102287. [Google Scholar] [CrossRef]
- Sbierski-Kind, J.; Grenkowitz, S.; Schlickeiser, S.; Sandforth, A.; Friedrich, M.; Kunkel, D.; Glauben, R.; Brachs, S.; Mai, K.; Thürmer, A. Effects of caloric restriction on the gut microbiome are linked with immune senescence. Microbiome 2022, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.S.; Luu, K.; Lagishetty, V.; Sedighian, F.; Woo, S.-L.; Dreskin, B.W.; Katzka, W.; Chang, C.; Zhou, Y.; Arias-Jayo, N. The intestinal microbiome predicts weight loss on a calorie-restricted diet and is associated with improved hepatic steatosis. Front. Nutr. 2021, 8, 718661. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Huang, M.; You, X.; Zhao, J.; Chen, L.; Wang, L.; Luo, Y.; Chen, Y. Gut microbiota mediates the anti-obesity effect of calorie restriction in mice. Sci. Rep. 2018, 8, 13037. [Google Scholar] [CrossRef] [PubMed]
- Ott, B.; Skurk, T.; Hastreiter, L.; Lagkouvardos, I.; Fischer, S.; Büttner, J.; Kellerer, T.; Clavel, T.; Rychlik, M.; Haller, D. Effect of caloric restriction on gut permeability, inflammation markers, and fecal microbiota in obese women. Sci. Rep. 2017, 7, 11955. [Google Scholar] [CrossRef]
- Chen, J.; Lin, Y.; Li, T.; Zhu, H.; Huang, F.; Yang, C.; Guo, F. Calorie restriction on normal body weight mice prevents body weight regain on a follow-up high-fat diet by shaping an obesity-resistant-like gut microbiota profile. Food Funct. 2022, 13, 7684–7696. [Google Scholar] [CrossRef]
- Dong, T.; Luu, K.; Lagishetty, V.; Sedighian, F.; Woo, S.; Dreskin, B.; Katzka, W.; Chang, C.; Zhou, Y.; Arias-Jayo, N. A high protein calorie restriction diet alters the gut microbiome in obesity. Nutrients 2020, 12, 3221. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, L.; Li, Q.; Song, C.; Han, N.; Yan, T.; Zhang, L.; Ren, D.; Zhao, Y.; Yang, X. Caloric restriction, friend or foe: Effects on metabolic status in association with the intestinal microbiome and metabolome. J. Agric. Food Chem. 2022, 70, 14061–14072. [Google Scholar] [CrossRef]
- Aragón-Vela, J.; Solis-Urra, P.; Ruiz-Ojeda, F.J.; Álvarez-Mercado, A.I.; Olivares-Arancibia, J.; Plaza-Diaz, J. Impact of exercise on gut microbiota in obesity. Nutrients 2021, 13, 3999. [Google Scholar] [CrossRef]
- Yu, C.; Liu, S.; Chen, L.; Shen, J.; Niu, Y.; Wang, T.; Zhang, W.; Fu, L. Effect of exercise and butyrate supplementation on microbiota composition and lipid metabolism. J. Endocrinol. 2019, 243, 125–135. [Google Scholar] [CrossRef]
- Nagano, T.; Yano, H. Effect of dietary cellulose nanofiber and exercise on obesity and gut microbiota in mice fed a high-fat-diet. Biosci. Biotechnol. Biochem. 2020, 84, 613–620. [Google Scholar] [CrossRef]
- Quiroga, R.; Nistal, E.; Estébanez, B.; Porras, D.; Juárez-Fernández, M.; Martínez-Flórez, S.; García-Mediavilla, M.V.; de Paz, J.A.; González-Gallego, J.; Sánchez-Campos, S. Exercise training modulates the gut microbiota profile and impairs inflammatory signaling pathways in obese children. Exp. Mol. Med. 2020, 52, 1048–1061. [Google Scholar] [CrossRef] [PubMed]
- Verheggen, R.J.; Konstanti, P.; Smidt, H.; Hermus, A.R.; Thijssen, D.H.; Hopman, M.T. Eight-week exercise training in humans with obesity: Marked improvements in insulin sensitivity and modest changes in gut microbiome. Obesity 2021, 29, 1615–1624. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Zuo, Y.; Wen, S.; Wang, X.; Liu, Y.; Li, T. Impact of exercise training on gut microbiome imbalance in obese individuals: A study based on Mendelian randomization analysis. Front. Physiol. 2024, 14, 1264931. [Google Scholar] [CrossRef] [PubMed]
- Torquati, L.; Gajanand, T.; Cox, E.; Willis, C.; Zaugg, J.; Keating, S.; Coombes, J. Effects of exercise intensity on gut microbiome composition and function in people with type 2 diabetes. Eur. J. Sport Sci. 2023, 23, 530–541. [Google Scholar] [CrossRef]
- Poursalehi, D.; Lotfi, K.; Mirzaei, S.; Asadi, A.; Akhlaghi, M.; Saneei, P. Association between methyl donor nutrients and metabolic health status in overweight and obese adolescents. Sci. Rep. 2022, 12, 17045. [Google Scholar] [CrossRef]
- Du, J.; Zhang, P.; Luo, J.; Shen, L.; Zhang, S.; Gu, H.; He, J.; Wang, L.; Zhao, X.; Gan, M. Dietary betaine prevents obesity through gut microbiota-drived microRNA-378a family. Gut Microbes 2021, 13, 1862612. [Google Scholar] [CrossRef]
- Jin, L.; Duan, B.; Liu, J.; Zhang, X.; Li, X.; Zhu, X.; Xin, Y.; Li, Y.; Alsareii, S.A.; Wang, Q. Dietary Betaine Attenuates Obesity: Involvement of Hepatic MicroRNAs. Sci. Adv. Mater. 2021, 13, 2319–2326. [Google Scholar] [CrossRef]
- Kushwaha, V.; Rai, P.; Varshney, S.; Gupta, S.; Khandelwal, N.; Kumar, D.; Gaikwad, A.N. Sodium butyrate reduces endoplasmic reticulum stress by modulating CHOP and empowers favorable anti-inflammatory adipose tissue immune-metabolism in HFD fed mice model of obesity. Food Chem. Mol. Sci. 2022, 4, 100079. [Google Scholar] [CrossRef]
- Cavaliere, G.; Catapano, A.; Trinchese, G.; Cimmino, F.; Penna, E.; Pizzella, A.; Cristiano, C.; Lama, A.; Crispino, M.; Mollica, M.P. Butyrate improves neuroinflammation and mitochondrial impairment in cerebral cortex and synaptic fraction in an animal model of diet-induced obesity. Antioxidants 2022, 12, 4. [Google Scholar] [CrossRef]
- Zhu, W.; Peng, K.; Zhao, Y.; Xu, C.; Tao, X.; Liu, Y.; Huang, Y.; Yang, X. Sodium butyrate attenuated diet-induced obesity, insulin resistance and inflammation partly by promoting fat thermogenesis via intro-adipose sympathetic innervation. Front. Pharmacol. 2022, 13, 938760. [Google Scholar] [CrossRef]
- Wang, X.; Duan, C.; Li, Y.; Lu, H.; Guo, K.; Ge, X.; Chen, T.; Shang, Y.; Liu, H.; Zhang, D. Sodium butyrate reduces overnutrition-induced microglial activation and hypothalamic inflammation. Int. Immunopharmacol. 2022, 111, 109083. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yi, C.-X.; Katiraei, S.; Kooijman, S.; Zhou, E.; Chung, C.K.; Gao, Y.; van den Heuvel, J.K.; Meijer, O.C.; Berbée, J.F. Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut 2018, 67, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.V.; Frassetto, A.; Kowalik Jr, E.J.; Nawrocki, A.R.; Lu, M.M.; Kosinski, J.R.; Hubert, J.A.; Szeto, D.; Yao, X.; Forrest, G. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 2012, 7, e35240. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Jia, Y.; Pan, S.; Jia, L.; Li, H.; Han, Z.; Cai, D.; Zhao, R. Butyrate alleviates high fat diet-induced obesity through activation of adiponectin-mediated pathway and stimulation of mitochondrial function in the skeletal muscle of mice. Oncotarget 2016, 7, 56071–56082. [Google Scholar] [CrossRef]
- Arnoldussen, I.A.; Wiesmann, M.; Pelgrim, C.E.; Wielemaker, E.M.; van Duyvenvoorde, W.; Amaral-Santos, P.; Verschuren, L.; Keijser, B.J.; Heerschap, A.; Kleemann, R. Butyrate restores HFD-induced adaptations in brain function and metabolism in mid-adult obese mice. Int. J. Obes. 2017, 41, 935–944. [Google Scholar] [CrossRef]
- Christiansen, C.B.; Gabe, M.B.N.; Svendsen, B.; Dragsted, L.O.; Rosenkilde, M.M.; Holst, J.J. The impact of short-chain fatty acids on GLP-1 and PYY secretion from the isolated perfused rat colon. Am. J. Physiol.-Gastrointest. Liver Physiol. 2018, 315, G53–G65. [Google Scholar] [CrossRef]
- Mollica, M.P.; Mattace Raso, G.; Cavaliere, G.; Trinchese, G.; De Filippo, C.; Aceto, S.; Prisco, M.; Pirozzi, C.; Di Guida, F.; Lama, A. Butyrate regulates liver mitochondrial function, efficiency, and dynamics in insulin-resistant obese mice. Diabetes 2017, 66, 1405–1418. [Google Scholar] [CrossRef]
- Fang, W.; Xue, H.; Chen, X.; Chen, K.; Ling, W. Supplementation with sodium butyrate modulates the composition of the gut microbiota and ameliorates high-fat diet-induced obesity in mice. J. Nutr. 2019, 149, 747–754. [Google Scholar] [CrossRef]
- Beisner, J.; Filipe Rosa, L.; Kaden-Volynets, V.; Stolzer, I.; Günther, C.; Bischoff, S.C. Prebiotic inulin and sodium butyrate attenuate obesity-induced intestinal barrier dysfunction by induction of antimicrobial peptides. Front. Immunol. 2021, 12, 678360. [Google Scholar] [CrossRef]
- Tang, X.; Sun, Y.; Li, Y.; Ma, S.; Zhang, K.; Chen, A.; Lyu, Y.; Yu, R. Sodium butyrate protects against oxidative stress in high-fat-diet-induced obese rats by promoting GSK-3β/Nrf2 signaling pathway and mitochondrial function. J. Food Biochem. 2022, 46, e14334. [Google Scholar] [CrossRef]
- Fu, Q.; Li, T.; Zhang, C.; Ma, X.; Meng, L.; Liu, L.; Shao, K.; Wu, G.; Zhu, X.; Zhao, X. Butyrate mitigates metabolic dysfunctions via the ERα-AMPK pathway in muscle in OVX mice with diet-induced obesity. Cell Commun. Signal. 2023, 21, 95. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Zheng, M.; Hu, M.; Fang, X.; Geng, D.; Liu, S.; Wang, L.; Zhang, J.; Guan, L.; Zheng, P. Butyrate ameliorates quinolinic acid–induced cognitive decline in obesity models. J. Clin. Investig. 2023, 133, e154612. [Google Scholar] [CrossRef] [PubMed]
- Shon, J.; Han, Y.; Song, S.; Kwon, S.Y.; Na, K.; Lindroth, A.M.; Park, Y.J. Anti-obesity effect of butyrate links to modulation of gut microbiome and epigenetic regulation of muscular circadian clock. J. Nutr. Biochem. 2024, 127, 109590. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Gao, M.; Cheng, X.; Kang, G.; Cao, X.; Huang, H. Engineered butyrate-producing bacteria prevents high fat diet-induced obesity in mice. Microb. Cell Factories 2020, 19, 94. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cheng, X.; Bai, L.; Gao, M.; Kang, G.; Cao, X.; Huang, H. Positive interventional effect of engineered butyrate-producing bacteria on metabolic disorders and intestinal flora disruption in obese mice. Microbiol. Spectr. 2022, 10, e0114721. [Google Scholar] [CrossRef]
- González Hernández, M.A.; Canfora, E.E.; Jocken, J.W.; Blaak, E.E. The short-chain fatty acid acetate in body weight control and insulin sensitivity. Nutrients 2019, 11, 1943. [Google Scholar] [CrossRef]
- Jiao, A.; Yu, B.; He, J.; Yu, J.; Zheng, P.; Luo, Y.; Luo, J.; Yan, H.; Wang, Q.; Wang, H. Sodium acetate, propionate, and butyrate reduce fat accumulation in mice via modulating appetite and relevant genes. Nutrition 2021, 87–88, 111198. [Google Scholar] [CrossRef]
- Den Besten, G.; Bleeker, A.; Gerding, A.; van Eunen, K.; Havinga, R.; van Dijk, T.H.; Oosterveer, M.H.; Jonker, J.W.; Groen, A.K.; Reijngoud, D.-J. Short-chain fatty acids protect against high-fat diet–induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes 2015, 64, 2398–2408. [Google Scholar] [CrossRef]
- Lu, Y.; Fan, C.; Li, P.; Lu, Y.; Chang, X.; Qi, K. Short chain fatty acids prevent high-fat-diet-induced obesity in mice by regulating G protein-coupled receptors and gut microbiota. Sci. Rep. 2016, 6, 37589. [Google Scholar] [CrossRef]
- Kondo, T.; Kishi, M.; Fushimi, T.; Kaga, T. Acetic acid upregulates the expression of genes for fatty acid oxidation enzymes in liver to suppress body fat accumulation. J. Agric. Food Chem. 2009, 57, 5982–5986. [Google Scholar] [CrossRef]
- Petersen, K.F.; Impellizeri, A.; Cline, G.W.; Shulman, G.I. The effects of increased acetate turnover on glucose-induced insulin secretion in lean and obese humans. J. Clin. Transl. Sci. 2019, 3, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Fan, X.; Lu, Y.; Chen, D.; Zhao, Y.; Qi, K. Dietary acetic acid suppress high-fat diet-induced obesity in mice by altering taurine conjugated bile acids metabolism. Curr. Res. Food Sci. 2022, 5, 1976–1984. [Google Scholar] [CrossRef] [PubMed]
- Mandaliya, D.K.; Patel, S.; Seshadri, S. The combinatorial effect of acetate and propionate on high-fat diet induced diabetic inflammation or metaflammation and T cell polarization. Inflammation 2021, 44, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Eslick, S.; Williams, E.J.; Berthon, B.S.; Wright, T.; Karihaloo, C.; Gately, M.; Wood, L.G. Weight loss and short-chain fatty acids reduce systemic inflammation in monocytes and adipose tissue macrophages from obese subjects. Nutrients 2022, 14, 765. [Google Scholar] [CrossRef]
- Lu, Y.; Fan, C.; Liang, A.; Fan, X.; Wang, R.; Li, P.; Qi, K. Effects of SCFA on the DNA methylation pattern of adiponectin and resistin in high-fat-diet-induced obese male mice. Br. J. Nutr. 2018, 120, 385–392. [Google Scholar] [CrossRef]
- Weitkunat, K.; Schumann, S.; Nickel, D.; Kappo, K.A.; Petzke, K.J.; Kipp, A.P.; Blaut, M.; Klaus, S. Importance of propionate for the repression of hepatic lipogenesis and improvement of insulin sensitivity in high-fat diet-induced obesity. Mol. Nutr. Food Res. 2016, 60, 2611–2621. [Google Scholar] [CrossRef]
- Han, K.; Meadows, A.M.; Rodman, M.J.; Russo, A.C.; Sharma, R.; Singh, K.; Hassanzadeh, S.; Dagur, P.K.; Huffstutler, R.D.; Krause, F.N. Propionate functions as a feeding state-dependent regulatory metabolite to counter proinflammatory signaling linked to nutrient load and obesity. J. Leukoc. Biol. 2024, 115, 738–749. [Google Scholar] [CrossRef]
- Miyamoto, J.; Ando, Y.; Yamano, M.; Nishida, A.; Murakami, K.; Kimura, I. Acidipropionibacterium acidipropionici, a propionate-producing bacterium, contributes to GPR41 signaling and metabolic regulation in high-fat diet-induced obesity in mice. Front. Nutr. 2025, 12, 1542196. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D′Angelo, C.; Massi-Benedetti, C.; Fallarino, F. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef]
- Shimada, Y.; Kinoshita, M.; Harada, K.; Mizutani, M.; Masahata, K.; Kayama, H.; Takeda, K. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS ONE 2013, 8, e80604. [Google Scholar] [CrossRef]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.-H.; Xin, F.-Z.; Xue, Y.; Hu, Z.; Han, Y.; Ma, F.; Zhou, D.; Liu, X.-L.; Cui, A.; Liu, Z. Indole-3-propionic acid inhibits gut dysbiosis and endotoxin leakage to attenuate steatohepatitis in rats. Exp. Mol. Med. 2019, 51, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chimerel, C.; Emery, E.; Summers, D.; Keyser, U.; Gribble, F.; Reimann, F. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep. 2014, 9, 1202–1208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Jiang, M.; Zhao, J.; Song, Y.; Du, W.; Shi, J. The mechanism underlying the influence of indole-3-propionic acid: A relevance to metabolic disorders. Front. Endocrinol. 2022, 13, 841703. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, S.; Liu, L.; Mao, A.; Kan, H.; Yu, F.; Ma, X.; Feng, L.; Zhou, T. The gut microbiota-derived metabolite indole-3-propionic acid enhances leptin sensitivity by targeting STAT3 against diet-induced obesity. Clin. Transl. Med. 2024, 14, e70053. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, G.; Bai, J.; Zhao, N.; Wang, Q.; Zhou, R.; Li, G.; Hu, C.; Li, X.; Tao, K. Role of indole-3-acetic acid in NAFLD amelioration after sleeve gastrectomy. Obes. Surg. 2021, 31, 3040–3052. [Google Scholar] [CrossRef]
- DiMattia, Z.; Damani, J.J.; Van Syoc, E.; Rogers, C.J. Effect of probiotic supplementation on intestinal permeability in overweight and obesity: A systematic review of randomized controlled trials and animal studies. Adv. Nutr. 2024, 15, 100162. [Google Scholar] [CrossRef]
- Judkins, T.C.; Archer, D.L.; Kramer, D.C.; Solch, R.J. Probiotics, nutrition, and the small intestine. Curr. Gastroenterol. Rep. 2020, 22, 2. [Google Scholar] [CrossRef]
- Musazadeh, V.; Zarezadeh, M.; Ghalichi, F.; Ahrabi, S.S.; Jamilian, P.; Jamilian, P.; Ghoreishi, Z. Anti-obesity properties of probiotics; a considerable medical nutrition intervention: Findings from an umbrella meta-analysis. Eur. J. Pharmacol. 2022, 928, 175069. [Google Scholar] [CrossRef]
- Wang, X.; Ba, T.; Cheng, Y.; Zhang, P.; Chang, X. Probiotics alleviate adipose inflammation in high-fat diet–induced obesity by restoring adipose invariant natural killer T cells. Nutrition 2021, 89, 111285. [Google Scholar] [CrossRef]
- Ondee, T.; Pongpirul, K.; Janchot, K.; Kanacharoen, S.; Lertmongkolaksorn, T.; Wongsaroj, L.; Somboonna, N.; Ngamwongsatit, N.; Leelahavanichkul, A. Lactiplantibacillus plantarum dfa1 outperforms enterococcus faecium dfa1 on anti-obesity in high fat-induced obesity mice possibly through the differences in gut dysbiosis attenuation, despite the similar anti-inflammatory properties. Nutrients 2021, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- Vähämiko, S.; Laiho, A.; Lund, R.; Isolauri, E.; Salminen, S.; Laitinen, K. The impact of probiotic supplementation during pregnancy on DNA methylation of obesity-related genes in mothers and their children. Eur. J. Nutr. 2019, 58, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, X.-K.; Tang, M.; Lei, L.; Li, J.-R.; Sun, H.; Jiang, J.; Dong, B.; Li, H.-Y.; Jiang, J.-D. Bacteroides thetaiotaomicron ameliorates mouse hepatic steatosis through regulating gut microbial composition, gut-liver folate and unsaturated fatty acids metabolism. Gut Microbes 2024, 16, 2304159. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Ye, H.; Yang, X.; Shen, L.; Dang, X.; Liu, X.; Gong, Y.; Wu, Q.; Wang, L.; Ge, X. Probiotic Clostridium butyricum ameliorates cognitive impairment in obesity via the microbiota-gut-brain axis. Brain Behav. Immun. 2024, 115, 565–587. [Google Scholar] [CrossRef]
- Choi, Y.; Choi, S.I.; Kim, N.; Nam, R.H.; Jang, J.Y.; Na, H.Y.; Shin, C.M.; Lee, D.H.; Min, H.; Kim, Y.-R. Effect of Clostridium butyricum on high-fat diet-induced intestinal inflammation and production of short-chain fatty acids. Dig. Dis. Sci. 2023, 68, 2427–2440. [Google Scholar] [CrossRef]
- Vu, V.; Muthuramalingam, K.; Singh, V.; Hyun, C.; Kim, Y.M.; Unno, T.; Cho, M. Effects of β-glucan, probiotics, and synbiotics on obesity-associated colitis and hepatic manifestations in C57BL/6J mice. Eur. J. Nutr. 2022, 61, 793–807. [Google Scholar] [CrossRef]
- Cai, H.; Wen, Z.; Zhao, L.; Yu, D.; Meng, K.; Yang, P. Lactobacillus plantarum FRT4 alleviated obesity by modulating gut microbiota and liver metabolome in high-fat diet-induced obese mice. Food Nutr. Res. 2022, 66, 7974. [Google Scholar] [CrossRef]
- Won, S.-M.; Chen, S.; Lee, S.Y.; Lee, K.E.; Park, K.W.; Yoon, J.-H. Lactobacillus sakei ADM14 induces anti-obesity effects and changes in gut microbiome in high-fat diet-induced obese mice. Nutrients 2020, 12, 3703. [Google Scholar] [CrossRef]
- Joung, H.; Chu, J.; Kim, B.-K.; Choi, I.-S.; Kim, W.; Park, T.-S. Probiotics ameliorate chronic low-grade inflammation and fat accumulation with gut microbiota composition change in diet-induced obese mice models. Appl. Microbiol. Biotechnol. 2021, 105, 1203–1213. [Google Scholar] [CrossRef]
- Benvenuti, L.; D′Antongiovanni, V.; Pellegrini, C.; Fornai, M.; Bernardini, N.; Ippolito, C.; Segnani, C.; Di Salvo, C.; Colucci, R.; Martelli, A. Dietary supplementation with the probiotic SF68 reinforces intestinal epithelial barrier in obese mice by improving butyrate bioavailability. Mol. Nutr. Food Res. 2023, 67, 2200442. [Google Scholar] [CrossRef]
- Yavorov-Dayliev, D.; Milagro, F.I.; López-Yoldi, M.; Clemente, I.; Riezu-Boj, J.I.; Ayo, J.; Oneca, M.; Aranaz, P. Pediococcus acidilactici (pA1c®) alleviates obesity-related dyslipidemia and inflammation in Wistar rats by activating beta-oxidation and modulating the gut microbiota. Food Funct. 2023, 14, 10855–10867. [Google Scholar] [CrossRef] [PubMed]
- Hosomi, K.; Saito, M.; Park, J.; Murakami, H.; Shibata, N.; Ando, M.; Nagatake, T.; Konishi, K.; Ohno, H.; Tanisawa, K. Oral administration of Blautia wexlerae ameliorates obesity and type 2 diabetes via metabolic remodeling of the gut microbiota. Nat. Commun. 2022, 13, 4477. [Google Scholar] [CrossRef] [PubMed]
- Crovesy, L.; El-Bacha, T.; Rosado, E.L. Modulation of the gut microbiota by probiotics and symbiotics is associated with changes in serum metabolite profile related to a decrease in inflammation and overall benefits to metabolic health: A double-blind randomized controlled clinical trial in women with obesity. Food Funct. 2021, 12, 2161–2170. [Google Scholar] [PubMed]
- Chen, A.-C.; Fang, T.-J.; Ho, H.-H.; Chen, J.-F.; Kuo, Y.-W.; Huang, Y.-Y.; Tsai, S.-Y.; Wu, S.-F.; Lin, H.-C.; Yeh, Y.-T. A multi-strain probiotic blend reshaped obesity-related gut dysbiosis and improved lipid metabolism in obese children. Front. Nutr. 2022, 9, 922993. [Google Scholar] [CrossRef]
- Kang, M.; Choe, D.; Kim, K.; Cho, B.-K.; Cho, S. Synthetic biology approaches in the development of engineered therapeutic microbes. Int. J. Mol. Sci. 2020, 21, 8744. [Google Scholar] [CrossRef]
- Patra, D. Synthetic Biology-Enabled Engineering of Probiotics for Precision and Targeted Therapeutic Delivery Applications. Exon 2024, 1, 54–66. [Google Scholar] [CrossRef]
- Nazir, A.; Hussain, F.H.N.; Raza, A. Advancing microbiota therapeutics: The role of synthetic biology in engineering microbial communities for precision medicine. Front. Bioeng. Biotechnol. 2024, 12, 1511149. [Google Scholar] [CrossRef]
- Bober, J.R.; Beisel, C.L.; Nair, N.U. Synthetic biology approaches to engineer probiotics and members of the human microbiota for biomedical applications. Annu. Rev. Biomed. Eng. 2018, 20, 277–300. [Google Scholar] [CrossRef]
- Dou, J.; Bennett, M.R. Synthetic biology and the gut microbiome. Biotechnol. J. 2018, 13, 1700159. [Google Scholar] [CrossRef]
- Chua, K.J.; Kwok, W.C.; Aggarwal, N.; Sun, T.; Chang, M.W. Designer probiotics for the prevention and treatment of human diseases. Curr. Opin. Chem. Biol. 2017, 40, 8–16. [Google Scholar] [CrossRef]
- Goh, Y.J.; Barrangou, R. Harnessing CRISPR-Cas systems for precision engineering of designer probiotic lactobacilli. Curr. Opin. Biotechnol. 2019, 56, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Maftei, N.-M.; Raileanu, C.R.; Balta, A.A.; Ambrose, L.; Boev, M.; Marin, D.B.; Lisa, E.L. The potential impact of probiotics on human health: An update on their health-promoting properties. Microorganisms 2024, 12, 234. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Yoshikawa, S.; Ikeda, Y.; Taniguchi, K.; Sawamura, H.; Tsuji, A.; Matsuda, S. Encouraging tactics with genetically modified probiotics to improve immunity for the prevention of immune-related diseases including cardio-metabolic disorders. Biomolecules 2022, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Mays, Z.J.; Nair, N.U. Synthetic biology in probiotic lactic acid bacteria: At the frontier of living therapeutics. Curr. Opin. Biotechnol. 2018, 53, 224–231. [Google Scholar] [CrossRef]
- Zhu, L.-B.; Zhang, Y.-C.; Huang, H.-H.; Lin, J. Prospects for clinical applications of butyrate-producing bacteria. World J. Clin. Pediatr. 2021, 10, 84–92. [Google Scholar] [CrossRef]
- Duan, F.F.; Liu, J.H.; March, J.C. Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes 2015, 64, 1794–1803. [Google Scholar] [CrossRef]
- Dosoky, N.S.; Chen, Z.; Guo, Y.; McMillan, C.; Flynn, C.R.; Davies, S.S. Two-week administration of engineered Escherichia coli establishes persistent resistance to diet-induced obesity even without antibiotic pre-treatment. Appl. Microbiol. Biotechnol. 2019, 103, 6711–6723. [Google Scholar] [CrossRef]
- Chen, X.; Gao, M.; Wang, L.; Qiang, G.; Wu, Y.; Huang, H.; Kang, G. A synthetic microbial consortium protects against obesity by regulating vitamin B6 metabolism. Gut Microbes 2024, 16, 2304901. [Google Scholar] [CrossRef]
- Rasaei, N.; Heidari, M.; Esmaeili, F.; Khosravi, S.; Baeeri, M.; Tabatabaei-Malazy, O.; Emamgholipour, S. The effects of prebiotic, probiotic or synbiotic supplementation on overweight/obesity indicators: An umbrella review of the trials’ meta-analyses. Front. Endocrinol. 2024, 15, 1277921. [Google Scholar] [CrossRef]
- Guo, M.; Li, J.; Zhang, L.; Chen, C.; Wei, Y.; Shen, Z.-a. Effects of oral supplementation of probiotics on body weight and visceral fat in obese patients: A meta-analysis and systematic review. Sci. Rep. 2025, 15, 6355. [Google Scholar] [CrossRef]
- Bevilacqua, A.; Campaniello, D.; Speranza, B.; Racioppo, A.; Sinigaglia, M.; Corbo, M.R. An update on prebiotics and on their health effects. Foods 2024, 13, 446. [Google Scholar] [CrossRef] [PubMed]
- Megur, A.; Daliri, E.B.-M.; Baltriukienė, D.; Burokas, A. Prebiotics as a tool for the prevention and treatment of obesity and diabetes: Classification and ability to modulate the gut microbiota. Int. J. Mol. Sci. 2022, 23, 6097. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhao, Y.; Zhou, C.; Zhao, Q.; Zhong, H.; Zhu, X.; Fu, T.; Pan, L.; Shang, Q.; Yu, G. Dietary polysaccharide from Enteromorpha clathrata attenuates obesity and increases the intestinal abundance of butyrate-producing bacterium, Eubacterium xylanophilum, in mice fed a high-fat diet. Polymers 2021, 13, 3286. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Lyu, Y.; Xu, H.; Luo, H.; Yin, X.; Zheng, H. Raspberry polysaccharides attenuate hepatic inflammation and oxidative stress in diet-induced obese mice by enhancing butyrate-mediated intestinal barrier function. Int. J. Biol. Macromol. 2024, 262, 130007. [Google Scholar] [CrossRef]
- Naudhani, M.; Thakur, K.; Ni, Z.-J.; Zhang, J.-G.; Wei, Z.-J. Formononetin reshapes the gut microbiota, prevents progression of obesity and improves host metabolism. Food Funct. 2021, 12, 12303–12324. [Google Scholar] [CrossRef]
- Wen, J.-J.; Li, M.-Z.; Hu, J.-L.; Wang, J.; Wang, Z.-Q.; Chen, C.-H.; Yang, J.-R.; Huang, X.-J.; Xie, M.-Y.; Nie, S.-P. Different dietary fibers unequally remodel gut microbiota and charge up anti-obesity effects. Food Hydrocoll. 2023, 140, 108617. [Google Scholar] [CrossRef]
- Zheng, H.; Ji, H.; Fan, K.; Xu, H.; Huang, Y.; Zheng, Y.; Xu, Q.; Li, C.; Zhao, L.; Li, Y. Targeting gut microbiota and host metabolism with Dendrobium officinale dietary fiber to prevent obesity and improve glucose homeostasis in diet-induced obese mice. Mol. Nutr. Food Res. 2022, 66, 2100772. [Google Scholar] [CrossRef]
- Wang, B.; Yu, H.; He, Y.; Wen, L.; Gu, J.; Wang, X.; Miao, X.; Qiu, G.; Wang, H. Effect of soybean insoluble dietary fiber on prevention of obesity in high-fat diet fed mice via regulation of the gut microbiota. Food Funct. 2021, 12, 7923–7937. [Google Scholar] [CrossRef]
- Xu, C.; Liu, J.; Gao, J.; Wu, X.; Cui, C.; Wei, H.; Peng, J.; Zheng, R. The effect of functional fiber on microbiota composition in different intestinal segments of obese mice. Int. J. Mol. Sci. 2021, 22, 6525. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, N.; Yang, L.; Hong, Z.; Cai, B.; Le, Q.; Yang, T.; Shi, L.; He, J.; Cui, C.-B. Insoluble dietary fiber derived from brown seaweed Laminaria japonica ameliorate obesity-related features via modulating gut microbiota dysbiosis in high-fat diet–fed mice. Food Funct. 2021, 12, 587–601. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, M.; Wang, H.; Li, N.; Lu, Z.; Li, L.; Hui, S.; Xu, H. Gut microbiota and short chain fatty acids partially mediate the beneficial effects of inulin on metabolic disorders in obese ob/ob mice. J. Food Biochem. 2022, 46, e14063. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Han, J.; Li, C.; Tang, J.; Wang, X.; Ma, Y.; Chen, Y.; Xiao, D.; Guo, X. Tibetan highland barley fiber improves obesity and regulates gut microbiota in high-fat diet-fed mice. Food Biosci. 2023, 53, 102620. [Google Scholar] [CrossRef]
- Thompson, S.V.; Bailey, M.A.; Taylor, A.M.; Kaczmarek, J.L.; Mysonhimer, A.R.; Edwards, C.G.; Reeser, G.E.; Burd, N.A.; Khan, N.A.; Holscher, H.D. Avocado consumption alters gastrointestinal bacteria abundance and microbial metabolite concentrations among adults with overweight or obesity: A randomized controlled trial. J. Nutr. 2021, 151, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, Y.; Li, J.; Wu, T. Swimming combined with corn peptide counteracts high-fat diet-induced obesity through rescuing imbalanced gut flora. Adv. Exerc. Health Sci. 2024, 1, 260–269. [Google Scholar] [CrossRef]
- Shih, M.-K.; Tain, Y.-L.; Chen, Y.-W.; Hsu, W.-H.; Yeh, Y.-T.; Chang, S.K.; Liao, J.-X.; Hou, C.-Y. Resveratrol butyrate esters inhibit obesity caused by perinatal exposure to bisphenol a in female offspring rats. Molecules 2021, 26, 4010. [Google Scholar] [CrossRef]
- Qiao, Y.; Zhang, Z.; Zhai, Y.; Yan, X.; Zhou, W.; Liu, H.; Guan, L.; Peng, L. Apigenin alleviates obesity-associated metabolic syndrome by regulating the composition of the gut microbiome. Front. Microbiol. 2022, 12, 805827. [Google Scholar] [CrossRef]
- Li, Y.; Cui, Y.; Lu, F.; Wang, X.; Liao, X.; Hu, X.; Zhang, Y. Beneficial effects of a chlorophyll-rich spinach extract supplementation on prevention of obesity and modulation of gut microbiota in high-fat diet-fed mice. J. Funct. Foods 2019, 60, 103436. [Google Scholar] [CrossRef]
- Liu, X.; Hu, G.; Wang, A.; Long, G.; Yang, Y.; Wang, D.; Zhong, N.; Jia, J. Black tea reduces diet-induced obesity in mice via modulation of gut microbiota and gene expression in host tissues. Nutrients 2022, 14, 1635. [Google Scholar] [CrossRef]
- Youn, H.-Y.; Seo, K.-H.; Kim, H.-J.; Kim, Y.-S.; Kim, H. Effect of postbiotics derived from kefir lactic acid bacteria-mediated bioconversion of citrus pomace extract and whey on high-fat diet-induced obesity and gut dysbiosis. Food Res. Int. 2022, 162, 111930. [Google Scholar] [CrossRef]
- Yao, H.; Fan, C.; Lu, Y.; Fan, X.; Xia, L.; Li, P.; Wang, R.; Tang, T.; Wang, Y.; Qi, K. Alteration of gut microbiota affects expression of adiponectin and resistin through modifying DNA methylation in high-fat diet-induced obese mice. Genes Nutr. 2020, 15, 12. [Google Scholar] [CrossRef]
- Nandy, D.; Craig, S.J.; Cai, J.; Tian, Y.; Paul, I.M.; Savage, J.S.; Marini, M.E.; Hohman, E.E.; Reimherr, M.L.; Patterson, A.D. Metabolomic profiling of stool of two-year old children from the INSIGHT study reveals links between butyrate and child weight outcomes. Pediatr. Obes. 2022, 17, e12833. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Tun, H.M.; Ng, S.C.; Wai, H.K.-F.; Zhang, X.; Parks, J.; Field, C.J.; Mandhane, P.; Moraes, T.J.; Simons, E. Maternal smoking during pregnancy increases the risk of gut microbiome-associated childhood overweight and obesity. Gut Microbes 2024, 16, 2323234. [Google Scholar] [CrossRef] [PubMed]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef] [PubMed]
- Choo, J.M.; Leong, L.E.; Rogers, G.B. Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 2015, 5, 16350. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).