The Athlete Gut Microbiome: A Narrative Review of Multi-Omics Insights and Next-Generation Probiotic Strategies
Abstract
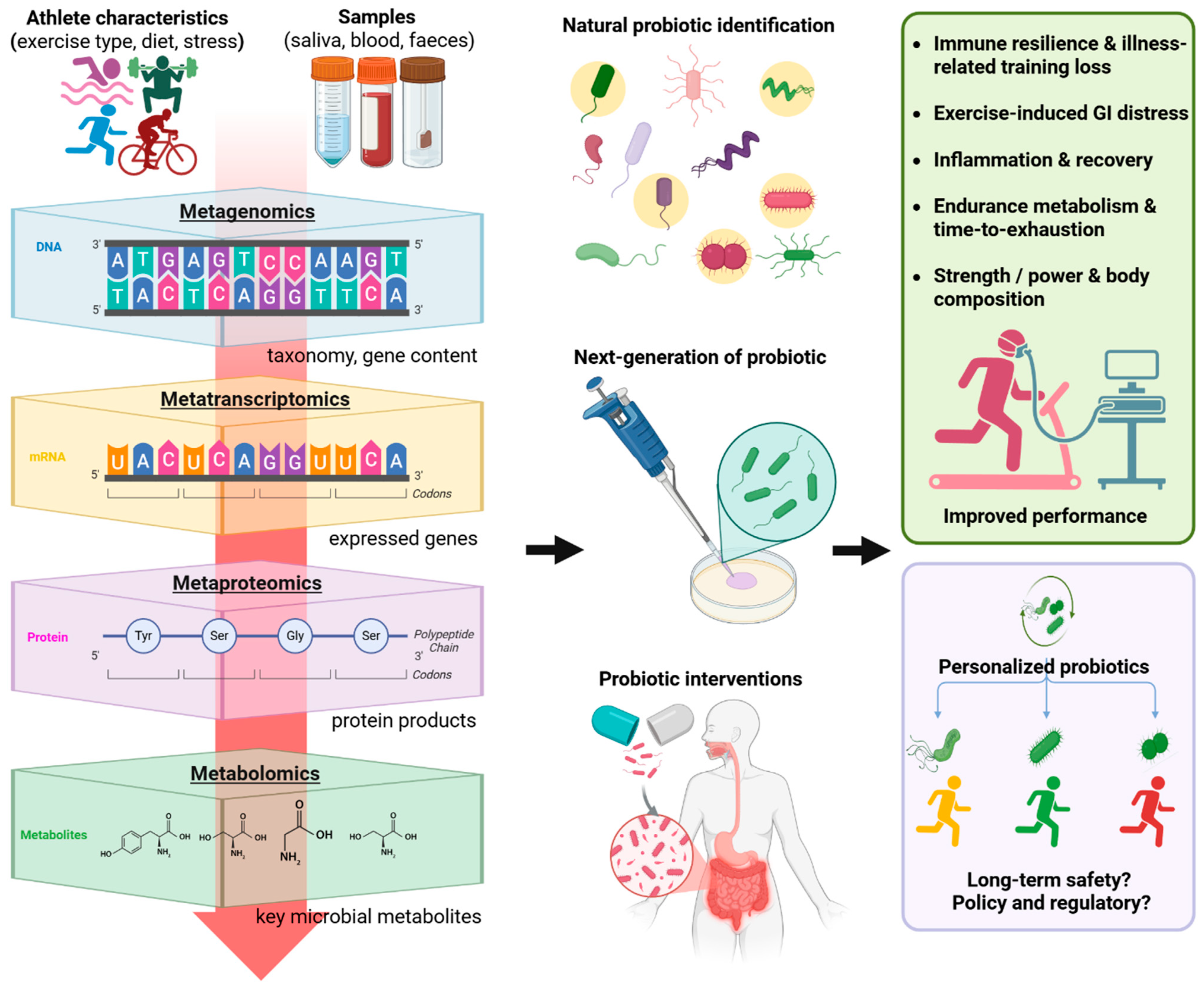
1. Introduction
2. Omics Studies of the Athlete Gut Microbiome
2.1. Metagenomics: Taxonomic and Functional Gene Profiles
2.2. Metatranscriptomics and Metaproteomics: Active Pathways and Expressed Proteins
2.3. Metabolomics: SCFAs, Amino Acid Metabolites, and Others
3. Mechanistic Connections Between Gut Microbiota and Athletic Performance
3.1. Immune Resilience and Illness-Related Training Loss
3.2. Inflammation and Recovery
3.3. GI Integrity
3.4. Endurance Metabolism and Time-to-Exhaust
3.5. Strength, Power and Body Composition
4. Probiotic Interventions for Enhancing Athletic Performance
4.1. Candidate Microbes
| Functional Type | Representative Strain(s) | Potential Athletic Benefit(s) | Ref. |
|---|---|---|---|
| Lactic acid bacteria (LAB) | Lactobacillus, Bifidobacterium (multi-strain) | Immune modulation, gut-barrier support, thus reducing URTIs and GI symptoms, improving training consistency. | [36] |
| Lactiplantibacillus plantarum TWK10 | Enhances energy metabolism, promotes muscle glycogen storage, Improved endurance, muscle strength, favorable body composition. | [42] | |
| Bifidobacterium longum subsp. longum OLP-01 | Gut–muscle axis modulation, immune regulation, Improved running endurance and adaptation. | [43] | |
| Gut-barrier strengthening strains | Akkermansia muciniphila | Mucin layer reinforcement, tight-junction signaling, better gut integrity, nutrient absorption, stability under endurance stress. | [31,44] |
| Escherichia coli Nissle 1917 | Tight-junction upregulation, OMV signaling, help reducing intestinal permeability (“leaky gut”) under physical stress. | [46] | |
| Saccharomyces boulardii | Reinforces tight-junction structure and epithelial integrity, it may benefit physical performance. | [48,49] | |
| SCFA producers | Veillonella atypica | Converts lactate to propionate, enhanced endurance via lactate recycling and fueling. | [14] |
| Faecalibacterium prausnitzii | A cornerstone of a healthy gut microbiome, supports gut barrier integrity and modulates inflammation through butyrate production, thereby may enhance recovery and performance resilience in athletes. | [51] | |
| Roseburia spp. | Enriched in active and athletic populations, contributes to butyrate production that fuels colonocytes, strengthens the gut barrier, and exerts anti-inflammatory effects, thereby supporting recovery and gastrointestinal stability during endurance training. | [52,54] | |
| Carbohydrate & amino acid metabolism strains | Prevotella copri | Carbohydrate fermentation, amino acid metabolism, association with endurance and carb utilization, reduced muscular fatigue; context-dependent inflammation. | [20] |
| Methanobrevibacter smithii | H2 scavenging, fermentation balance, potentially greater energy harvest; enriched in elite cyclists | [20] | |
| Bacillus subtilis; Bacillus clausii; Bacillus coagulans | Enhanced protein digestion, anti-DOMS activity, better protein utilization, reduced muscle damage, faster recovery revealed by RCTs | [55,56] | |
| Bile acid transformation | Clostridium scindens, Eubacterium spp. | 7α-dehydroxylation; FXR/TGR5 signaling, improved lipid and glucose metabolism, insulin sensitivity, mitochondrial activity, and gut repair | [57,58] |
4.2. Next-Generation Probiotics (NGPs)
5. Limitations, Challenges, and Research Gaps
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Sommer, F.; Bäckhed, F. The Gut Microbiota—Masters of Host Development and Physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Chen, J.; Dharmasiddhi, I.P.W.; Chen, S.; Liu, Y.; Liu, H. Review of the Potential of Probiotics in Disease Treatment: Mechanisms, Engineering, and Applications. Processes 2024, 12, 316. [Google Scholar] [CrossRef]
- Mohr, A.E.; Jäger, R.; Carpenter, K.C.; Kerksick, C.M.; Purpura, M.; Townsend, J.R.; West, N.P.; Black, K.; Gleeson, M.; Pyne, D.B.; et al. The Athletic Gut Microbiota. J. Int. Soc. Sports Nutr. 2020, 17, 24. [Google Scholar] [CrossRef]
- Duan, D.; Wang, M.; Han, J.; Li, M.; Wang, Z.; Zhou, S.; Xin, W.; Li, X. Advances in Multi-Omics Integrated Analysis Methods Based on the Gut Microbiome and Their Applications. Front. Microbiol. 2025, 15, 1509117. [Google Scholar] [CrossRef]
- Elrayess, M.A.; Botrè, F.; Palermo, A. Editorial: OMICS-Based Approaches in Sports Research. Front. Mol. Biosci. 2022, 9, 870728. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.; Reczek, S.; Omozusi, O.; Hogue, T.; Cook, M.D.; Hampton-Marcell, J. Machine Learning Reveals Microbial Taxa Associated with a Swim across the Pacific Ocean. Biomedicines 2024, 12, 2309. [Google Scholar] [CrossRef]
- Molavian, R.; Fatahi, A.; Abbasi, H.; Khezri, D. Artificial Intelligence Approach in Biomechanics of Gait and Sport: A Systematic Literature Review. J. Biomed. Phys. Eng. 2023, 13, 383. [Google Scholar] [CrossRef]
- Wosinska, L.; Cotter, P.D.; O’Sullivan, O.; Guinane, C. The Potential Impact of Probiotics on the Gut Microbiome of Athletes. Nutrients 2019, 11, 2270. [Google Scholar] [CrossRef]
- Yadav, R.; Kumar, V.; Baweja, M.; Shukla, P. Gene Editing and Genetic Engineering Approaches for Advanced Probiotics: A Review. Crit. Rev. Food Sci. Nutr. 2018, 58, 1735–1746. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-Omics Data Integration, Interpretation, and Its Application. Bioinforma. Biol. Insights 2020, 14, 117793221989905. [Google Scholar] [CrossRef]
- Lee, J.-Y. The Principles and Applications of High-Throughput Sequencing Technologies. Dev. Reprod. 2023, 27, 9–24. [Google Scholar] [CrossRef]
- Han, D.; Gao, P.; Li, R.; Tan, P.; Xie, J.; Zhang, R.; Li, J. Multicenter Assessment of Microbial Community Profiling Using 16S rRNA Gene Sequencing and Shotgun Metagenomic Sequencing. J. Adv. Res. 2020, 26, 111–121. [Google Scholar] [CrossRef]
- Mancin, L.; Paoli, A.; Berry, S.; Gonzalez, J.T.; Collins, A.J.; Lizarraga, M.A.; Mota, J.F.; Nicola, S.; Rollo, I. Standardization of Gut Microbiome Analysis in Sports. Cell Rep. Med. 2024, 5, 101759. [Google Scholar] [CrossRef]
- Scheiman, J.; Luber, J.M.; Chavkin, T.A.; MacDonald, T.; Tung, A.; Pham, L.-D.; Wibowo, M.C.; Wurth, R.C.; Punthambaker, S.; Tierney, B.T.; et al. Meta-Omics Analysis of Elite Athletes Identifies a Performance-Enhancing Microbe That Functions via Lactate Metabolism. Nat. Med. 2019, 25, 1104–1109. [Google Scholar] [CrossRef]
- Fontana, F.; Longhi, G.; Tarracchini, C.; Mancabelli, L.; Lugli, G.A.; Alessandri, G.; Turroni, F.; Milani, C.; Ventura, M. The Human Gut Microbiome of Athletes: Metagenomic and Metabolic Insights. Microbiome 2023, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Aya, V.; Pardo-Rodriguez, D.; Vega, L.C.; Cala, M.P.; Ramírez, J.D. Integrating Metagenomics and Metabolomics to Study the Gut Microbiome and Host Relationships in Sports across Different Energy Systems. Sci. Rep. 2025, 15, 15356. [Google Scholar] [CrossRef] [PubMed]
- Bashiardes, S.; Zilberman-Schapira, G.; Elinav, E. Use of Metatranscriptomics in Microbiome Research. Bioinforma. Biol. Insights 2016, 10, BBI.S34610. [Google Scholar] [CrossRef] [PubMed]
- Starr, A.E.; Deeke, S.A.; Li, L.; Zhang, X.; Daoud, R.; Ryan, J.; Ning, Z.; Cheng, K.; Nguyen, L.V.H.; Abou-Samra, E.; et al. Proteomic and Metaproteomic Approaches to Understand Host–Microbe Interactions. Anal. Chem. 2018, 90, 86–109. [Google Scholar] [CrossRef]
- Ojala, T.; Häkkinen, A.-E.; Kankuri, E.; Kankainen, M. Current Concepts, Advances, and Challenges in Deciphering the Human Microbiota with Metatranscriptomics. Trends Genet. 2023, 39, 686–702. [Google Scholar] [CrossRef]
- Petersen, L.M.; Bautista, E.J.; Nguyen, H.; Hanson, B.M.; Chen, L.; Lek, S.H.; Sodergren, E.; Weinstock, G.M. Community Characteristics of the Gut Microbiomes of Competitive Cyclists. Microbiome 2017, 5, 98. [Google Scholar] [CrossRef]
- Martin, D.; Bonneau, M.; Orfila, L.; Horeau, M.; Hazon, M.; Demay, R.; Lecommandeur, E.; Boumpoutou, R.; Guillotel, A.; Guillemot, P.; et al. Atypical Gut Microbial Ecosystem from Athletes with Very High Exercise Capacity Improves Insulin Sensitivity and Muscle Glycogen Store in Mice. Cell Rep. 2025, 44, 115448. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-Gut Microbiota Metabolic Interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, S.; Biancucci, F.; Menotta, M.; Nasoni, M.G.; Luchetti, F. Untargeted Metabolomics Profiling of 24-h Ultramarathon Runners: New Insights into the Biochemical Alterations and the Identification of Performance Biomarkers. Life Sci. 2025, 378, 123853. [Google Scholar] [CrossRef] [PubMed]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The Microbiome of Professional Athletes Differs from That of More Sedentary Subjects in Composition and Particularly at the Functional Metabolic Level. Gut 2017, 67, 625–633. [Google Scholar] [CrossRef]
- Nieman, D.C. Upper Respiratory Tract Infections and Exercise. Thorax 1995, 50, 1229–1231. [Google Scholar] [CrossRef]
- Cuthbertson, L.; Turner, S.E.G.; Jackson, A.; Ranson, C.; Loosemore, M.; Kelleher, P.; Moffatt, M.F.; Cookson, W.O.C.; Hull, J.H.; Shah, A. Evidence of Immunometabolic Dysregulation and Airway Dysbiosis in Athletes Susceptible to Respiratory Illness. eBioMedicine 2022, 79, 104024. [Google Scholar] [CrossRef] [PubMed]
- Purton, T.; Staskova, L.; Lane, M.M.; Dawson, S.L.; West, M.; Firth, J.; Clarke, G.; Cryan, J.F.; Berk, M.; O’Neil, A.; et al. Prebiotic and Probiotic Supplementation and the Tryptophan-Kynurenine Pathway: A Systematic Review and Meta Analysis. Neurosci. Biobehav. Rev. 2021, 123, 1–13. [Google Scholar] [CrossRef]
- Strasser, B.; Geiger, D.; Schauer, M.; Gostner, J.; Gatterer, H.; Burtscher, M.; Fuchs, D. Probiotic Supplements Beneficially Affect Tryptophan–Kynurenine Metabolism and Reduce the Incidence of Upper Respiratory Tract Infections in Trained Athletes: A Randomized, Double-Blinded, Placebo-Controlled Trial. Nutrients 2016, 8, 752. [Google Scholar] [CrossRef]
- Walsh, N.P. Nutrition and Athlete Immune Health: New Perspectives on an Old Paradigm. Sports Med. 2019, 49, 153–168. [Google Scholar] [CrossRef]
- Liu, X.; Xu, M.; Wang, H.; Zhu, L. Role and Mechanism of Short-Chain Fatty Acids in Skeletal Muscle Homeostasis and Exercise Performance. Nutrients 2025, 17, 1463. [Google Scholar] [CrossRef]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.B.; Wood-Martin, R.; et al. Exercise and Associated Dietary Extremes Impact on Gut Microbial Diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef]
- Li, Y.; Cheng, M.; Zha, Y.; Yang, K.; Tong, Y.; Wang, S.; Lu, Q.; Ning, K. Gut Microbiota and Inflammation Patterns for Specialized Athletes: A Multi-Cohort Study across Different Types of Sports. mSystems 2023, 8, e0025923. [Google Scholar] [CrossRef] [PubMed]
- Ribichini, E.; Scalese, G.; Cesarini, A.; Mocci, C.; Pallotta, N.; Severi, C.; Corazziari, E.S. Exercise-Induced Gastrointestinal Symptoms in Endurance Sports: A Review of Pathophysiology, Symptoms, and Nutritional Management. Dietetics 2023, 2, 289–307. [Google Scholar] [CrossRef]
- Keohane, D.M.; Woods, T.; O’Connor, P.; Underwood, S.; Cronin, O.; Whiston, R.; O’Sullivan, O.; Cotter, P.; Shanahan, F.; Molloy, M.G.M. Four Men in a Boat: Ultra-Endurance Exercise Alters the Gut Microbiome. J. Sci. Med. Sport 2019, 22, 1059–1064. [Google Scholar] [CrossRef]
- Hintikka, J.E.; Munukka, E.; Valtonen, M.; Luoto, R.; Ihalainen, J.K.; Kallonen, T.; Waris, M.; Heinonen, O.J.; Ruuskanen, O.; Pekkala, S. Gut Microbiota and Serum Metabolome in Elite Cross-Country Skiers: A Controlled Study. Metabolites 2022, 12, 335. [Google Scholar] [CrossRef]
- Pugh, J.N.; Sparks, A.S.; Doran, D.A.; Fleming, S.C.; Langan-Evans, C.; Kirk, B.; Fearn, R.; Morton, J.P.; Close, G.L. Four Weeks of Probiotic Supplementation Reduces GI Symptoms during a Marathon Race. Eur. J. Appl. Physiol. 2019, 119, 1491–1501. [Google Scholar] [CrossRef]
- Przewłócka, K.; Folwarski, M.; Kaźmierczak-Siedlecka, K.; Skonieczna-Żydecka, K.; Kaczor, J.J. Gut-Muscle Axis Exists and May Affect Skeletal Muscle Adaptation to Training. Nutrients 2020, 12, 1451. [Google Scholar] [CrossRef] [PubMed]
- Mörkl, S.; Lackner, S.; Müller, W.; Gorkiewicz, G.; Kashofer, K.; Oberascher, A.; Painold, A.; Holl, A.; Holzer, P.; Meinitzer, A.; et al. Gut Microbiota and Body Composition in Anorexia Nervosa Inpatients in Comparison to Athletes, Overweight, Obese, and Normal Weight Controls. Int. J. Eat. Disord. 2017, 50, 1421–1431. [Google Scholar] [CrossRef]
- Sivamaruthi, B. A Comprehensive Review on Clinical Outcome of Probiotic and Synbiotic Therapy for Inflammatory Bowel Diseases. Asian Pac. J. Trop. Biomed. 2018, 8, 179. [Google Scholar] [CrossRef]
- Kearns, R.P.; Dooley, J.S.G.; Matthews, M.; McNeilly, A.M. Do Probiotics Mitigate GI-Induced Inflammation and Perceived Fatigue in Athletes? A Systematic Review. J. Int. Soc. Sports Nutr. 2024, 21, 2388085. [Google Scholar] [CrossRef]
- Miles, M.P. Probiotics and Gut Health in Athletes. Curr. Nutr. Rep. 2020, 9, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-C.; Liao, Y.-C.; Lee, M.-C.; Cheng, Y.-C.; Chiou, S.-Y.; Lin, J.-S.; Huang, C.-C.; Watanabe, K. Different Impacts of Heat-Killed and Viable Lactiplantibacillus Plantarum TWK10 on Exercise Performance, Fatigue, Body Composition, and Gut Microbiota in Humans. Microorganisms 2022, 10, 2181. [Google Scholar] [CrossRef]
- Lin, C.-L.; Hsu, Y.-J.; Ho, H.-H.; Chang, Y.-C.; Kuo, Y.-W.; Yeh, Y.-T.; Tsai, S.-Y.; Chen, C.-W.; Chen, J.-F.; Huang, C.-C.; et al. Bifidobacterium Longum Subsp. Longum OLP-01 Supplementation during Endurance Running Training Improves Exercise Performance in Middle- and Long-Distance Runners: A Double-Blind Controlled Trial. Nutrients 2020, 12, 1972. [Google Scholar] [CrossRef]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; De Vos, W.M. Akkermansia Muciniphila Gen. Nov., Sp. Nov., a Human Intestinal Mucin-Degrading Bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef]
- Kang, C.-H.; Jung, E.-S.; Jung, S.-J.; Han, Y.-H.; Chae, S.-W.; Jeong, D.Y.; Kim, B.-C.; Lee, S.-O.; Yoon, S.-J. Pasteurized Akkermansia Muciniphila HB05 (HB05P) Improves Muscle Strength and Function: A 12-Week, Randomized, Double-Blind, Placebo-Controlled Clinical Trial. Nutrients 2024, 16, 4037. [Google Scholar] [CrossRef]
- Mooren, F.C.; Maleki, B.H.; Pilat, C.; Ringseis, R.; Eder, K.; Teschler, M.; Krüger, K. Effects of Escherichia Coli Strain Nissle 1917 on Exercise-Induced Disruption of Gastrointestinal Integrity. Eur. J. Appl. Physiol. 2020, 120, 1591–1599. [Google Scholar] [CrossRef]
- Alvarez, C.-S.; Giménez, R.; Cañas, M.-A.; Vera, R.; Díaz-Garrido, N.; Badia, J.; Baldomà, L. Extracellular Vesicles and Soluble Factors Secreted by Escherichia Coli Nissle 1917 and ECOR63 Protect against Enteropathogenic E. Coli-Induced Intestinal Epithelial Barrier Dysfunction. BMC Microbiol. 2019, 19, 166. [Google Scholar] [CrossRef] [PubMed]
- Dahan, S.; Dalmasso, G.; Imbert, V.; Peyron, J.-F.; Rampal, P.; Czerucka, D. Saccharomyces Boulardii Interferes with Enterohemorrhagic Escherichia Coli-Induced Signaling Pathways in T84 Cells. Infect. Immun. 2003, 71, 766–773. [Google Scholar] [CrossRef]
- Soares, A.D.N.; Wanner, S.P.; Morais, E.S.S.; Hudson, A.S.R.; Martins, F.S.; Cardoso, V.N. Supplementation with Saccharomyces Boulardii Increases the Maximal Oxygen Consumption and Maximal Aerobic Speed Attained by Rats Subjected to an Incremental-Speed Exercise. Nutrients 2019, 11, 2352. [Google Scholar] [CrossRef]
- Bongiovanni, T.; Yin, M.O.L.; Heaney, L.M. The Athlete and Gut Microbiome: Short-Chain Fatty Acids as Potential Ergogenic Aids for Exercise and Training. Int. J. Sports Med. 2021, 42, 1143–1158. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Miquel, S.; Benevides, L.; Bridonneau, C.; Robert, V.; Hudault, S.; Chain, F.; Berteau, O.; Azevedo, V.; Chatel, J.M.; et al. Functional Characterization of Novel Faecalibacterium Prausnitzii Strains Isolated from Healthy Volunteers: A Step Forward in the Use of F. Prausnitzii as a Next-Generation Probiotic. Front. Microbiol. 2017, 8, 1226. [Google Scholar] [CrossRef]
- Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia Spp.: A Marker of Health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, S.S.; Ribeiro, F.M.; Sousa Neto, I.V.; Franco, O.L.; Petriz, B. Effects of Physical Exercise on Akkermansia Muciniphila: A Systematic Review of Human and Animal Studies. Benef. Microbes 2024, 15, 565–587. [Google Scholar] [CrossRef] [PubMed]
- Çıplak, M.E.; Genç, A.; Bilici, M.F.; Güder, F.; Acar, H.; Tutkun, E. Roseburia Species in Intestinal Flora in Amateur and Professional Male Football Players: Roseburia Intestinal Flora Football Players. Prog. Nutr. 2020, 22, 515–520. [Google Scholar] [CrossRef]
- Elshaghabee, F.M.F.; Rokana, N.; Gulhane, R.D.; Sharma, C.; Panwar, H. Bacillus as Potential Probiotics: Status, Concerns, and Future Perspectives. Front. Microbiol. 2017, 8, 1490. [Google Scholar] [CrossRef]
- Williams, N.; Weir, T.L. Spore-Based Probiotic Bacillus Subtilis: Current Applications in Humans and Future Perspectives. Fermentation 2024, 10, 78. [Google Scholar] [CrossRef]
- Kulecka, M.; Fraczek, B.; Mikula, M.; Zeber-Lubecka, N.; Karczmarski, J.; Paziewska, A.; Ambrozkiewicz, F.; Jagusztyn-Krynicka, K.; Cieszczyk, P.; Ostrowski, J. The Composition and Richness of the Gut Microbiota Differentiate the Top Polish Endurance Athletes from Sedentary Controls. Gut Microbes 2020, 11, 1374–1384. [Google Scholar] [CrossRef] [PubMed]
- Wahlström, A.; Sayin, S.I.; Marschall, H.-U.; Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef]
- De Filippis, F.; Pasolli, E.; Tett, A.; Tarallo, S.; Naccarati, A.; De Angelis, M.; Neviani, E.; Cocolin, L.; Gobbetti, M.; Segata, N.; et al. Distinct Genetic and Functional Traits of Human Intestinal Prevotella Copri Strains Are Associated with Different Habitual Diets. Cell Host Microbe 2019, 25, 444–453.e3. [Google Scholar] [CrossRef]
- Stecker, R.A.; Moon, J.M.; Russo, T.J.; Ratliff, K.M.; Mumford, P.W.; Jäger, R.; Purpura, M.; Kerksick, C.M. Bacillus Coagulans GBI-30, 6086 Improves Amino Acid Absorption from Milk Protein. Nutr. Metab. 2020, 17, 93. [Google Scholar] [CrossRef]
- De Paiva, A.K.F.; De Oliveira, E.P.; Mancini, L.; Paoli, A.; Mota, J.F. Effects of Probiotic Supplementation on Performance of Resistance and Aerobic Exercises: A Systematic Review. Nutr. Rev. 2023, 81, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, C.; Tamir, S.; Golan, R.; Weinstein, A.; Weinstein, Y. The Effect of Probiotic Supplementation on Performance, Inflammatory Markers and Gastro-intestinal Symptoms in Elite Road Cyclists. J. Int. Soc. Sports Nutr. 2021, 18, 36. [Google Scholar] [CrossRef]
- Yang, M.; Hutchinson, N.; Ye, N.; Timek, H.; Jennings, M.; Yin, J.; Guan, M.; Wang, Z.; Chen, P.; Yang, S.; et al. Engineered Bacillus Subtilis as Oral Probiotics to Enhance Clearance of Blood Lactate. ACS Synth. Biol. 2025, 14, 101–112. [Google Scholar] [CrossRef]
- Bai, Y.; Mansell, T.J. Production and Sensing of Butyrate in a Probiotic E. Coli Strain. Int. J. Mol. Sci. 2020, 21, 3615. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, J.; Raj, K.; Baerg, L.; Nathan, N.; Philpott, D.J.; Mahadevan, R. A Universal Strategy to Promote Secretion of G+/G– Bacterial Extracellular Vesicles and Its Application in Host Innate Immune Responses. ACS Synth. Biol. 2023, 12, 319–328. [Google Scholar] [CrossRef]
- Chen, J.; Liu, M.; Chen, S.; Chou, C.P.; Liu, H.; Wu, D.; Liu, Y. Engineered Therapeutic Bacteria with High-Yield Membrane Vesicle Production Inspired by Eukaryotic Membrane Curvature for Treating Inflammatory Bowel Disease. ACS Nano 2025, 19, 2405–2418. [Google Scholar] [CrossRef]
- Hassan-Casarez, C.; Ryan, V.; Shuster, B.M.; Oliver, J.W.K.; Abbott, Z.D. Engineering a Probiotic Bacillus Subtilis for Acetaldehyde Removal: A Hag Locus Integration to Robustly Express Acetaldehyde Dehydrogenase. PLoS ONE 2024, 19, e0312457. [Google Scholar] [CrossRef]
- Kavanagh, K. First CRISPR Horses Spark Controversy: What’s next for Gene-Edited Animals? Nature 2025, 645, 565. [Google Scholar] [CrossRef] [PubMed]
- Gemmell, M.R.; Jayawardana, T.; Koentgen, S.; Brooks, E.; Kennedy, N.; Berry, S.; Lees, C.; Hold, G.L. Optimised Human Stool Sample Collection for Multi-Omic Microbiota Analysis. Sci. Rep. 2024, 14, 16816. [Google Scholar] [CrossRef]
- Szopinska-Tokov, J.; Bloemendaal, M.; Boekhorst, J.; Hermes, G.D.; Ederveen, T.H.; Vlaming, P.; Buitelaar, J.K.; Franke, B.; Arias-Vasquez, A. A Comparison of Bioinformatics Pipelines for Compositional Analysis of the Human Gut Microbiome. bioRxiv 2023. [Google Scholar] [CrossRef]
| Omics Layer | Main Focus/Connection with Other Omics | Athlete-Sample Matrix | Core Instrumentation | Cost Level * | Functions |
|---|---|---|---|---|---|
| Metagenomics | Microbial gene content and potential functions; Provides the blueprint confirmed by transcriptomics and proteomics | Fecal stool | Illumina NextSeq 2000/NovaSeq for shotgun; PacBio Vega or ONT PromethION for long-reads | Moderate ($$) | Reveals which microbes are present and the functional genes they encode (e.g., SCFA production, lactate utilization, vitamin biosynthesis), and how these features correlate with performance metrics such as VO2 max, lactate threshold, and recovery. |
| Metatranscriptomics | Microbial gene expression profiles; Links gene presence (genomics) to active function (proteomics and metabolomics) | Frozen or preservative-stabilized stool samples; serial sampling around endurance events | Illumina NextSeq 2000 stranded RNA-seq; optional PacBio Iso-Seq | High ($$$) | Shows which microbial genes are actively expressed under training stress, capturing dynamic pathways such as carbohydrate and SCFA metabolism that support glycogen replenishment and gut-barrier adaptation. |
| Metaproteomics | Protein abundance and enzyme activity; Bridges transcriptional activity and metabolite production | Stool protein extracts | LC-MS/MS on Orbitrap or Q-TOF | High ($$$) | Identifies microbial and host proteins produced, confirming functional shifts in enzymes, transporters, IgA, and stress proteins that reflect gut-barrier strain and host–microbe interactions. |
| Metabolomics | Small-molecule metabolites; Provides functional readout validating upstream omics layers | Stool, plasma/serum, dried-blood spots, urine; dense peri-exercise time-series | LC-MS/MS, RP-MS, GC-MS, GC×GC-MS, complemented by NMR | Low ($) | Measures a wide range of small metabolites (e.g., SCFAs, bile acids, BCAAs, acyl-carnitines) that provide real-time markers of training load, fatigue, adaptation, and overtraining risk. |
| No. | First Author (Year) | Omics Approaches | Cohort & Sport | Headline Gut-Microbiota Findings | Ref. |
|---|---|---|---|---|---|
| Immune Resilience | |||||
| 1 | Cuthbertson (2022) | 16S rRNA sequencing Metabolomics | 121 elite athletes | RTI-susceptible athletes displayed upper airway microbiome dysbiosis (reduced bacterial biomass and diversity), along with immunometabolic dysregulation in the sphingolipid pathway and reduced circulating memory T-regulatory cells, indicating a perturbed mucosal microbial–immune ecosystem linked to lowered immune resilience. | [26] |
| Inflammation and recovery | |||||
| 2 | Clarke (2014) | 16S rRNA sequencing | 40 rugby players, 46 controls | Early landmark showing athletes’ higher α-diversity that positively correlates with creatine kinase and lower systemic inflammation, and Akkermansia abundance inversely correlates with metabolic disorders. | [31] |
| 3 | Barton (2017) | 16S rRNA sequencing Metabolomics | 40 pro rugby players, 46 controls | Athletes showed greater gut microbial diversity and enriched SCFA-producing pathways (Roseburia), with higher fecal SCFAs linked to protein/fiber intake, creatine kinase, and reduced systemic inflammation, distinguishing them metabolically from controls. | [24] |
| 4 | Li (2023) | 16S rRNA | 543 multi-sport athletes | Sport-type shaped ten discrete microbial subgroups; a Prevotella-centric enterotype tracked systemic inflammation markers, and sex modified the exercise–microbiota relationship. Created athlete microbiota catalog; identified inflammation-risk signatures per sport. | [32] |
| GI integrity | |||||
| 5 | Keohane (2019) | 16S rRNA sequencing | 4 well-trained male athletes completing a trans-Atlantic, ultra-endurance rowing race | Ultra-endurance rowing increased gut α-diversity, abundance of certain species such as Prevotella copri, Dorea longicatena, and some butyrate producing species (Roseburia and Subdoligranulum) that linked to reduced inflammation and strengthened gut barrier function. | [34] |
| 6 | Hintikka (2022) | 16S rRNA sequencing Metabolomics | 27 elite skiers, 27 controls | Skiers exhibited a low-diversity microbiome characterized by depletion of mucin-degrading taxa (Akkermansia, Bacteroides, Bifidobacterium, and Ruminococcus), possibly reflecting GI stress during heavy training; Butyricicoccus was positively associated with higher HDL and HDL2 cholesterol and larger HDL particle size, suggesting its potential as a beneficial probiotic. | [35] |
| Endurance metabolism | |||||
| 7 | Petersen (2017) | Metagenomics Metatranscriptomics | 33 competitive cyclists (18 professional, 15 amateur) | High-fitness athletes enriched for Prevotella species; Prevotella-dominant microbiomes associated with enhanced branched-chain amino acid and carbohydrate metabolism; exercise volume correlated with microbiota composition. | [20] |
| 8 | Scheiman (2019) | 16S rRNA sequencing Metagenomics Metatranscriptomics Metabolomics | 15 marathoners (pre/post), mouse validation | Post-race bloom of Veillonella atypica; strain isolated and shown to enhance treadmill endurance in mice via lactate-to-propionate metabolism | [14] |
| 9 | Martin (2025) | Metagenomics Metaproteomics Metabolomics | 18 elite footballers/cyclists | Despite low overall diversity, athletes carried a protein-rich SCFA-producing guild; transplanting these communities into mice boosted muscle glycogen and insulin sensitivity, underscoring a causal link between microbial SCFA output and energy metabolism. | [21] |
| Strength, power and body composition | |||||
| 10 | Mörkl (2017) | 16S rRNA sequencing | 20 female athletes, 86 comparators | Athletes exhibited the highest gut α-diversity; diversity mirrored leanness and anti-inflammatory markers, highlighting exercise as a driver of beneficial gut ecology across body-composition strata. | [38] |
| 11 | Aya (2025) | Metagenomics Metabolomics Lipidomics | 16 elite weightlifters, 13 cyclists | Weightlifters carried carnitine-utilizing guilds (enriched Alistipes, Blautia) and branched-chain-amino-acid gene modules, correlating with peak power; cyclists showed Prevotella/Veillonella-driven SCFA & triglyceride-hydrolysis pathways, tracking VO2max. | [16] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Li, Y.; Wang, Y.; Chen, J.; Liu, Y. The Athlete Gut Microbiome: A Narrative Review of Multi-Omics Insights and Next-Generation Probiotic Strategies. Nutrients 2025, 17, 3260. https://doi.org/10.3390/nu17203260
Li Z, Li Y, Wang Y, Chen J, Liu Y. The Athlete Gut Microbiome: A Narrative Review of Multi-Omics Insights and Next-Generation Probiotic Strategies. Nutrients. 2025; 17(20):3260. https://doi.org/10.3390/nu17203260
Chicago/Turabian StyleLi, Zhiwei, Youqiang Li, Yufei Wang, Jinjin Chen, and Yilan Liu. 2025. "The Athlete Gut Microbiome: A Narrative Review of Multi-Omics Insights and Next-Generation Probiotic Strategies" Nutrients 17, no. 20: 3260. https://doi.org/10.3390/nu17203260
APA StyleLi, Z., Li, Y., Wang, Y., Chen, J., & Liu, Y. (2025). The Athlete Gut Microbiome: A Narrative Review of Multi-Omics Insights and Next-Generation Probiotic Strategies. Nutrients, 17(20), 3260. https://doi.org/10.3390/nu17203260







