Arctigenin from Saussurea medusa Maxim. Targets the PI3K/AKT Pathway to Inhibit Hepatocellular Carcinoma Proliferation and Induces Apoptosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Cell
2.1.2. Reagents and Instruments
2.1.3. Experimental Animals
2.2. Network Pharmacology
2.3. Molecular Docking
2.4. Molecular Dynamics (MD) Simulation
2.5. In Vitro Studies
2.5.1. MTT Assay for Cell Inhibition
2.5.2. Flow Cytometry for Apoptosis Detection
2.5.3. Western Blotting Analysis of Protein Expression
2.6. In Vivo Studies
2.6.1. Preparation of the Model
2.6.2. Grouping and Administration
2.6.3. Observation Indicators
2.7. Statistical Analysis
3. Results
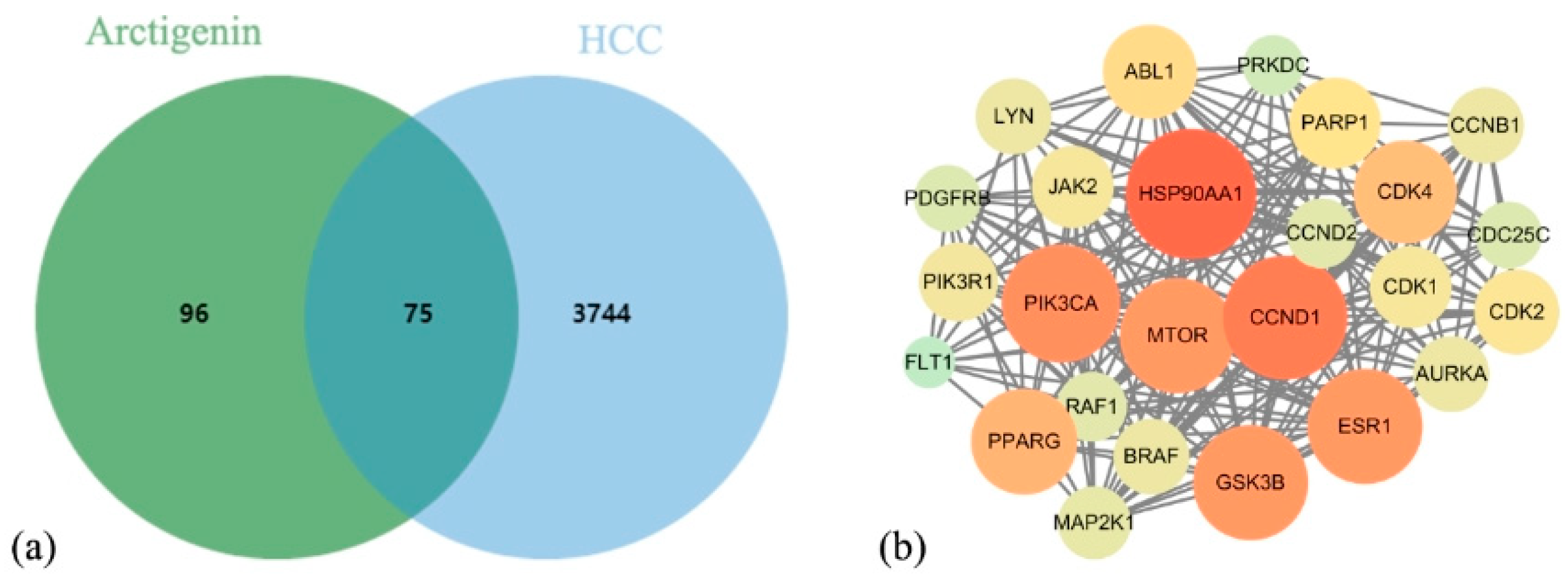
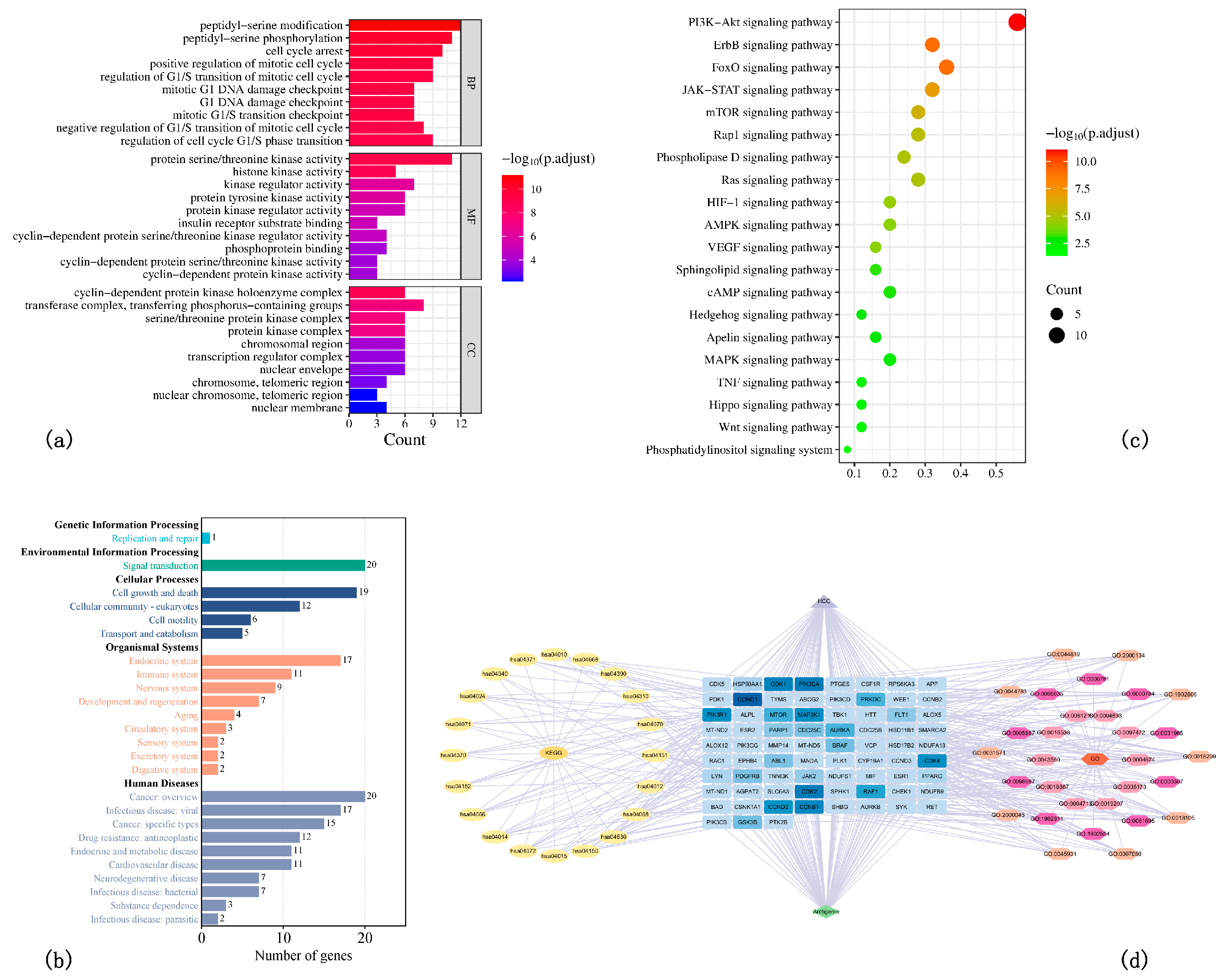
3.1. Network Pharmacology Results
3.2. Molecular Docking Results
3.3. Molecular Dynamics Simulation Results
3.4. In Vitro Results
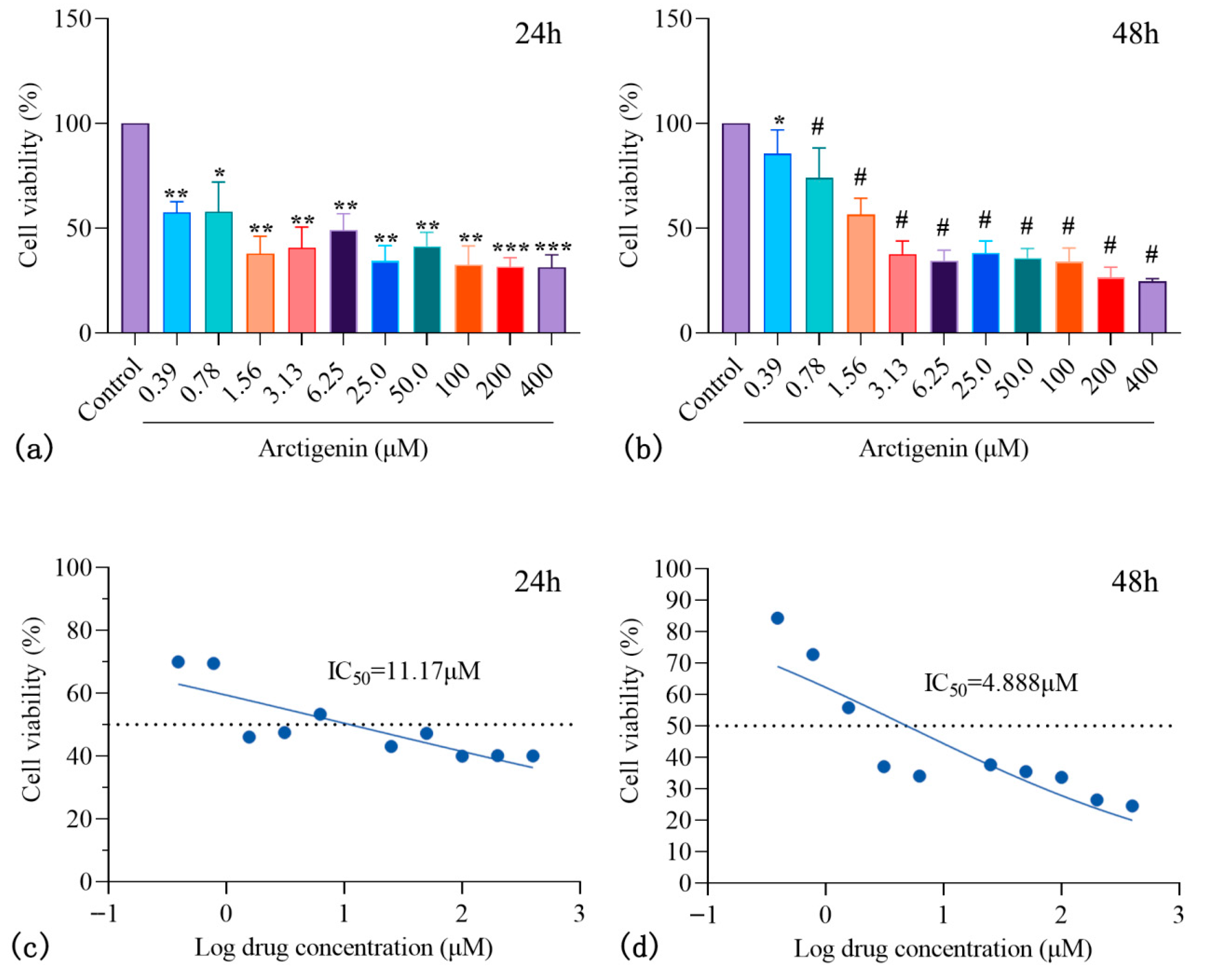
3.4.1. ARC Inhibits HepG2 Proliferation
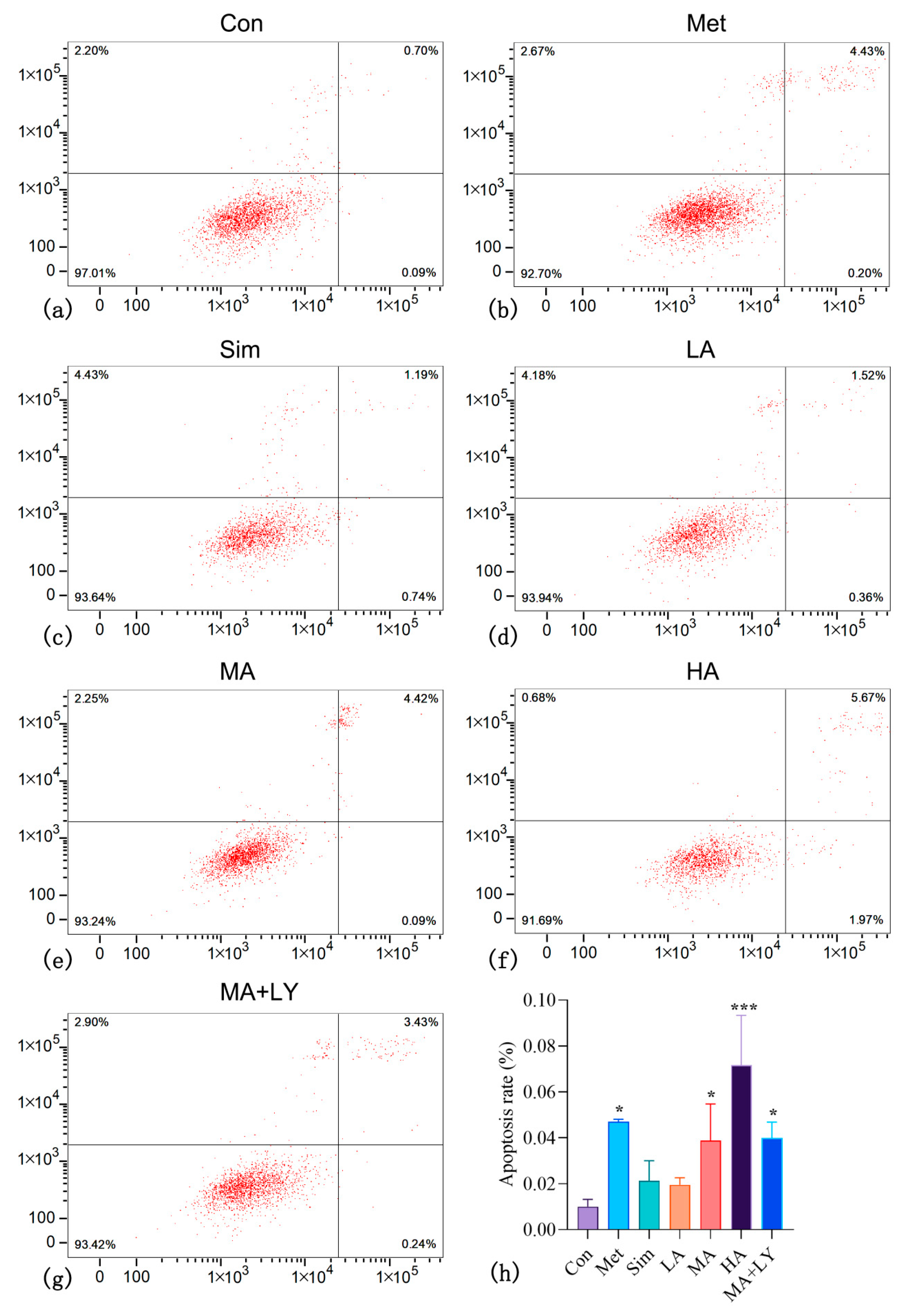
3.4.2. The Effect of ARC on HepG2 Cell Apoptosis
3.4.3. The Effect of ARC on the Expression of PI3K/AKT Pathway-Related Proteins in HepG2
3.5. In Vivo Results
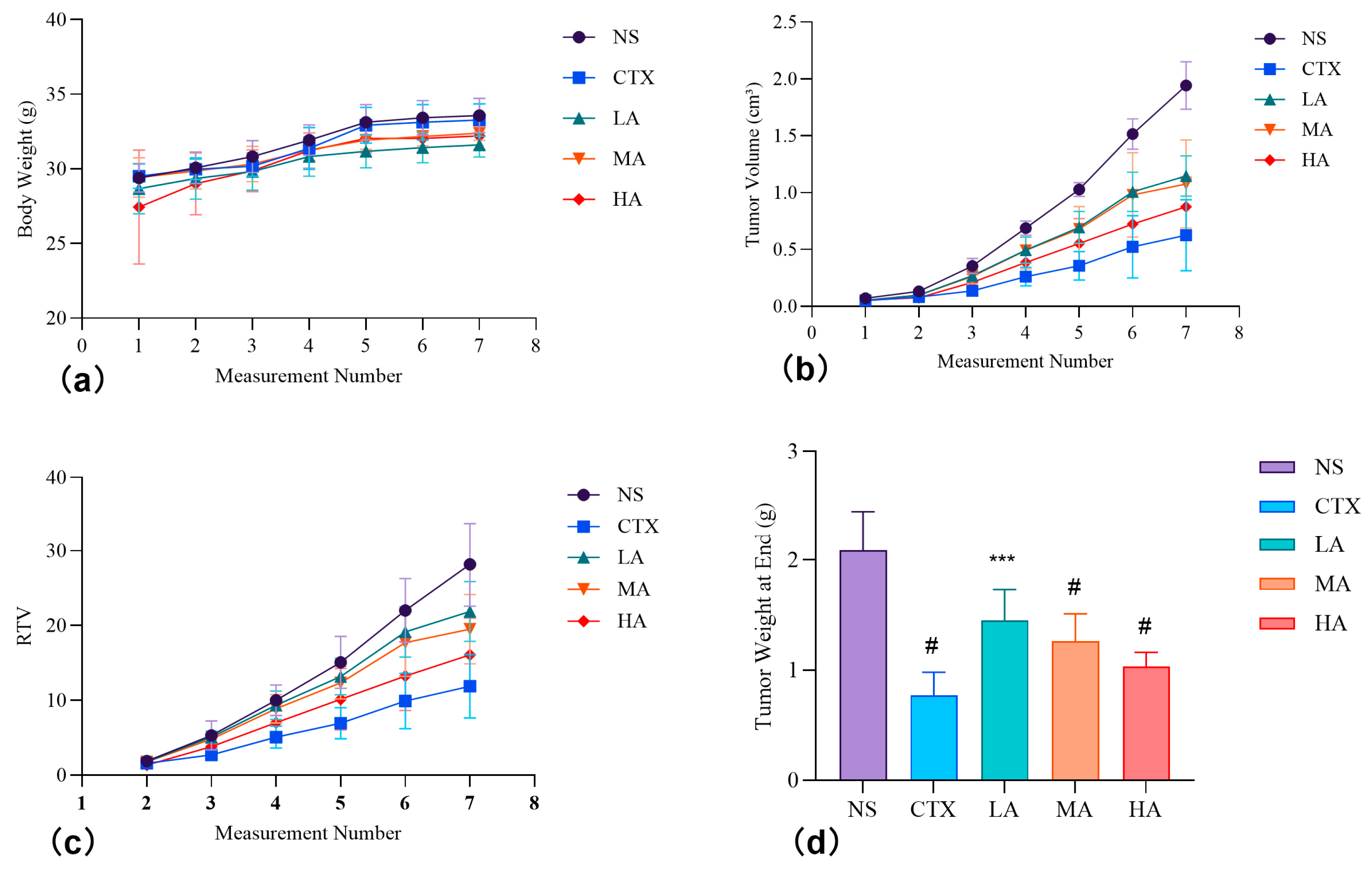
3.5.1. Effect on the Body Weight of Test Animals
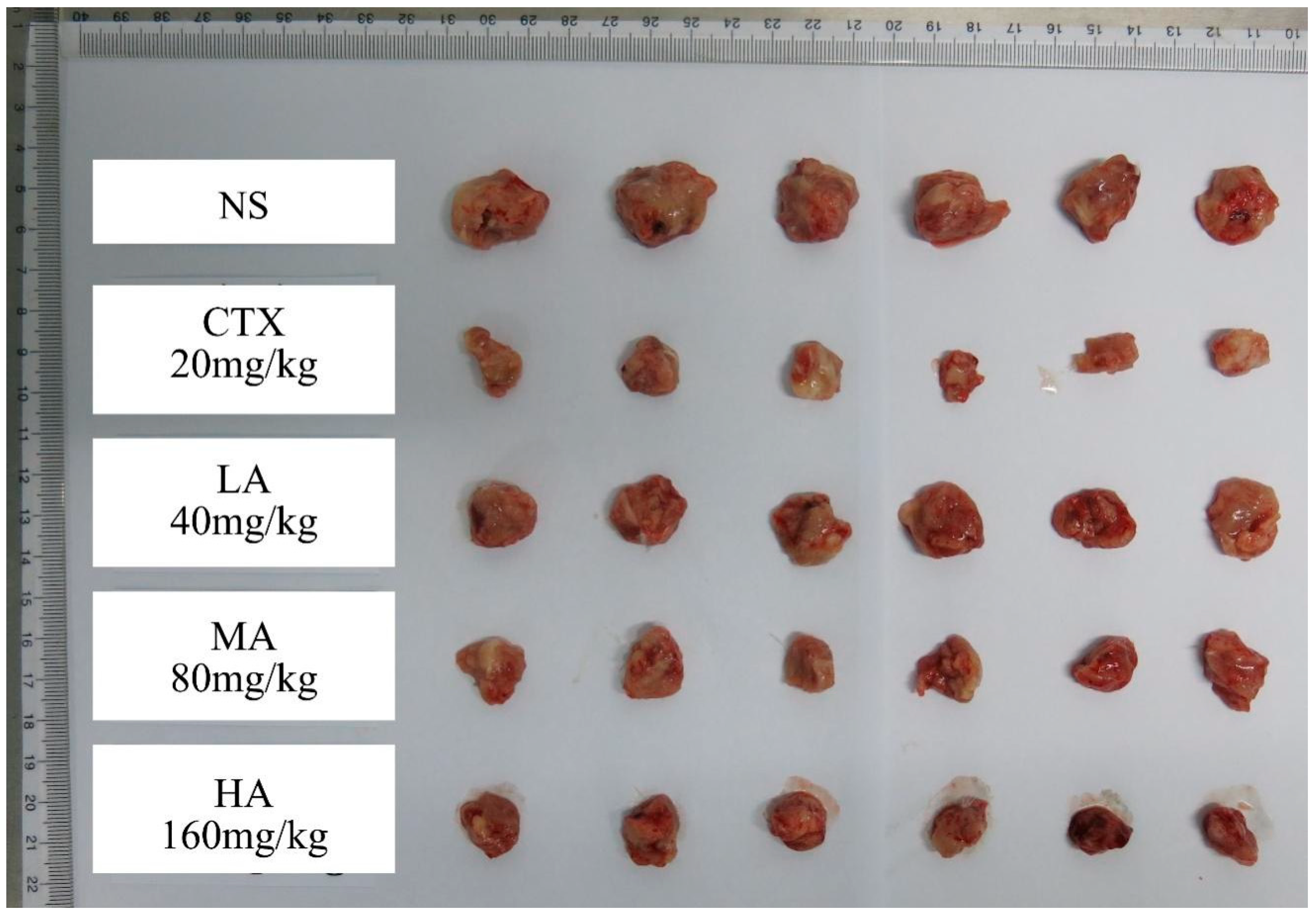
3.5.2. Changes in Tumor Volume and Weight, and Tumor Inhibition Effects
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AKT | Protein Kinase B |
| Annexin V-FITC/PI | Fluorescein Isothiocyanate Annexin V And Propidium Iodide |
| AP-1 | Activator Protein 1 |
| ARC | Arctigenin |
| AURKA | Aurora Kinase A |
| BCA | Bicinchoninic Acid Protein Assay |
| BP | Biological Process |
| BRAF | Serine/Threonine-Protein Kinase |
| CC | Cellular Component |
| CCNB1 | G2/Mitotic-Specific Cyclin-B1 |
| CCND1 | G1/S-Specific Cyclin-D1 |
| CCND2 | G1/S-Specific Cyclin-D2 |
| CDK1 | Cyclin-Dependent Kinase 1 |
| CDK2 | Cyclin-Dependent Kinase 2 |
| CDK4 | Cyclin-Dependent Kinase 4 |
| CMC-Na | Carboxymethylcellulose Sodium |
| Con | Blank Control Group (no treatment) |
| COU | Coulomb Force |
| CTX | Cyclophosphamide |
| EMT | Epithelial–Mesenchymal Transition |
| ErbB | Erythroblastic Leukemia Viral Oncogene Homolog |
| FoxO | Forkhead Box O |
| GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase |
| GM-CSF | Granulocyte-Macrophage Colony-Stimulating Factor |
| GO | Gene Ontology |
| GSK3B | Glycogen Synthase Kinase 3 Beta |
| HA | High-Dose Arctigenin Treated Group |
| HCC | Hepatocellular Carcinoma |
| HepG2 | Human Hepatocellular Carcinoma Cell Line |
| HPLC | High Performance Liquid Chromatography |
| IARC | International Agency For Research On Cancer |
| IC50 | Half Inhibitory Concentration |
| JAK-STAT | Janus Kinase-Signal Transducer And Activator Of Transcription |
| KEGG | Kyoto Encyclopedia Of Genes And Genomes |
| LA | Low-Dose Arctigenin Treated Group |
| MA | Medium-Dose Arctigenin Treated Group |
| MA + LY | Medium-Dose Arctigenin + Ly294002 Treated Group |
| MAP2K1 | Mitogen-Activated Protein Kinase Kinase 1 |
| MAPK | Mitogen-Activated Protein Kinases |
| MD | Molecular Dynamics |
| Met | Positive Control Group (Treated with Metformin) |
| MF | Molecular Function |
| MM | Molecular Interaction Force |
| mTOR | Serine/Threonine-Protein Kinase |
| NC | Network Centrality |
| NF-κB | Nuclear Factor Kappa-Light-Chain-Enhancer Of Activated B Cells |
| NS | Model Control |
| PB | Polar Solvation Energy |
| PI3K | Phosphoinositol-3 Kinase |
| PIK3CA | Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha |
| PIK3R1 | Phosphoinositol-3-Kinase Regulatory Subunit Alpha |
| PPI | Protein–Protein Interaction |
| RAF1 | Rapidly Accelerated Fibrosarcoma Kinase |
| RMSD | Root Mean Square Deviation |
| ROS | Reactive Oxygen Species |
| RTV | Relative Tumor Volume |
| SA | Nonpolar Solvation Energy |
| Sim | Positive Control Group (Treated with Simvastatin) |
| SMILES | Simplified Molecular-Input Line-Entry System |
| SMM | Saussurea medusa Maxim. |
| STAT3 | Signal Transducer And Activator Of Transcription 3 |
| T/C | Relative Tumor Proliferation Rate |
| TACE | Transcatheter Arterial Chemoembolization |
| TGF-β | Transforming Growth Factor Beta |
| TSLP | Thymic Stromal Lymphopoietin |
| TV | Tumor Volume |
| -TΔS | Entropy Change |
| VDW | Van Der Waals Force |
| ΔG | Binding Free Energy |
| ΔH | Enthalpy Change |
Appendix A
| Group | 1st Measurement | 2nd Measurement | 3rd Measurement | 4th Measurement | 5th Measurement | 6th Measurement | 7th Measurement |
|---|---|---|---|---|---|---|---|
| NS | 29.4 ± 1.0 | 30.1 ± 1.1 | 30.8 ± 1.1 | 31.9 ± 1.0 | 33.1 ± 1.2 | 33.4 ± 1.2 | 33.6 ± 1.2 |
| CTX | 29.5 ± 0.8 | 30.0 ± 0.7 | 30.2 ± 0.7 | 31.4 ± 1.4 | 32.9 ± 1.2 | 33.1 ± 1.2 | 33.3 ± 1.1 |
| LA | 28.7 ± 1.7 | 29.4 ± 1.4 | 29.8 ± 1.3 | 30.8 ± 1.3 | 31.2 ± 1.1 * | 31.4 ± 1.0 * | 31.6 ± 0.8 * |
| MA | 29.4 ± 1.3 | 29.9 ± 1.2 | 30.3 ± 1.2 | 31.3 ± 0.7 | 31.9 ± 0.6 | 32.2 ± 0.6 | 32.4 ± 0.5 |
| HA | 27.5 ± 3.8 * | 29.0 ± 2.1 | 29.9 ± 1.4 | 31.2 ± 1.2 | 32.1 ± 1.0 | 32.1 ± 1.0 | 32.2 ± 0.9 |
| Group | 1st Measurement | 2nd Measurement | 3rd Measurement | 4th Measurement | 5th Measurement | 6th Measurement | 7th Measurement |
|---|---|---|---|---|---|---|---|
| NS | 0.072 ± 0.021 | 0.132 ± 0.023 | 0.355 ± 0.068 | 0.689 ± 0.064 | 1.028 ± 0.060 | 1.516 ± 0.134 | 1.944 ± 0.210 |
| CTX | 0.052 ± 0.010 | 0.082 ± 0.020 | 0.137 ± 0.032 | 0.261 ± 0.083 # | 0.357 ± 0.124 # | 0.524 ± 0.273 # | 0.626 ± 0.312 # |
| LA | 0.053 ± 0.010 | 0.099 ± 0.034 | 0.270 ± 0.051 | 0.495 ± 0.115 | 0.695 ± 0.140 ** | 1.008 ± 0.172 # | 1.148 ± 0.177 # |
| MA | 0.055 ± 0.010 | 0.098 ± 0.028 | 0.264 ± 0.066 | 0.493 ± 0.164 | 0.679 ± 0.199 ** | 0.980 ± 0.372 # | 1.076 ± 0.388 # |
| HA | 0.055 ± 0.008 | 0.075 ± 0.019 | 0.212 ± 0.070 | 0.387 ± 0.150 ** | 0.554 ± 0.220 # | 0.723 ± 0.248 # | 0.876 ± 0.262 # |
| Group | 2nd Measurement | 3rd Measurement | 4th Measurement | 5th Measurement | 6th Measurement | 7th Measurement |
|---|---|---|---|---|---|---|
| NS | 1.88 ± 0.23 | 5.31 ± 1.94 | 10.02 ± 2.02 | 15.08 ± 3.49 | 22.03 ± 4.28 | 28.20 ± 5.62 |
| CTX | 1.59 ± 0.33 | 2.70 ± 0.75 | 5.08 ± 1.48 * | 6.94 ± 2.09 # | 9.92 ± 3.69 # | 11.89 ± 4.24 # |
| LA | 1.81 ± 0.38 | 5.10 ± 0.80 | 9.36 ± 1.91 | 13.17 ± 2.46 | 19.15 ± 3.36 | 21.89 ± 4.02 ** |
| MA | 1.77 ± 0.27 | 4.81 ± 0.71 | 8.88 ± 1.90 | 12.36 ± 2.17 | 17.72 ± 4.51 * | 19.50 ± 4.65 # |
| HA | 1.36 ± 0.19 | 3.82 ± 1.01 | 7.02 ± 2.54 | 10.15 ± 4.12 * | 13.26 ± 4.62 # | 16.07 ± 4.90 # |
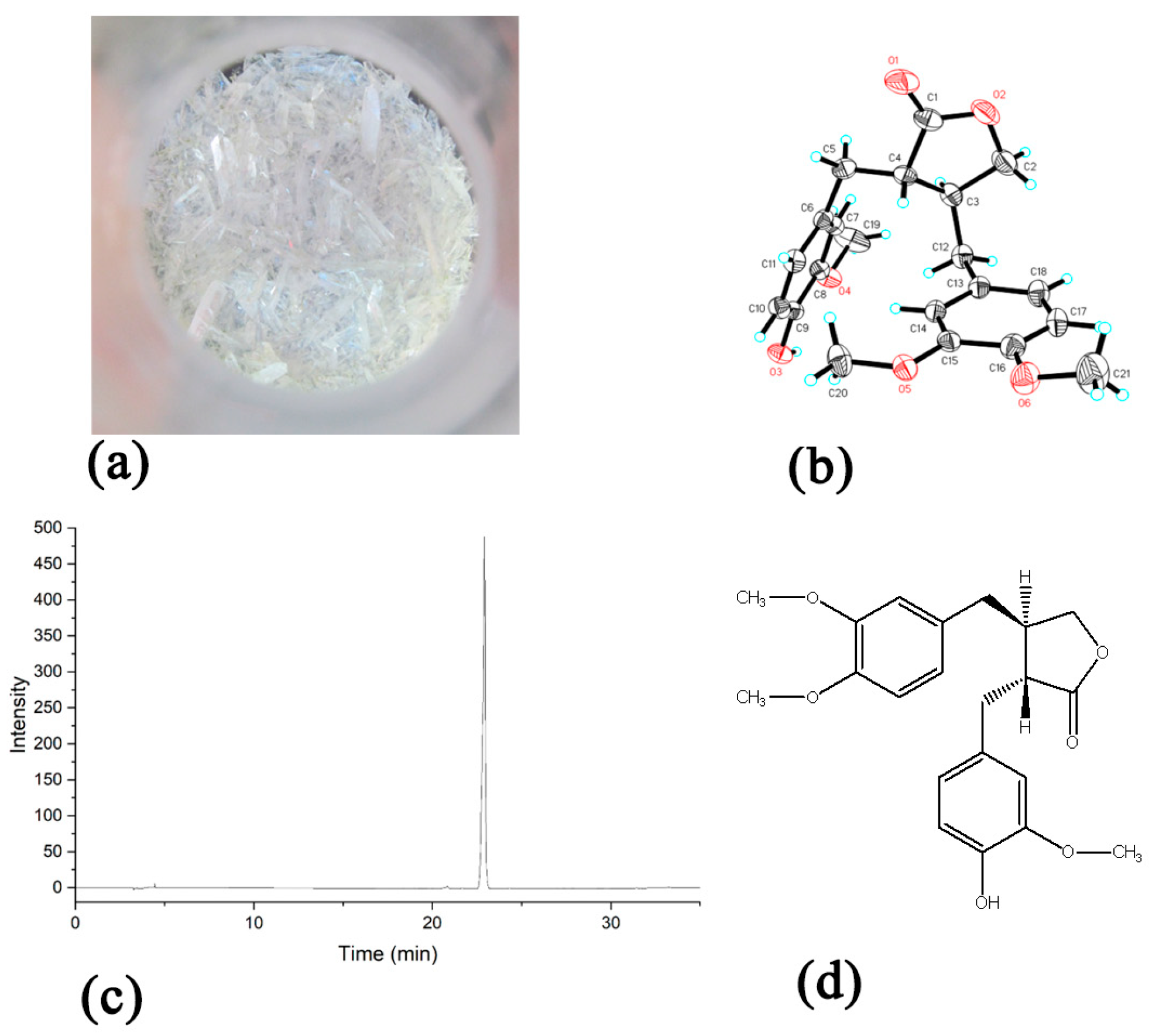

References
- Zhou, Y.; Wang, Z.; Ren, S.; Li, W. Mechanism of action of protopanaxadiol ginsenosides on hepatocellular carcinoma and network pharmacological analysis. Chin. Herb. Med. 2024, 16, 548–557. [Google Scholar] [CrossRef]
- Llovet, J.M.; Pinyol, R.; Kelley, R.K.; El-Khoueiry, A.; Reeves, H.L.; Wang, X.W.; Gores, G.J.; Villanueva, A. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nat. Cancer 2022, 3, 386–401. [Google Scholar] [CrossRef]
- Meng, L.Z.; Li, H.; Ji, Y.J.; Yu, P.; Wang, Z.Z.; Cao, L.; Shi, B.; Shao, Y.L.; Yan, J.; Gao, Y.J.; et al. Efficacy, safety, and biomarker analysis of TACE combined with lenvatinib plus sintilimab in unresectable hepatocellular carcinoma: A real-world study. Cancer Immunol. Immunother. 2024, 74, 13. [Google Scholar] [CrossRef]
- Editorial Board of Flora of China. Flora of China; Science Press: Beijing, China, 2004; Volume 20. [Google Scholar]
- Cao, J.Y.; Dong, Q.; Wang, Z.Y.; Mei, L.J.; Tao, Y.D.; Yu, R.T. Three Pairs of Novel Enantiomeric 8-O-4′ Type Neolignans from Saussurea medusa and Their Anti-inflammatory Effects In Vitro. Int. J. Mol. Sci. 2022, 23, 14062. [Google Scholar] [CrossRef]
- Li, J.; Li, X.; Ren, Y.S.; Lv, Y.Y.; Zhang, J.S.; Xu, X.L.; Wang, X.Z.; Yao, J.C.; Zhang, G.M.; Liu, Z. Elucidation of Arctigenin Pharmacokinetics and Tissue Distribution after Intravenous, Oral, Hypodermic and Sublingual Administration in Rats and Beagle Dogs: Integration of In Vitro and In Vivo Findings. Front. Pharmacol. 2017, 8, 376. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Yang, M.; Zuo, Z. Overview of the anti-inflammatory effects, pharmacokinetic properties and clinical efficacies of arctigenin and arctiin from Arctium lappa L. Acta Pharmacol. Sin. 2018, 39, 787–801. [Google Scholar] [CrossRef]
- Ikeda, M.; Sato, A.; Mochizuki, N.; Toyosaki, K.; Miyoshi, C.; Fujioka, R.; Mitsunaga, S.; Ohno, I.; Hashimoto, Y.; Takahashi, H.; et al. Phase I trial of GBS-01 for advanced pancreatic cancer refractory to gemcitabine. Cancer Sci. 2016, 107, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Jiang, X.X.; Zeng, L.P.; Liu, L.; Zhou, H.; Liu, Y.B. Arctigenin, a Natural Lignan Compound, Induces Apoptotic Death of Hepatocellular Carcinoma Cells via Suppression of PI3-K/Akt Signaling. Anti Tumor Pharm. 2015, 5, 430–435. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Hsieh, P.-L.; Liao, Y.-W.; Peng, C.-Y.; Yu, C.-C.; Lu, M.-Y. Arctigenin Reduces Myofibroblast Activities in Oral Submucous Fibrosis by LINC00974 Inhibition. Int. J. Mol. Sci. 2019, 20, 1328. [Google Scholar] [CrossRef]
- Lu, Z.; Cao, S.b.; Zhou, H.b.; Hua, L.; Zhang, S.s.; Cao, J.y. Mechanism of Arctigenin-Induced Specific Cytotoxicity Against Human Hepatocellular Carcinoma Cell Lines: Hep G2 and SMMC7721. PLoS ONE 2015, 10, e0125727. [Google Scholar] [CrossRef]
- Maxwell, T.; Chun, S.-Y.; Lee, K.-S.; Kim, S.; Nam, K.-S. The anti-metastatic effects of the phytoestrogen arctigenin on human breast cancer cell lines regardless of the status of ER expression. Int. J. Oncol. 2017, 50, 727–735. [Google Scholar] [CrossRef]
- Hsieh, C.-J.; Kuo, P.-L.; Hsu, Y.-C.; Huang, Y.-F.; Tsai, E.-M.; Hsu, Y.-L. Arctigenin, a dietary phytoestrogen, induces apoptosis of estrogen receptor-negative breast cancer cells through the ROS/p38 MAPK pathway and epigenetic regulation. Free. Radic. Biol. Med. 2014, 67, 159–170. [Google Scholar] [CrossRef]
- Li, A.; Wang, J.; Wu, M.J.; Zhang, X.X.; Zhang, H.Z. The inhibition of activated hepatic stellate cells proliferation by arctigenin through G0/G1 phase cell cycle arrest: Persistent p27Kip1 induction by interfering with PI3K/Akt/FOXO3a signaling pathway. Eur. J. Pharmacol. 2015, 747, 71–87. [Google Scholar] [CrossRef]
- Xiong, R.; Lei, J.; Wang, L.; Zhang, S.p.; Liu, H.x.; Wang, H.p.; Liu, T.; Lai, X.d. Efficient analysis of adverse drug events and toxicological mechanisms of newly marketed drugs by integrating pharmacovigilance and network toxicology: Selumetinib as an example. Front. Pharmacol. 2024, 15, 1432759. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, X.Q.; Li, G.W. Integrated bioinformatics and network pharmacology to identify the therapeutic target and molecular mechanisms of Huangqin decoction on ulcerative Colitis. Sci. Rep. 2022, 12, 159. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Wu, Y.; Gu, Y.; Lv, Q.; Qi, F.F.; Gong, S.R.; Chen, X.P. Analysis of the molecular mechanism of Pudilan (PDL) treatment for COVID-19 by network pharmacology tools. Biomed. Pharmacother. 2020, 128, 110316. [Google Scholar] [CrossRef]
- Chen, H.M.; Shi, J.; Wang, Z.Y.; Tang, Y.; Yao, X.L. Study on the Mechanism of Chrysanthemum-Buddleja Officinalis Herb Pair in the Treatment of Corneal Injury Based on Network Pharmacology and Molecular docking. Clin. J. Tradit. Chin. Med. 2023, 35, 302–309. [Google Scholar] [CrossRef]
- Sun, J.G.; Li, H.J.; Bao, W.P.; Qing, Y.L.; Jiang, L.X.; Liu, D.M.; Li, D.Y.; Pan, Z.; Wei, D.X. Key ingredients and mechanism of iridoid glycoside in Gentiana straminea against rheumatoid arthritis by network pharmacology, molecular dynamics and experimental verification. Chin. Tradit. Herb. Drugs 2024, 55, 5853–5866. [Google Scholar]
- Chen, J.h.; Wu, X.k.; Yu, R.t. Unraveling the Therapeutic Mechanism of Saussurea involucrata against Rheumatoid Arthritis: A Network Pharmacology and Molecular Modeling-Based Investigation. Nutrients 2023, 15, 4294. [Google Scholar] [CrossRef]
- Vidya, N.; Vadivukkarasi, B.; Manivannan, G.; Anbarasu, K. Molecular modeling and docking studies of glutamate racemase in Vibrio vulnificus CMCP6. Silico Biol. 2008, 8, 471–483. [Google Scholar] [CrossRef]
- Jiménez, J.; Doerr, S.; Martínez-Rosell, G.; Rose, A.S.; De Fabritiis, G. DeepSite: Protein-binding site predictor using 3D-convolutional neural networks. Bioinformatics 2017, 33, 3036–3042. [Google Scholar] [CrossRef]
- Wang, Y. Study on Mechanism of Five Flavonoids Inhibiting hIAPP Aggregation and NtMGAM Activity Based on Molecular Simulation. Master’s Thesis, Chongqing University, Chongqing, China, 2020. [Google Scholar]
- Lu, T. Sobtop 1.0 (dev3.1). 2022. Available online: http://sobereva.com/soft/Sobtop/ (accessed on 15 August 2023).
- Yedjou, C.G.; Tchounwou, P.B. Oxidative Stress in Human Leukemia (HL-60), Human Liver Carcinoma (HepG2), and Human (Jurkat-T) Cells Exposed to Arsenic Trioxide. Met. Ions Biol. Med. 2006, 9, 298–303. [Google Scholar]
- Cao, P.; Huang, G.; Yang, Q.; Guo, J.; Su, Z. The effect of chitooligosaccharides on oleic acid-induced lipid accumulation in HepG2 cells. Saudi Pharm. J. 2016, 24, 292–298. [Google Scholar] [CrossRef]
- Ran, J. Effect of Metformin on Hepatocellular Carcinoma Hepg2 Cells and Its Mechanism. Ph.D. Thesis, Anhui Medical University, Hefei, China, 2020. [Google Scholar]
- Gulzar, M.; Ali, S.; Khan, F.I.; Khan, P.; Taneja, P.; Hassan, M.I. Binding mechanism of caffeic acid and simvastatin to the integrin linked kinase for therapeutic implications: A comparative docking and MD simulation studies. J. Biomol. Struct. Dyn. 2019, 37, 4327–4337. [Google Scholar] [CrossRef]
- Jiang, W.; Yang, H.; Liu, C.J.; Pan, X.M.; Yu, N.; Chen, J.H. Effect and mechanism of tilianin on cholesterol metabolism of HepG2 cells. Chin. J. Clin. Pharmacol. 2022, 38, 118–122. [Google Scholar] [CrossRef]
- Chan, J.K.; Hamilton, C.A.; Cheung, M.K.; Karimi, M.; Baker, J.; Gall, J.M.; Schulz, S.; Thorne, S.H.; Teng, N.N.; Contag, C.H. Enhanced killing of primary ovarian cancer by retargeting autologous cytokine-induced killer cells with bispecific antibodies: A preclinical study. Clin. Cancer Res. 2006, 12, 1859–1867. [Google Scholar] [CrossRef] [PubMed]
- Maiti, S.I.; Saikia, S.K. An approach for quantifying western blots: The case of signal intensity and the statistical analysis. Acta Fytotech. Zootech. 2021, 24. [Google Scholar] [CrossRef]
- Pantsar, T.; Poso, A. Binding Affinity via Docking: Fact and Fiction. Molecules 2018, 23, 1899. [Google Scholar] [CrossRef]
- Saranya, P.; Karunya, R.; Keerthi Varshini, G.; Kowsikan, K.; Prathiksha, R. In-silico docking studies of selected phytochemicals against papain like protease of SARS-CoV-2. Vegetos 2023, 36, 188–194. [Google Scholar] [CrossRef]
- Maliepaard, M.; Carree, W.; van Bussel, M.T.J. Dose selection and tolerability of anticancer agents evaluated by the European Medicines Agency in the period 2015–2020. ESMO Open 2021, 6, 100301. [Google Scholar] [CrossRef]
- Shah, M.; Rahman, A.; Theoret Marc, R.; Pazdur, R. The Drug-Dosing Conundrum in Oncology—When Less Is More. N. Engl. J. Med. 2021, 385, 1445–1447. [Google Scholar] [CrossRef]
- Ashique, S.; Bhowmick, M.; Pal, R.; Khatoon, H.; Kumar, P.; Sharma, H.; Garg, A.; Kumar, S.; Das, U. Multi drug resistance in Colorectal Cancer- approaches to overcome, advancements and future success. Adv. Cancer Biol. Metastasis 2024, 10, 100114. [Google Scholar] [CrossRef]
- Dean, M. ABC transporters, drug resistance, and cancer stem cells. J. Mammary Gland. Biol. Neoplasia 2009, 14, 3–9. [Google Scholar] [CrossRef]
- Shi, H.; Zhao, L.P.; Guo, X.L.; Fang, R.P.; Zhang, H.; Dong, G.J.; Fu, J.; Yan, F.L.; Zhang, J.F.; Ning, Z.C.; et al. Arctigenin Attenuates Breast Cancer Progression through Decreasing GM-CSF/TSLP/STAT3/β-Catenin Signaling. Int. J. Mol. Sci. 2020, 21, 6357. [Google Scholar] [CrossRef]
- Chen, Y.G.; Li, Z.W.; Wang, X.F. Where PI3K/Akt meets Smads: The crosstalk determines human embryonic stem cell fate. Cell Stem Cell 2012, 10, 231–232. [Google Scholar] [CrossRef]
- Aleksandrova, K.V.; Vorobev, M.L.; Suvorova, I.I. mTOR pathway occupies a central role in the emergence of latent cancer cells. Cell Death Dis. 2024, 15, 176. [Google Scholar] [CrossRef]
- Ferrín, G.; Guerrero, M.; Amado, V.; Rodríguez-Perálvarez, M.; De la Mata, M. Activation of mTOR Signaling Pathway in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, 1266. [Google Scholar] [CrossRef]



| Group | Administration Route | Dose (mg/kg) | Volume Per Day (ml) |
|---|---|---|---|
| NS | Gavage | - | 0.1 mL/10 g |
| CTX | Intraperitoneal | 20 | 0.1 mL/10 g |
| LA | Gavage | 40 | 0.1 mL/10 g |
| MA | Gavage | 80 | 0.1 mL/10 g |
| HA | Gavage | 160 | 0.1 mL/10 g |
| No. | Receptor | Degree of Receptor | Structure | Affinity (kcal/mol) |
|---|---|---|---|---|
| 1 | CCND1 | 28 | 6P8E | −5.726 |
| 2 | PIK3CA | 24 | 4JPS | −7.55 |
| 3 | CDK2 | 24 | 6Q4G | −6.569 |
| 4 | CDK1 | 23 | 6GUK | −8.151 |
| 5 | CCNB1 | 22 | 6GU2 | −5.78 |
| 6 | MAP2K1 | 21 | 3VVH | −6.683 |
| 7 | CDK4 | 21 | 6P8E | −5.967 |
| 8 | PIK3R1 | 21 | 5GJI | −5.96 |
| 9 | RAF1 | 18 | 6VJJ | −6.269 |
| 10 | mTOR | 16 | 4DRI | −8.309 |
| 11 | AURKA | 15 | 5OS0 | −6.143 |
| 12 | GSK3B | 11 | 1Q5K | −7.116 |
| 13 | BRAF | 11 | 8JNA | −5.562 |
| Complex | Contribution | |||||||
|---|---|---|---|---|---|---|---|---|
| ΔG | −TΔS | ΔH | MM | PB | SA | COU | VDW | |
| GSK3B | −5.30 | 12.35 | −17.66 | −52.39 | 40.71 | −5.98 | −15.03 | −37.35 |
| PIK3CA | −19.53 | 4.68 | −24.21 | −50.55 | 32.18 | −5.84 | −8.65 | −41.90 |
| Group | Start of Experiment | End of Experiment | Relative Tumor Proliferation Rate (T/C) | Tumor Weight at End (g) | Tumor Inhibition Rate (%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Number of Animals | Body Weight (g) | Tumor Volume (cm3) | Number of Animals | Body Weight (g) | Tumor Volume (cm3) | ||||
| NS | 6 | 29.4 ± 1.0 | 0.072 ± 0.021 | 6 | 33.6 ± 1.2 | 1.944 ± 0.210 | - | 2.09 ± 0.36 | - |
| CTX | 6 | 29.5 ± 0.8 | 0.052 ± 0.010 | 6 | 33.3 ± 1.1 | 0.626 ± 0.312 # | 42.2% | 0.77 ± 0.21 # | 63.0% |
| LA | 6 | 28.7 ± 1.7 | 0.053 ± 0.010 | 6 | 31.6 ± 0.8 * | 1.148 ± 0.177 # | 77.6% | 1.45 ± 0.28 *** | 30.6% |
| MA | 6 | 29.4 ± 1.3 | 0.055 ± 0.010 | 6 | 32.4 ± 0.5 | 1.076 ± 0.388 # | 69.2% | 1.26 ± 0.25 # | 39.6% |
| HA | 6 | 27.5 ± 3.8 * | 0.055 ± 0.008 | 6 | 32.2 ± 0.9 | 0.876 ± 0.262 # | 57.0% | 1.03 ± 0.13 # | 50.6% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, R.; Chen, J.; Yu, R. Arctigenin from Saussurea medusa Maxim. Targets the PI3K/AKT Pathway to Inhibit Hepatocellular Carcinoma Proliferation and Induces Apoptosis. Nutrients 2025, 17, 3151. https://doi.org/10.3390/nu17193151
Yu R, Chen J, Yu R. Arctigenin from Saussurea medusa Maxim. Targets the PI3K/AKT Pathway to Inhibit Hepatocellular Carcinoma Proliferation and Induces Apoptosis. Nutrients. 2025; 17(19):3151. https://doi.org/10.3390/nu17193151
Chicago/Turabian StyleYu, Ruitao, Jinghua Chen, and Ruixue Yu. 2025. "Arctigenin from Saussurea medusa Maxim. Targets the PI3K/AKT Pathway to Inhibit Hepatocellular Carcinoma Proliferation and Induces Apoptosis" Nutrients 17, no. 19: 3151. https://doi.org/10.3390/nu17193151
APA StyleYu, R., Chen, J., & Yu, R. (2025). Arctigenin from Saussurea medusa Maxim. Targets the PI3K/AKT Pathway to Inhibit Hepatocellular Carcinoma Proliferation and Induces Apoptosis. Nutrients, 17(19), 3151. https://doi.org/10.3390/nu17193151





