Integrated Multi-Omics Analysis Reveals the Regulatory Mechanism of Peanut Skin Procyanidins on Lipid Metabolism in High-Fat-Diet-Induced Obese Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Chemicals and Reagents
2.1.2. Instruments and Equipment
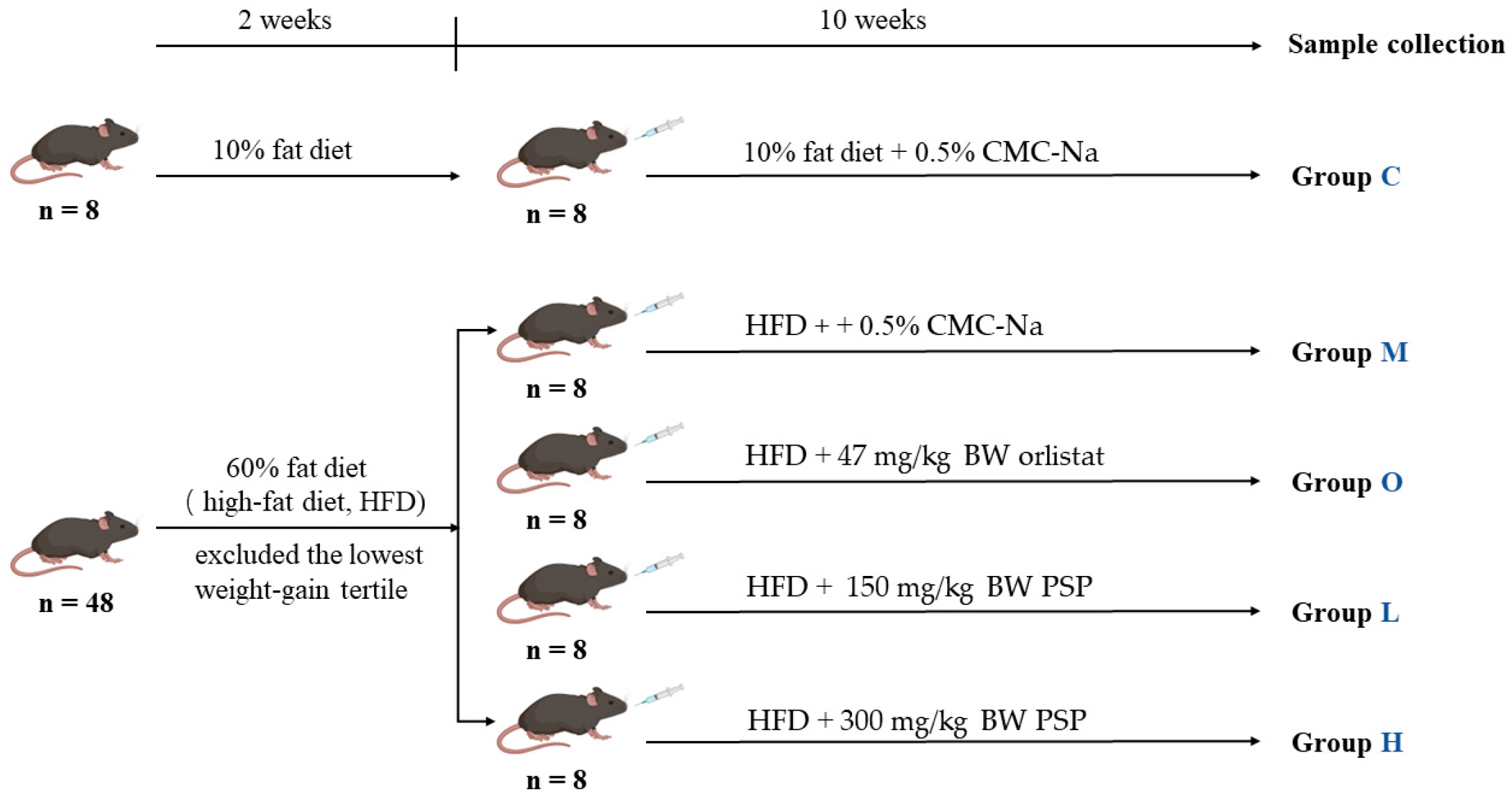
2.2. Animal Experimental Design
2.3. Biochemical Analysis
2.4. Hepatic Metabolomics Analysis
2.4.1. Sample Preparation
2.4.2. Lipidomics
2.4.3. Metabolomics
2.4.4. Data Processing
2.5. Network Pharmacology Analysis
2.6. Integrated Analysis of Metabolomics and Network Pharmacology
2.7. Quantitative Real-Time PCR
2.8. Gut Microbiota Analysis
2.9. Statistical Analysis
3. Results
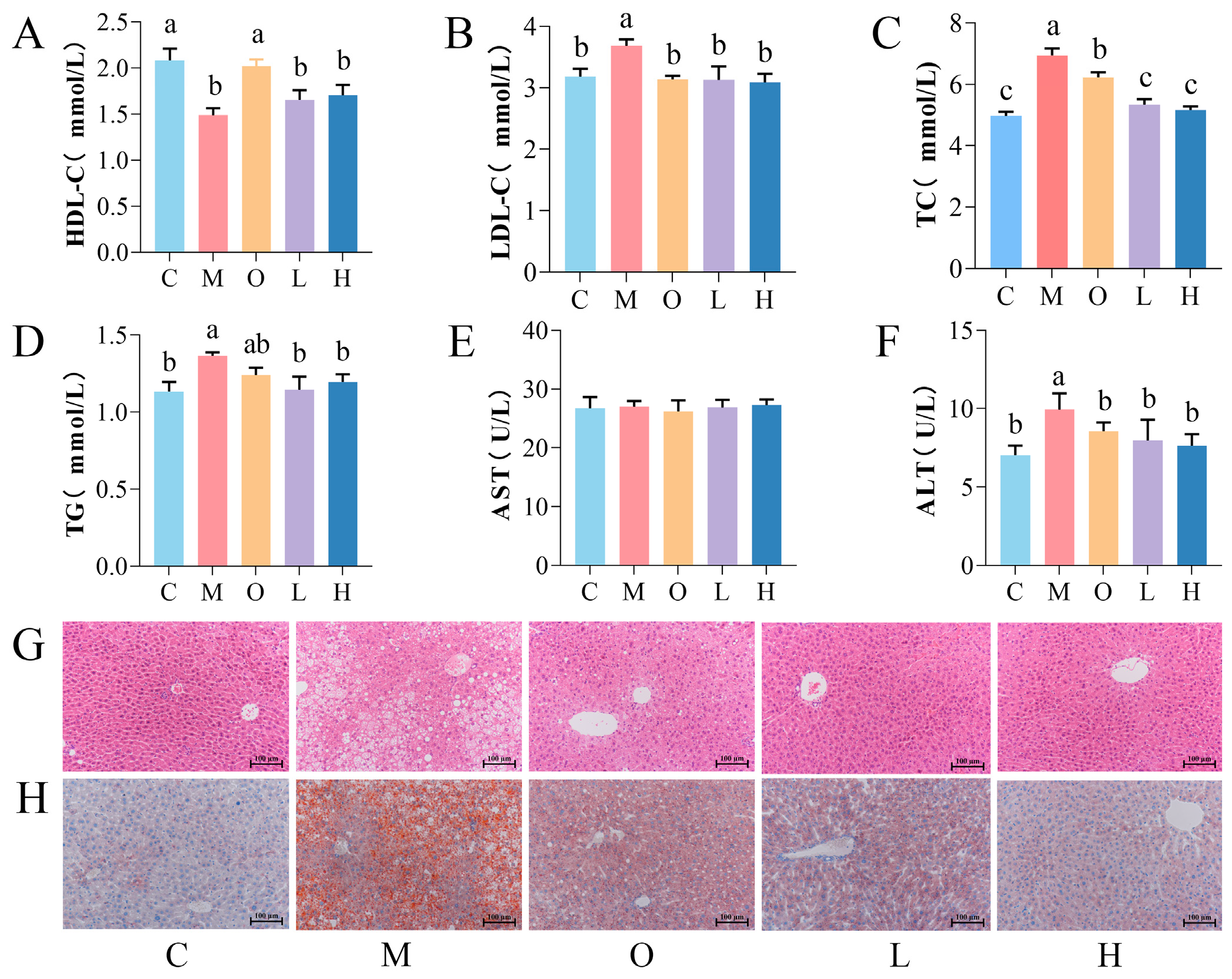
3.1. Effects of PSP on Body Weight, Lipid Profile, and Liver Function
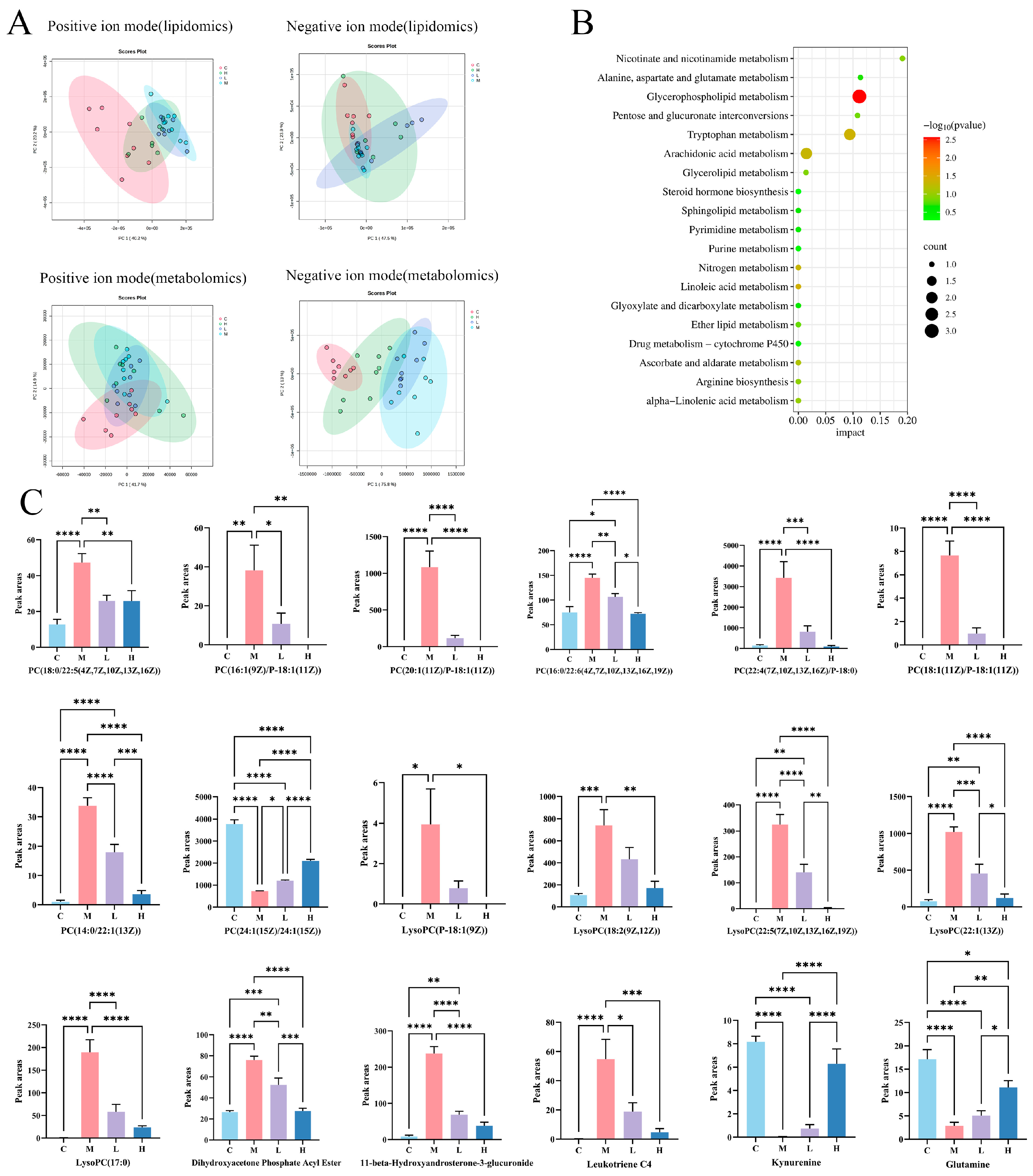
3.2. Hepatic Metabolomics Analysis
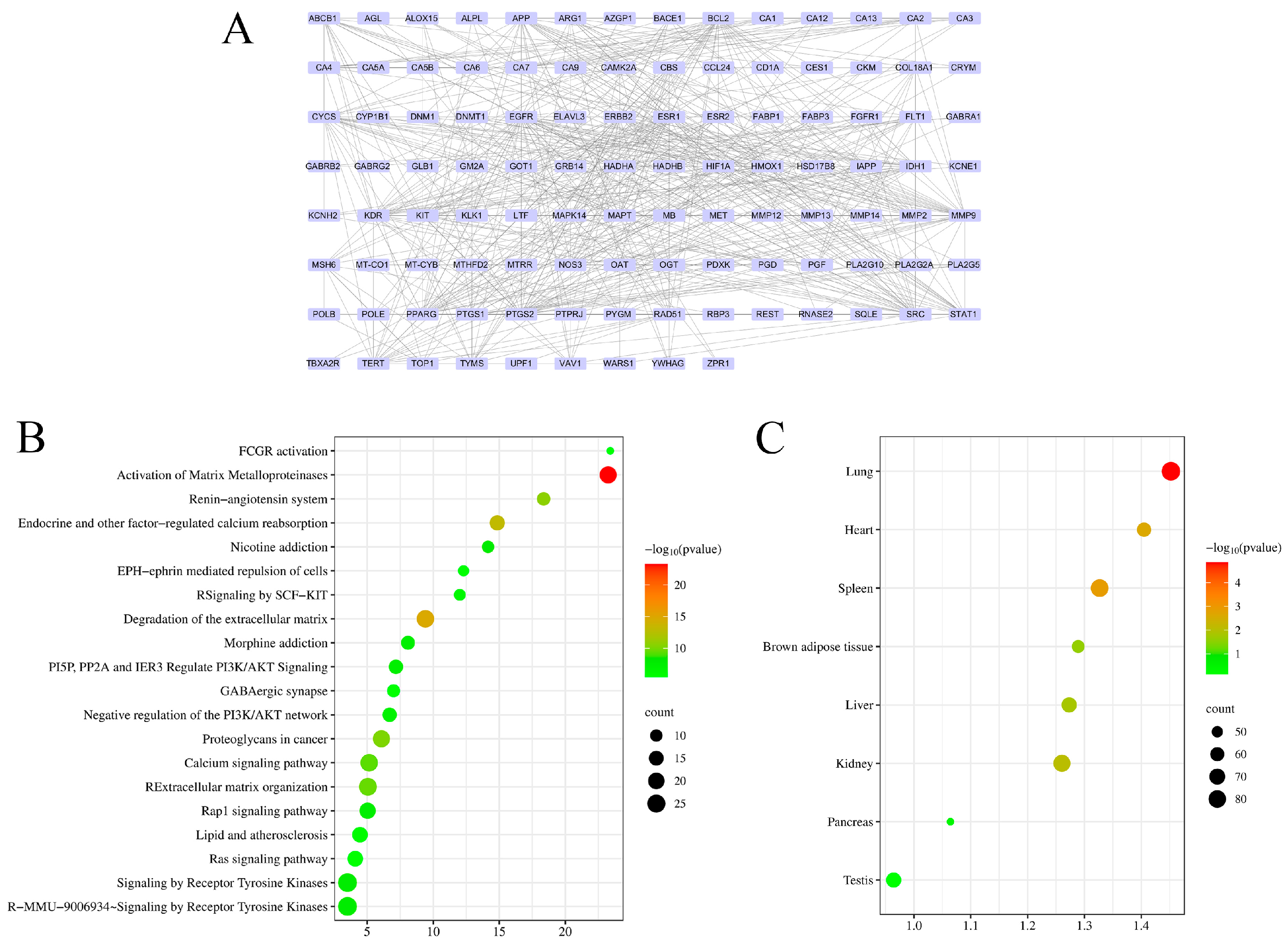
3.3. Network Pharmacology Analysis
3.4. Integrated Analysis of Metabolomics– and Network Pharmacology
3.5. Effect of PSP on Gut Microbiota Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AhR | Aryl hydrocarbon receptor |
| AMPK | Adenosine-monophosphate-activated protein kinase |
| BAT | Brown adipose tissue |
| CREG | Chemical–reaction–enzyme–gene |
| GLP-1 | Glucagon-like peptide-1 |
| HFD | High-fat diet |
| PPARγ | Peroxisome proliferator-activated receptor gamma |
| PSP | Peanut-skin-derived procyanidins |
| SCFA | Short-chain fatty acids |
Appendix A
| Feed Code | TP23300 | TP23302 |
|---|---|---|
| Protein (casein, L-cystine)/g | 276 | 194 |
| Carbohydrate (dextrin, sucrose)/g | 250 | |
| Carbohydrate (dextrin, sucrose, corn starch)/g | 673 | |
| Fat (soybean oil, lard)/g | 341 | 40 |
| Fiber (cellulose)/g | 68 | 48 |
| Mineral and vitamin mixture/g | 65 | 45 |
| Antioxidant (TBHQ)/g | 0.07 | 0.01 |
| Total/g | 1000 | 1000 |
| Gene Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Pla2g10 | GTGCAGGTGTGACGAGGAG | CACTTGGGAGAGTCCTTCTCA |
| Pla2g5 | CCAGGGGGCTTGCTAGAAC | AGCACCAATCAGTGCCATCC |
| Pla2g2a | TGGCTCAATACAGGACCAAGG | GTGGCATCCATAGAAGGCATAG |
| Cyp1b1 | CACCAGCCTTAGTGCAGACAG | GAGGACCACGGTTTCCGTTG |
| Number | HMDB | KEGG ID | Metabolites | Retention Time (s) |
|---|---|---|---|---|
| 1 | HMDB0000305 | C17276 | Vitamin A | 4.40_269.2272m/z |
| 2 | HMDB0000445 | C06768 | Alpha-tetrasaccharide | 8.59_672.2381m/z |
| 3 | HMDB0000641 | C00064 | Glutamine | 1.05_181.0387m/z |
| 4 | HMDB0000684 | C00328 | Kynurenine | 1.34_231.0733m/z |
| 5 | HMDB0000933 | C16308 | Traumatic acid | 1.95_246.1700m/z |
| 6 | HMDB0001045 | C16041 | Enkephalin L | 9.39_578.2563m/z |
| 7 | HMDB0001132 | C01185 | Nicotinic acid mononucleotide | 0.89_731.1126m/z |
| 8 | HMDB0001140 | C01545 | Octanal | 3.09_111.1175m/z |
| 9 | HMDB0001198 | C02166 | Leukotriene C4 | 8.76_606.2831m/z |
| 10 | HMDB0002937 | C10188 | Tamarixetin | 1.29_317.0681m/z |
| 11 | HMDB0003193 | C11134 | Testosterone glucuronide | 5.73_499.2103m/z |
| 12 | HMDB0004159 | C05793 | L-Urobilin | 4.12_617.3291m/z |
| 13 | HMDB0005398 | C00422 | TG(18:0/18:0/20:4(5Z,8Z,11Z,14Z)) | 18.72_893.7915m/z |
| 14 | HMDB0006061 | C05595 | 4-Hydroxyphenylacetylglutamic acid | 1.07_262.0725m/z |
| 15 | HMDB0006575 | C06372 | Lacto-N-biose I | 18.61_366.1412m/z |
| 16 | HMDB0007112 | C00165 | DG(16:0/20:4(5Z,8Z,11Z,14Z)/0:0) | 18.57_616.5089n |
| 17 | HMDB0007224 | C00165 | DG(18:1(9Z)/20:1(11Z)/0:0) | 12.44_683.5355m/z |
| 18 | HMDB0007228 | C00165 | DG(18:1(9Z)/20:4(5Z,8Z,11Z,14Z)/0:0) | 10.49_625.5188m/z |
| 19 | HMDB0007887 | C00157 | PC(14:0/22:1(13Z)) | 8.03_826.5726m/z |
| 20 | HMDB0007991 | C00157 | PC(16:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | 8.25_806.5698m/z |
| 21 | HMDB0008029 | C00157 | PC(16:1(9Z)/P-18:1(11Z)) | 9.76_722.5494m/z |
| 22 | HMDB0008055 | C00157 | PC(18:0/22:5(4Z,7Z,10Z,13Z,16Z)) | 7.09_818.6075m/z |
| 23 | HMDB0008095 | C00157 | PC(18:1(11Z)/P-18:1(11Z)) | 4.34_787.6343m/z |
| 24 | HMDB0008326 | C00157 | PC(20:1(11Z)/P-18:1(11Z)) | 11.53_778.6136m/z |
| 25 | HMDB0008653 | C00157 | PC(22:4(7Z,10Z,13Z,16Z)/P-18:0) | 10.84_802.6133m/z |
| 26 | HMDB0008816 | C00157 | PC(24:1(15Z)/24:1(15Z)) | 4.35_992.7452m/z |
| 27 | HMDB0010328 | C03033 | Tyramine glucuronide | 8.67_685.2440m/z |
| 28 | HMDB0010347 | C03033 | Octanoylglucuronide | 18.97_357.0953m/z |
| 29 | HMDB0010351 | C05643 | 11-beta-Hydroxyandrosterone-3-glucuronide | 8.86_481.2455m/z |
| 30 | HMDB0010386 | C04230 | LysoPC(18:2(9Z,12Z)) | 8.76_556.2811m/z |
| 31 | HMDB0010399 | C04230 | LysoPC(22:1(13Z)) | 10.86_577.4089n |
| 32 | HMDB0010403 | C04230 | LysoPC(22:5(7Z,10Z,13Z,16Z,19Z)) | 9.81_568.3387m/z |
| 33 | HMDB0010408 | C04230 | LysoPC(P-18:1(9Z)) | 9.08_542.3018m/z |
| 34 | HMDB0011750 | C03372 | Dihydroxyacetone phosphate acyl ester | 0.92_178.9747m/z |
| 35 | HMDB0012107 | C00550 | SM (d18:1/24:1(15Z)) | 18.61_813.6849m/z |
| 36 | HMDB0012108 | C04230 | LysoPC (17:0) | 8.90_509.3504n |
| 37 | HMDB0013046 | C02744 | Psychosine sulfate | 6.86_540.2838m/z |
| 38 | HMDB0014363 | C07663 | Moxifloxacin | 7.86_436.1453m/z |
| 39 | HMDB0014383 | C07263 | Nevirapine | 1.24_303.0645m/z |
| 40 | HMDB0014498 | C07777 | Buclizine | 5.34_467.2012m/z |
| 41 | HMDB0014700 | C12012 | Tigecycline | 7.66_566.2610m/z |
| 42 | HMDB0014878 | C07937 | Riluzole | 18.77_254.9817m/z |
| 43 | HMDB0014965 | C08052 | Cinoxacin | 1.07_283.0336m/z |
| 44 | HMDB0015090 | C07657 | Netilmicin | 8.19_475.3029n |
| 45 | HMDB0015107 | C07768 | Azelastine | 8.86_380.1543m/z |
| 46 | HMDB0015150 | C07315 | Sulfamethoxazole | 8.12_565.1193m/z |
| 47 | HMDB0015228 | C07341 | Oxamniquine | 9.45_617.3309m/z |
| 48 | HMDB0015232 | C07566 | Pimozide | 13.54_981.4670m/z |
| 49 | HMDB0015383 | C07543 | Ergonovine | 7.36_370.1516m/z |
| 50 | HMDB0029215 | C19157 | Thorium | 7.50_523.0885m/z |
| 51 | HMDB0029706 | C19180 | 2-Amino-3-methylimidazo [4,5-f]quinoline | 18.38_179.0734m/z |
| 52 | HMDB0030358 | C09041 | Aspidospermatine | 10.98_359.1728m/z |
| 53 | HMDB0032287 | C01913 | Ferrocyanide salts | 18.66_232.9280m/z |
| 54 | HMDB0033122 | C10688 | Glycosminine | 0.88_273.0425m/z |
| 55 | HMDB0035041 | C11865 | Gibberellin A51 | 9.41_297.1492m/z |
| 56 | HMDB0035798 | C09711 | Piperdial | 1.38_231.1381m/z |
| 57 | HMDB0037790 | C15588 | Polyethylene glycol | 4.32_210.1106m/z |
| 58 | HMDB0040851 | C08964 | Calenduloside E | 10.86_613.3715m/z |
| 59 | HMDB0040867 | C10373 | 5-O-Methylembelin | 19.02_309.2065m/z |
| 60 | HMDB0060669 | C16584 | p-Hydroxyfelbamate | 7.85_567.1954m/z |
| 61 | HMDB0060676 | C07493 | 10-Hydroxycarbazepine | 0.91_291.0529m/z |
| Number | Gene | Number | Gene | Number | Gene | Number | Gene | Number | Gene |
|---|---|---|---|---|---|---|---|---|---|
| 1 | PPARG | 24 | MTRR | 47 | CA1 | 70 | POLE | 93 | ITPK1 |
| 2 | MT-CYB | 25 | MGAT1 | 48 | CA5B | 71 | CA3 | 94 | RAD51 |
| 3 | KCNH2 | 26 | TERT | 49 | CA13 | 72 | PLA2G2A | 95 | OAT |
| 4 | NOS3 | 27 | STAT1 | 50 | CA7 | 73 | KLK1 | 96 | PDXK |
| 5 | MMP9 | 28 | AZGP1 | 51 | MMP12 | 74 | PTGS1 | 97 | REST |
| 6 | VEGFA | 29 | PGF | 52 | MAPT | 75 | GABRA1 | 98 | CD1A |
| 7 | ESR1 | 30 | COL18A1 | 53 | KDR | 76 | CAMK2A | 99 | ELAVL3 |
| 8 | IAPP | 31 | IDH1 | 54 | ALOX15 | 77 | HSD17B8 | 100 | MT-CO1 |
| 9 | FLT1 | 32 | ARG1 | 55 | MMP14 | 78 | KCNE1 | 101 | TOP1 |
| 10 | FGFR1 | 33 | DNMT1 | 56 | MSH6 | 79 | SQLE | 102 | DNM1 |
| 11 | HIF1A | 34 | ZPR1 | 57 | YWHAG | 80 | CYP1B1 | 103 | POLB |
| 12 | PTGS2 | 35 | GOT1 | 58 | CKM | 81 | OGT | 104 | MTHFD2 |
| 13 | HMOX1 | 36 | ABCB1 | 59 | CA9 | 82 | GABRB2 | 105 | PCMT1 |
| 14 | FABP1 | 37 | APP | 60 | KIT | 83 | CRYM | 106 | DGKZ |
| 15 | EGFR | 38 | ALPL | 61 | BACE1 | 84 | PYGM | 107 | MS4A1 |
| 16 | BCL2 | 39 | MET | 62 | CA12 | 85 | MB | 108 | GRK6 |
| 17 | ERBB2 | 40 | MMP13 | 63 | RBP3 | 86 | TYMS | 109 | VAV1 |
| 18 | MMP2 | 41 | MAPK14 | 64 | PGD | 87 | GABRG2 | 110 | AGL |
| 19 | SRC | 42 | CES1 | 65 | CCL24 | 88 | UPF1 | 111 | GLB1 |
| 20 | CBS | 43 | GRB14 | 66 | HADHB | 89 | PLA2G10 | 112 | PLA2G5 |
| 21 | HADHA | 44 | CA2 | 67 | CYCS | 90 | PTPRJ | 113 | RNASE2 |
| 22 | ESR2 | 45 | LTF | 68 | CA6 | 91 | WARS1 | 114 | TBXA2R |
| 23 | FABP3 | 46 | CA4 | 69 | CA5A | 92 | GM2A |
References
- Li, R.; Huang, X.; Liang, X.; Su, M.; Lai, K.P.; Chen, J. Integrated omics analysis reveals the alteration of gut microbe–metabolites in obese adults. Brief. Bioinform. 2021, 22, bbaa165. [Google Scholar] [CrossRef] [PubMed]
- People’s Medical Publishing House Co., Ltd. Report on Nutrition and Chronic Disease Status of Chinese Residents (2020); People’s Medical Publishing House Co., Ltd.: Beijing, China, 2020. [Google Scholar]
- Berná, G.; López-Bermudo, L.; Escudero-López, B.; Martín, F. Chapter four—We are what we eat: The role of lipids in metabolic diseases. Adv. Food Nutr. Res 2023, 105, 173–219. [Google Scholar]
- Townsend, L.K.; Steinberg, G.R. AMPK and the Endocrine Control of Metabolism. Endocr. Rev. 2023, 44, 910–933. [Google Scholar] [CrossRef]
- Yesian, A.R.; Chalom, M.M.; Knudsen, N.H.; Hyde, A.L.; Personnaz, J.; Cho, H.; Liou, Y.-H.; Starost, K.A.; Lee, C.-W.; Tsai, D.-Y. Preadipocyte IL-13/IL-13Rα1 signaling regulates beige adipogenesis through modulation of PPARγ activity. J. Clin. Investig. 2025, 135, e169152. [Google Scholar] [CrossRef]
- Rue, E.A.; Rush, M.D.; van Breemen, R.B. Procyanidins: A comprehensive review encompassing structure elucidation via mass spectrometry. Phytochem. Rev. 2018, 17, 1–16. [Google Scholar] [CrossRef]
- Liu, M.; Yun, P.; Hu, Y.; Yang, J.; Khadka, R.B.; Peng, X. Effects of grape seed proanthocyanidin extract on obesity. Obes. Facts. 2020, 13, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.J.; Huang, K.L.; Zhao, C.H.; Xu, W.T.; Sheng, Y.; Luo, Y.B.; He, X.Y. Procyanidin attenuates weight gain and modifies the gut microbiota in high fat diet induced obese mice. J. Funct. Foods 2018, 49, 362–368. [Google Scholar] [CrossRef]
- Zhu, W.; Oteiza, P.I. Proanthocyanidins at the gastrointestinal tract: Mechanisms involved in their capacity to mitigate obesity-associated metabolic disorders. Crit. Rev. Food Sci. Nutr. 2024, 64, 220–240. [Google Scholar] [CrossRef] [PubMed]
- Zineb, O.Y.; Rashwan, A.K.; Karim, N.; Lu, Y.; Tangpong, J.; Chen, W. Recent developments in procyanidins on metabolic diseases, their possible sources, pharmacokinetic profile, and clinical outcomes. Food Res. Int. 2023, 39, 5255–5278. [Google Scholar] [CrossRef]
- National data, National Bureau of Statistics. Available online: https://data.stats.gov.cn/easyquery.htm?cn=C01 (accessed on 7 March 2025).
- Zhang, C.; Yang, Q.; Yu, S.; Yu, L.; Bi, J.; Zhu, F. Purification Methods of Proanthocy Anidins from Peanut Skins. Chinese Patent CN101665482B, 5 October 2011. [Google Scholar]
- Liu, G.; Shi, A.; Wang, N.; Li, M.; He, X.; Yin, C.; Tu, Q.; Shen, X.; Tao, Y.; Wang, Q.; et al. Polyphenolic proanthocyanidin-B2 suppresses proliferation of liver cancer cells and hepatocellular carcinogenesis through directly binding and inhibiting AKT activity. Redox Biol. 2020, 37, 101701. [Google Scholar] [CrossRef]
- Liu, M.; Shen, J.X.; Zhu, X.L.; Ju, T.T.; Willing, B.P.; Wu, X.; Lu, Q.; Liu, R. Peanut skin procyanidins reduce intestinal glucose transport protein expression, regulate serum metabolites and ameliorate hyperglycemia in diabetic mice. Food Res. Int. 2023, 173, 113471. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Lv, C.; Wang, H.; Lu, Q.; Ye, M.; Zhu, X.; Liu, R. Peanut skin extract ameliorates high-fat diet-induced atherosclerosis by regulating lipid metabolism, inflammation reaction and gut microbiota in ApoE−/− mice. Food Res. Int. 2022, 154, 111014. [Google Scholar] [CrossRef]
- Rui, L. Energy metabolism in the liver. Compr Physiol. 2014, 4, 177–197. [Google Scholar] [CrossRef]
- Huang, Y.; Sulek, K.; Stinson, S.E.; Holm, L.A.; Kim, M.; Trost, K.; Hooshmand, K.; Lund, M.A.V.; Fonvig, C.E.; Juel, H.B.; et al. Lipid profiling identifies modifiable signatures of cardiometabolic risk in children and adolescents with obesity. Nat. Med. 2025, 31, 294–305. [Google Scholar] [CrossRef]
- Bertran, L.; Capellades, J.; Abelló, S.; Richart, C. Untargeted lipidomic analysis of metabolic dysfunction-associated steatohepatitis in women with morbid obesity. PLoS ONE 2025, 20, e0318557. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.; Tian, H.; Yang, X.; Feng, S.; Chen, S.; Zhong, C.; Gao, T.; Gang, X.; Liu, M. Identification of hub genes and candidate herbal treatment in obesity through integrated bioinformatic analysis and reverse network pharmacology. Sci. Rep. 2022, 12, 17113. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Zeng, Z.; Jiang, J.; Xu, A.; Li, S.; Li, Y.; Chen, Z.; Chen, W.; Zhang, J.; Bi, X. Network pharmacology analysis and experimental validation to explore the mechanism of Bushao Tiaozhi capsule (BSTZC) on hyperlipidemia. Sci. Rep. 2022, 12, 6992. [Google Scholar] [CrossRef]
- Sun, Y.-L.; Zhao, P.-P.; Zhu, C.-B.; Jiang, M.-C.; Li, X.-M.; Tao, J.-L.; Hu, C.-C.; Yuan, B. Integrating metabolomics and network pharmacology to assess the effects of quercetin on lung inflammatory injury induced by human respiratory syncytial virus. Sci. Rep. 2023, 13, 8051. [Google Scholar] [CrossRef]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef]
- Kapoor, P.; Tiwari, A.; Sharma, S.; Tiwari, V.; Sheoran, B.; Ali, U.; Garg, M. Effect of anthocyanins on gut health markers, Firmicutes-Bacteroidetes ratio and short-chain fatty acids: A systematic review via meta-analysis. Sci. Rep. 2023, 13, 1729. [Google Scholar] [CrossRef]
- Ouyang, J.; Li, X.; Liu, C.; Lu, D.; Ouyang, J.; Zhou, F.; Liu, Q.; Huang, J.; Liu, Z. Junshanyinzhen tea extract prevents obesity by regulating gut microbiota and metabolic endotoxemia in high-fat diet fed rats. Food Sci. Hum. Well 2024, 13, 2036–2047. [Google Scholar] [CrossRef]
- Herrera-Balandrano, D.D.; Chai, Z.; Hutabarat, R.P.; Beta, T.; Feng, J.; Ma, K.; Li, D.; Huang, W. Hypoglycemic and hypolipidemic effects of blueberry anthocyanins by AMPK activation: In vitro and in vivo studies. Redox Biol. 2021, 46, 102100. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Tang, Z.; Herrera-Balandrano, D.D.; Qiu, Z.; Li, B.; Yang, Y.; Huang, W. In vitro fermentation characteristics of blueberry anthocyanins and their impacts on gut microbiota from obese human. Food Res. Int. 2024, 176, 113761. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Zhang, Y.; Shen, X.; Cao, Y.; Shi, J.; Ye, X.; Chen, S. Rethinking the mechanism of the health benefits of proanthocyanidins: Absorption, metabolism, and interaction with gut microbiota. Compr. Rev. Food Sci. Food Saf. 2019, 18, 971–985. [Google Scholar] [CrossRef]
- Liu, M.; Huang, B.J.; Wang, L.; Lu, Q.; Liu, R. Peanut skin procyanidins ameliorate insulin resistance via modulation of gut microbiota and gut barrier in type 2 diabetic mice. J. Sci. Food Agric. 2022, 102, 5935–5947. [Google Scholar] [CrossRef]
- Liu-Qing, S.; Yu-Xuan, Y.; Zheng-Yi, L.; Xi-Meng, Z.; En-Lian, M.; Meng-Fei, Z.; Ying-Ying, X. Bioinformatics analysis of key pharmacological pathways of proanthocyanidins combined with allicin against atherosclerosis. In Proceedings of the Second International Conference on Biomedical and Intelligent Systems (IC-BIS2023), Xiamen, China, 28 August 2023; p. 127240N. [Google Scholar]
- Chen, W. Gut Microbial Metabolism of Peanut Skin Flavan-3-ols and the Effect of Metabolite on Glucose and Lipid Metabolism of Obese Mice. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2021. [Google Scholar]
- Huang, B.; Wang, L.; Liu, M.; Wu, X.; Lu, Q.; Liu, R. The underlying mechanism of A-type procyanidins from peanut skin on DSS-induced ulcerative colitis mice by regulating gut microbiota and metabolism. J. Food Biochem. 2022, 46, e14103. [Google Scholar] [CrossRef]
- Wen, J.-J.; Li, M.-Z.; Chen, C.-H.; Hong, T.; Yang, J.-R.; Huang, X.-J.; Geng, F.; Hu, J.-L.; Nie, S.-P. Tea polyphenol and epigallocatechin gallate ameliorate hyperlipidemia via regulating liver metabolism and remodeling gut microbiota. Food Chem. 2023, 404, 134591. [Google Scholar] [CrossRef]
- Zhao, L.; Yan, F.F.; Lu, Q.; Tang, C.E.; Wang, X.H.; Liu, R. UPLC-Q-TOF-MS and NMR identification of structurally different A-type procyanidins from peanut skin and their inhibitory effect on acrylamide. J. Sci. Food Agric. 2022, 102, 7062–7071. [Google Scholar] [CrossRef]
- Iqbal, J.; Hussain, M.M. Intestinal lipid absorption. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1183–E1194. [Google Scholar] [CrossRef]
- Han, X.; Zhao, W.; Zhou, Q.; Chen, H.; Yuan, J.; Xiaofu, Z.; Zhang, Z. Procyanidins from hawthorn (Crataegus pinnatifida) alleviate lipid metabolism disorder via inhibiting insulin resistance and oxidative stress, normalizing the gut microbiota structure and intestinal barrier, and further suppressing hepatic inflammation and lipid accumulation. Food Funct. 2022, 13, 7901–7917. [Google Scholar] [CrossRef]
- Homan, R.; Hamelehle, K.L. Phospholipase A2 relieves phosphatidylcholine inhibition of micellar cholesterol absorption and transport by human intestinal cell line Caco-21. J. Lipid Res. 1998, 39, 1197–1209. [Google Scholar] [CrossRef]
- Engelking, L.R. Chapter 60—Lipid Digestion. In Textbook of Veterinary Physiological Chemistry, 3rd ed.; Engelking, L.R., Ed.; Academic Press: Boston, MA, USA, 2015; pp. 384–389. [Google Scholar]
- Bu, K.-B.; Kim, M.; Shin, M.K.; Lee, S.-H.; Sung, J.-S. Regulation of benzo[a]pyrene-induced hepatic lipid accumulation through CYP1B1-induced mTOR-mediated lipophagy. Int. J. Mol. Sci. 2024, 25, 1324. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.L.; Zhang, L.; Foroozesh, M.; Alworth, W.L.; Jefcoate, C.R. Metabolism-based polycyclic aromatic acetylene inhibition of CYP1B1 in 10T1/2 cells potentiates aryl hydrocarbon receptor activity. Toxicol. Appl. Pharmacol. 1999, 161, 123–139. [Google Scholar] [CrossRef]
- Dong, F.; Annalora, A.J.; Murray, I.A.; Tian, Y.; Marcus, C.B.; Patterson, A.D.; Perdew, G.H. Endogenous tryptophan-derived Ah receptor ligands are dissociated from CYP1A1/1B1-dependent negative-feedback. Int. J. Tryptophan Res. 2023, 16, 11786469231182508. [Google Scholar] [CrossRef]
- Opitz, C.A.; Litzenburger, U.M.; Sahm, F.; Ott, M.; Tritschler, I.; Trump, S.; Schumacher, T.; Jestaedt, L.; Schrenk, D.; Weller, M.; et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 2011, 478, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhao, S.; Wang, J.; Shi, J.; Sun, Y.; Wang, W.; Ning, G.; Hong, J.; Liu, R. Grape seed proanthocyanidin extract ameliorates inflammation and adiposity by modulating gut microbiota in high-fat diet mice. Mol. Nutr. Food Res. 2017, 61, 1601082. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.S.; Cho, C.H.; Yun, M.S.; Jang, S.J.; You, H.J.; Kim, J.-h.; Han, D.; Cha, K.H.; Moon, S.H.; Lee, K.; et al. Akkermansia muciniphila secretes a glucagon-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice. Nat. Microbiol. 2021, 6, 563–573. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The healthy human microbiome. Genome Med. 2016, 8, 51. [Google Scholar] [CrossRef]
- Nie, Q.; Luo, X.; Wang, K.; Ding, Y.; Jia, S.; Zhao, Q.; Li, M.; Zhang, J.; Zhuo, Y.; Lin, J.; et al. Gut symbionts alleviate MASH through a secondary bile acid biosynthetic pathway. Cell 2024, 187, 2717–2734.e2733. [Google Scholar] [CrossRef]
- Lukovac, S.; Belzer, C.; Pellis, L.; Keijser Bart, J.; de Vos Willem, M.; Montijn Roy, C.; Roeselers, G. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral Lipid metabolism and histone acetylation in mouse gut organoids. mBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Perez, V.M.; Gabell, J.; Behrens, M.; Wase, N.; DiRusso, C.C.; Black, P.N. Deletion of fatty acid transport protein 2 (FATP2) in the mouse liver changes the metabolic landscape by increasing the expression of PPARα-regulated genes. J. Biol. Chem. 2020, 295, 5737–5750. [Google Scholar] [CrossRef]
- Redondo-Castillejo, R.; Garcimartín, A.; Hernández-Martín, M.; López-Oliva, M.E.; Bocanegra, A.; Macho-González, A.; Bastida, S.; Benedí, J.; Sánchez-Muniz, F.J. Proanthocyanidins: Impact on gut microbiota and intestinal action mechanisms in theprevention and treatment of metabolic syndrome. Int. J. Mol. Sci. 2023, 24, 5369. [Google Scholar] [CrossRef]
- Delgado, T.C.; de las Heras, J.; Martínez-Chantar, M.L. Understanding gut-liver axis nitrogen metabolism in Fatty Liver Disease. Front. Endocrinol. 2022, 13, 1058101. [Google Scholar] [CrossRef]
- Pierzynowski, S.; Pierzynowska, K. Alpha-ketoglutarate, a key molecule involved in nitrogen circulation in both animals and plants, in the context of human gut microbiota and protein metabolism. Adv. Med. Sci. 2022, 67, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Sprenger, N.; Yap, I.; Wang, Y.; Bibiloni, R.; Rochat, F.; Rezzi, S.; Cherbut, C.; Kochhar, S.; Lindon, J.; et al. Panorganismal gut microbiome-host metabolic crosstalk. J. Proteome Res. 2009, 8, 2090–2105. [Google Scholar] [CrossRef]
- Gong, S.; Ma, J.; Li, F.; Li, T.; Liu, G.; Tan, B.; Huang, X.; Yin, J.; Yin, Y. Recent advances in the interaction between gut microbiota/metabolites and host lipid metabolism. Sci. Sin. Vitae 2021, 51, 375–383. [Google Scholar] [CrossRef]
- Won, T.H.; Arifuzzaman, M.; Parkhurst, C.N.; Miranda, I.C.; Zhang, B.; Hu, E.; Kashyap, S.; Letourneau, J.; Jin, W.-B.; Fu, Y.; et al. Host metabolism balances microbial regulation of bile acid signalling. Nature 2025, 638, 216–224. [Google Scholar] [CrossRef]
- Wang, P.; Wu, P.-F.; Wang, H.-J.; Liao, F.; Wang, F.; Chen, J.-G. Gut microbiome-derived ammonia modulates stress vulnerability in the host. Nat. Metab. 2023, 5, 1986–2001. [Google Scholar] [CrossRef]
- Yang, H.; Wang, T.; Qian, C.; Wang, H.; Yu, D.; Shi, M.; Fu, M.; Liu, X.; Pan, M.; Rong, X.; et al. Gut microbial-derived phenylacetylglutamine accelerates host cellular senescence. Nat. Aging 2025, 5, 401–418. [Google Scholar] [CrossRef]
- Xiao, L.; Tang, R.; Wang, J.; Wan, D.; Yin, Y.; Xie, L. Gut microbiota bridges the iron homeostasis and host health. Sci. China Life Sci. 2023, 66, 1952–1975. [Google Scholar] [CrossRef] [PubMed]
- Mouskeftara, T.; Deda, O.; Papadopoulos, G.; Chatzigeorgiou, A.; Gika, H. Lipidomic analysis of liver and adipose tissue in a high-fat diet-induced non-alcoholic fatty liver disease mice model reveals alterations in lipid metabolism by weight loss and aerobic exercise. Molecules 2024, 29, 1494. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yang, Q.; Zhou, J. Mechanisms of abnormal lipid metabolism in the pathogenesis of disease. Int. J. Mol. Sci. 2024, 25, 8465. [Google Scholar] [CrossRef] [PubMed]
- Galatou, E.; Mourelatou, E.; Hatziantoniou, S.; Vizirianakis, I.S. Nonalcoholic steatohepatitis (NASH) and atherosclerosis: Explaining their pathophysiology, association and the role of incretin-based drugs. Antioxidants 2022, 11, 1060. [Google Scholar] [CrossRef]
- Dutta, A.; Sharma-Walia, N. Curbing Lipids: Impacts ON Cancer and Viral Infection. Int. J. Mol. Sci. 2019, 20, 644. [Google Scholar] [CrossRef]
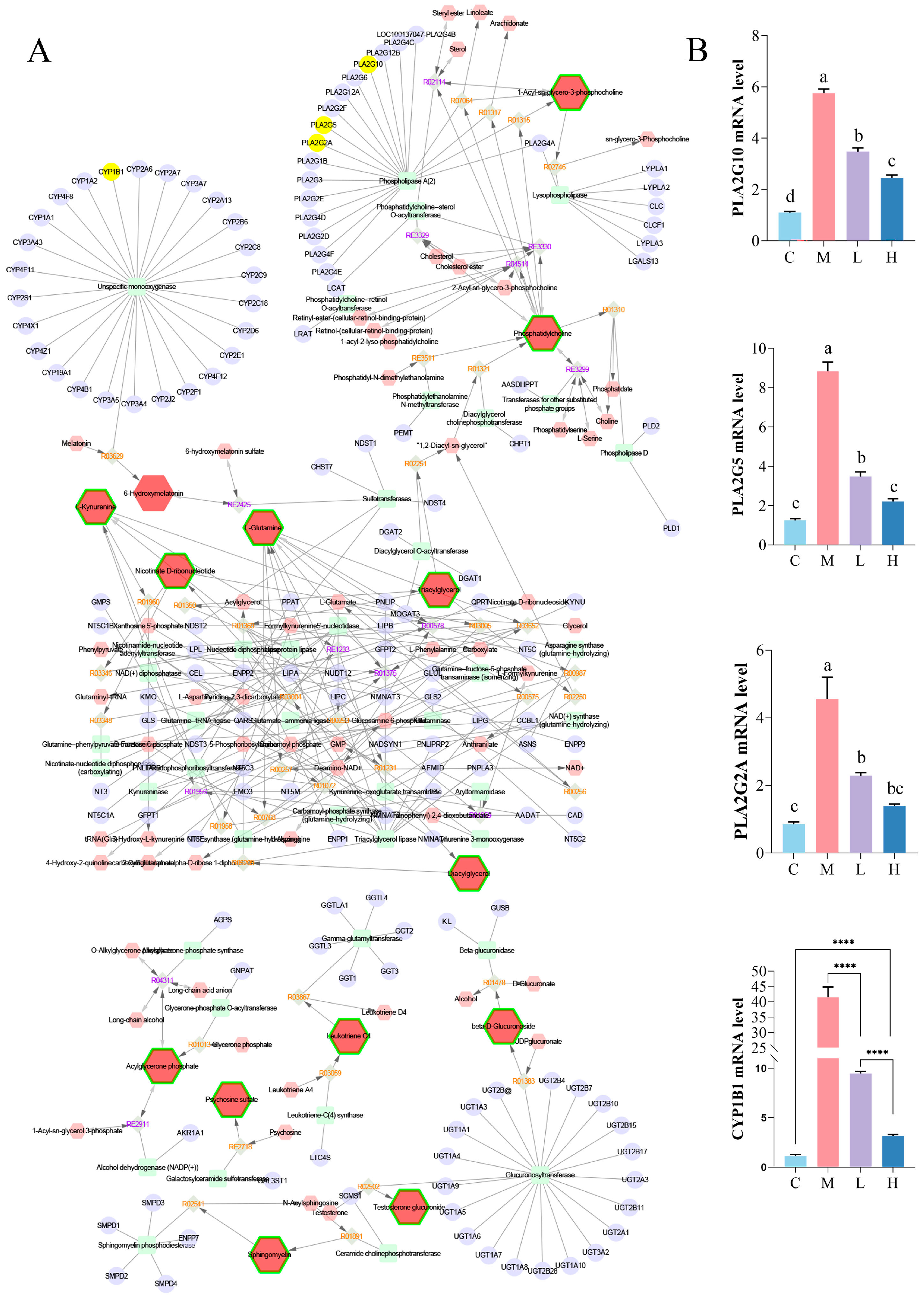
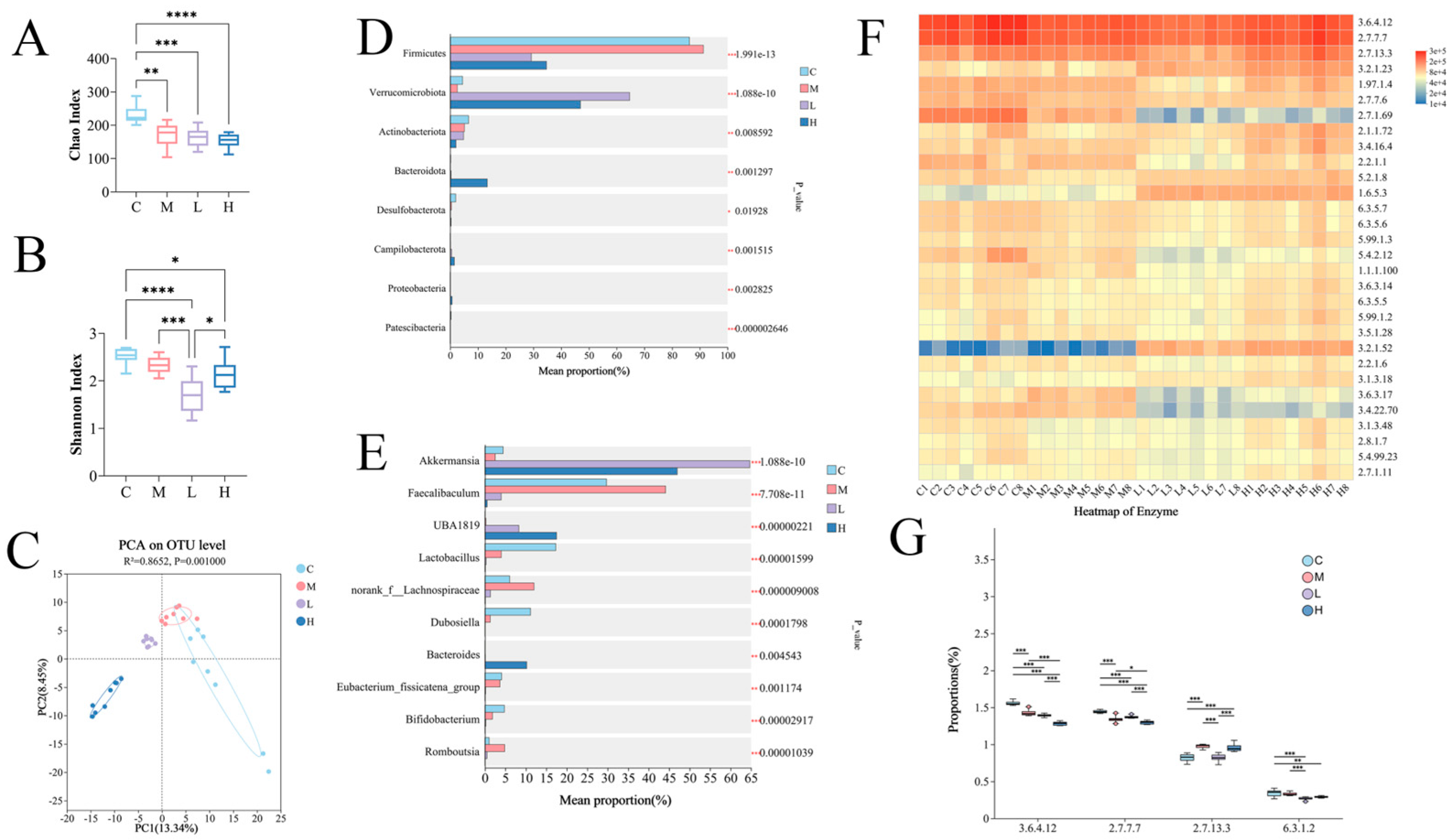
| Group | C | M | O | L | H |
|---|---|---|---|---|---|
| Initial body weight/g | 21.51 ± 0.69 b | 24.36 ± 1.37 a | 23.94 ± 0.77 a | 24.03 ± 0.45 a | 24.21 ± 0.19 a |
| Final body weight/g | 25.94 ± 1.30 b | 32.94 ± 5.40 a | 28.74 ± 1.24 b | 28.08 ± 1.70 b | 27.53 ± 0.67 b |
| Body weight gain/g | 4.43 ± 0.71 b | 8.57 ± 4.19 a | 4.80 ± 0.89 b | 4.05 ± 1.68 b | 3.31 ± 0.72 b |
| Energy intake (kcal/d/g) | 9.37 ± 0.50 c | 11.10 ± 0.08 b | 12.63 ± 1.01 a | 10.53 ± 0.10 b | 10.91 ± 0.92 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, J.; Zhou, Y.; Yang, D.; Liu, R.; Zhu, X.; Liu, R. Integrated Multi-Omics Analysis Reveals the Regulatory Mechanism of Peanut Skin Procyanidins on Lipid Metabolism in High-Fat-Diet-Induced Obese Mice. Nutrients 2025, 17, 2228. https://doi.org/10.3390/nu17132228
Shen J, Zhou Y, Yang D, Liu R, Zhu X, Liu R. Integrated Multi-Omics Analysis Reveals the Regulatory Mechanism of Peanut Skin Procyanidins on Lipid Metabolism in High-Fat-Diet-Induced Obese Mice. Nutrients. 2025; 17(13):2228. https://doi.org/10.3390/nu17132228
Chicago/Turabian StyleShen, Jinxin, Yi Zhou, Daijun Yang, Ruonan Liu, Xiaoling Zhu, and Rui Liu. 2025. "Integrated Multi-Omics Analysis Reveals the Regulatory Mechanism of Peanut Skin Procyanidins on Lipid Metabolism in High-Fat-Diet-Induced Obese Mice" Nutrients 17, no. 13: 2228. https://doi.org/10.3390/nu17132228
APA StyleShen, J., Zhou, Y., Yang, D., Liu, R., Zhu, X., & Liu, R. (2025). Integrated Multi-Omics Analysis Reveals the Regulatory Mechanism of Peanut Skin Procyanidins on Lipid Metabolism in High-Fat-Diet-Induced Obese Mice. Nutrients, 17(13), 2228. https://doi.org/10.3390/nu17132228





