Gut Microbiota as Targets for Preventing Ovalbumin-Induced Food Allergy
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
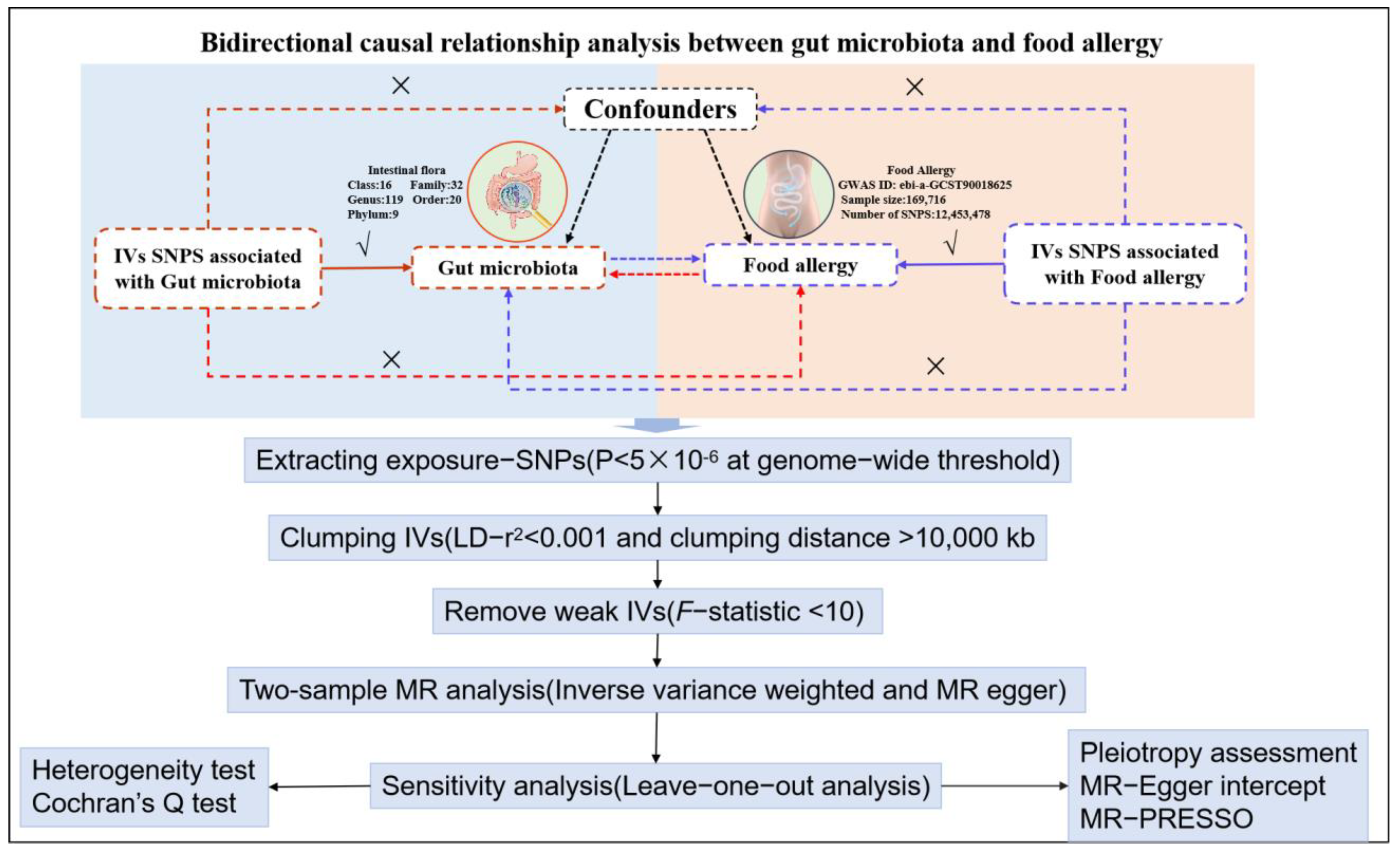
2.2.1. Two Sample Bidirectional MR Approach
2.2.2. Animal Experiments
2.2.3. DNA Extraction and PCR Amplification
2.3. Statistical Analysis
3. Results
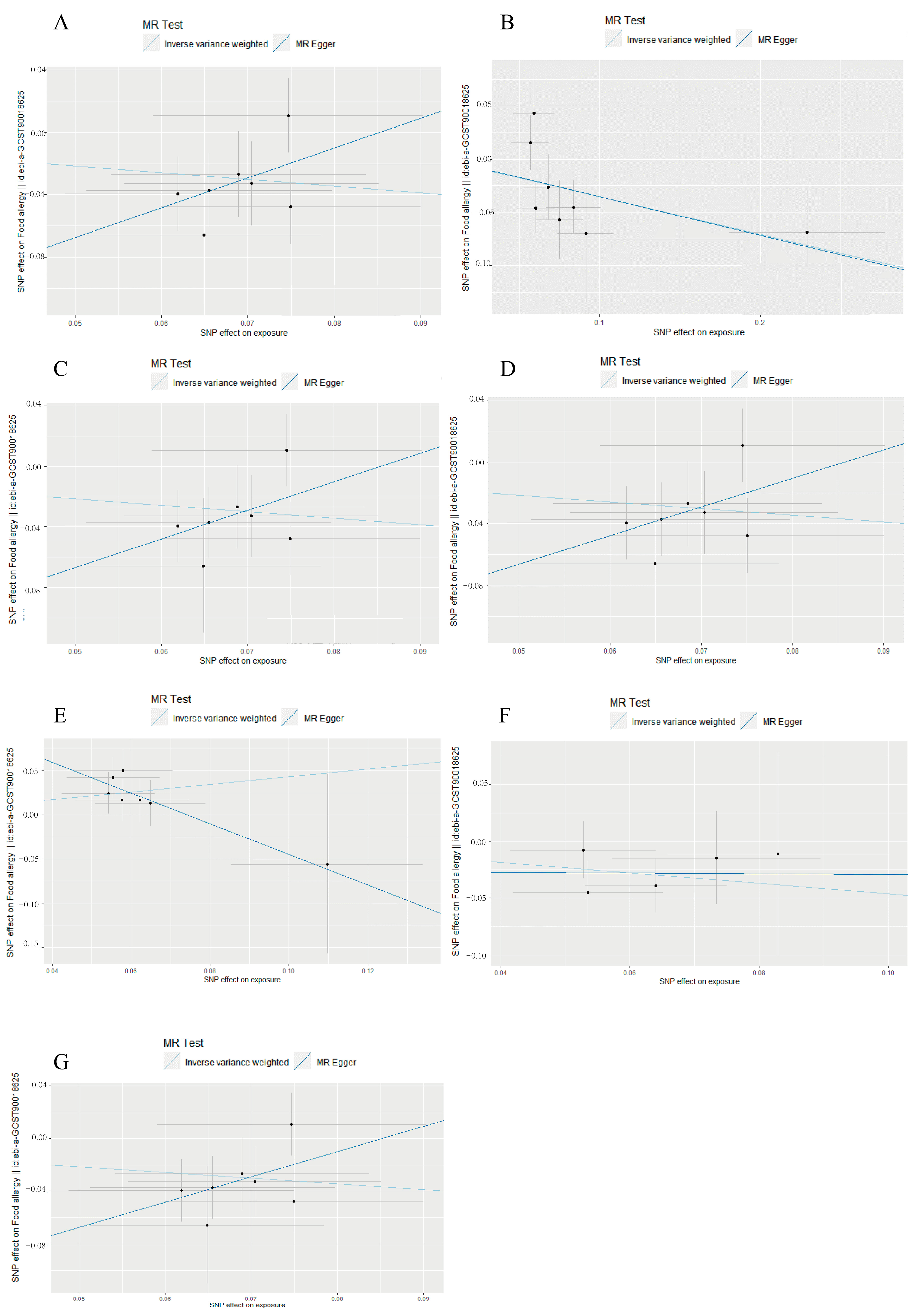
3.1. Causal Associations of Gut Microbiota with Food Allergy
3.2. Reverse Mendelian Randomization
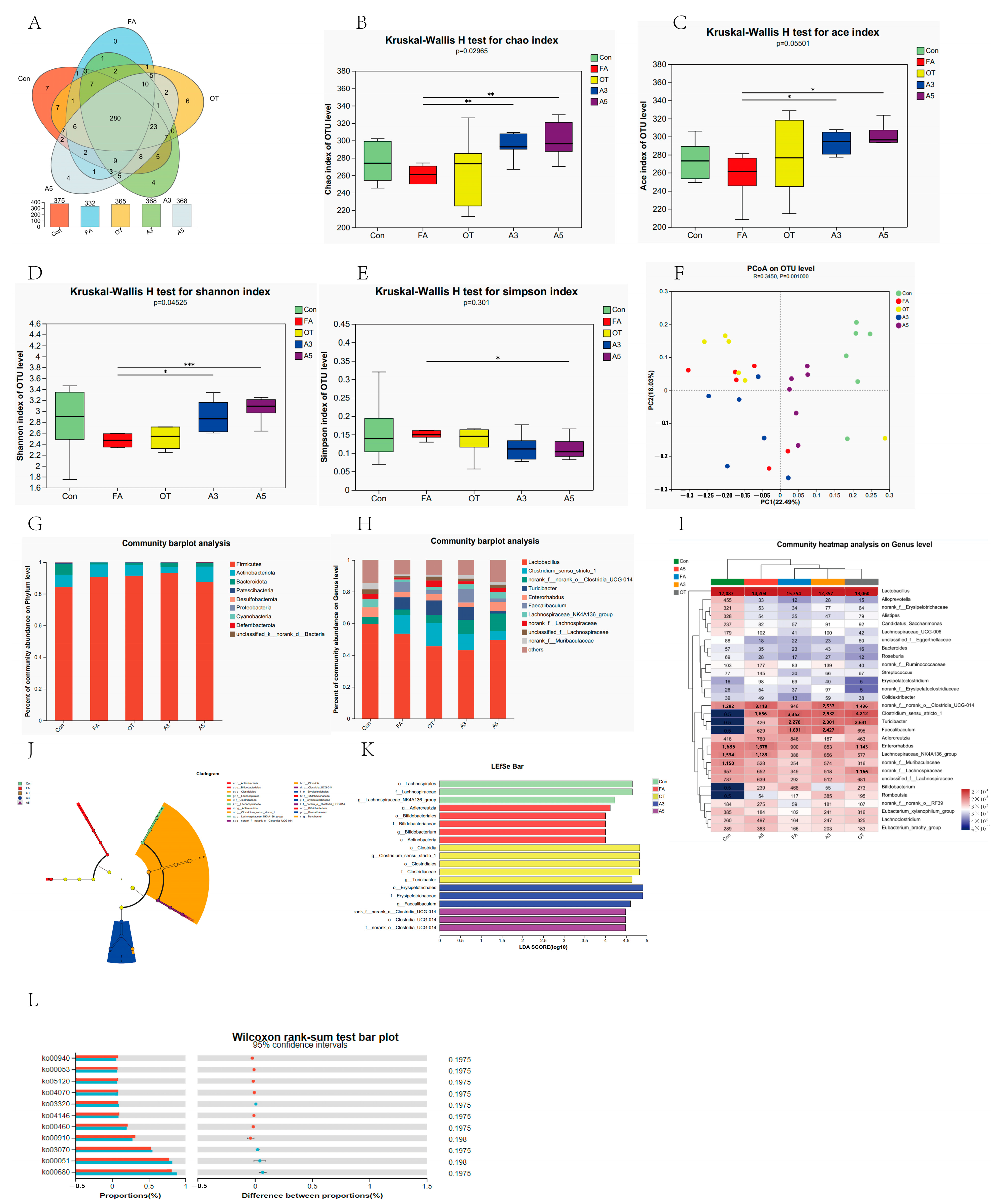
3.3. Gut Microbiota Composition and Diversity
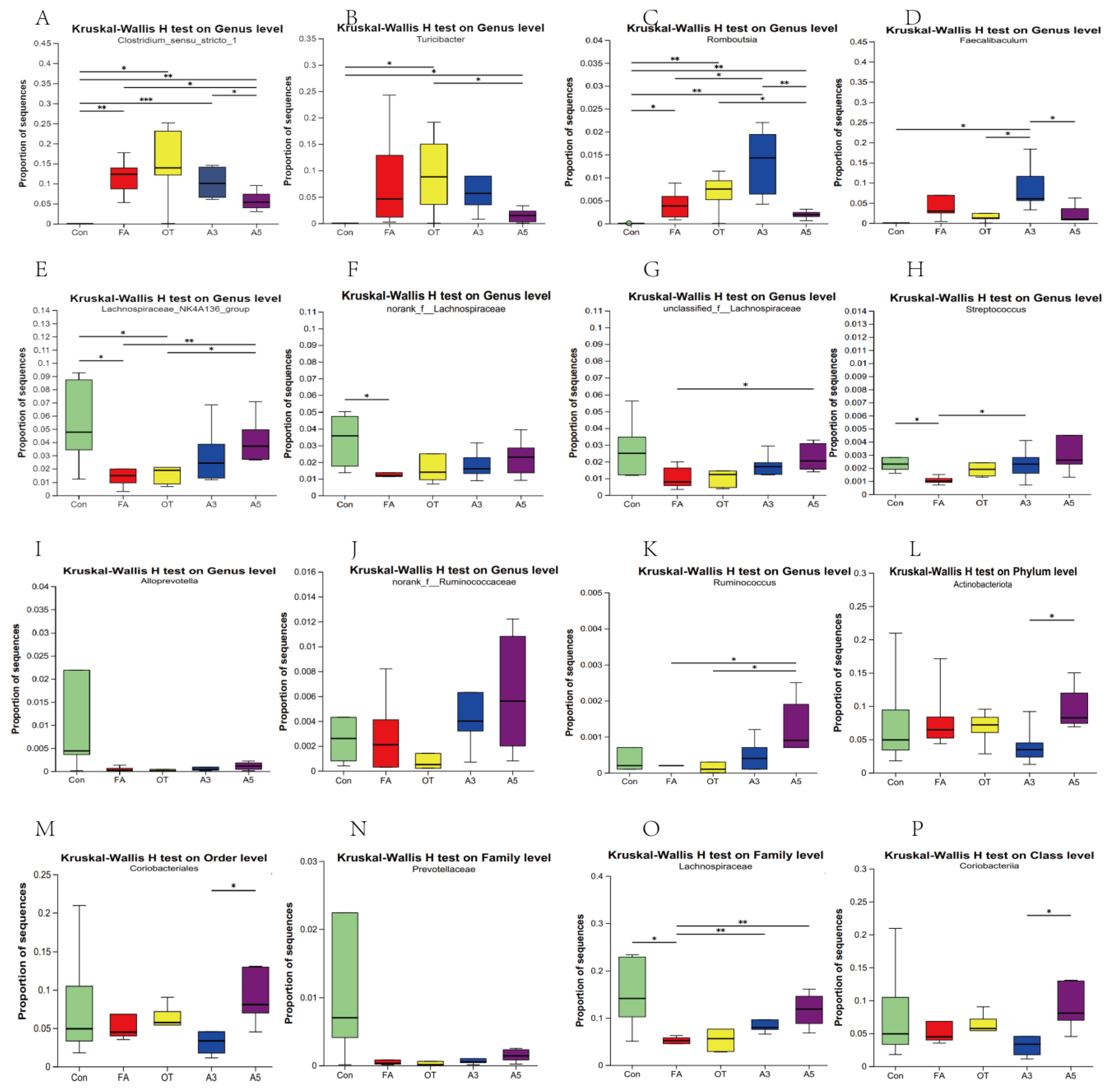
3.4. Changes in Target Gut Microbiota According to MR Analysis
3.5. Impacts of SCFAs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| OVA | Ovalbumin |
| WHO | The World Health Organization |
| MR | Mendelian randomization |
| IVW | Inverse-variance weighted |
| CI | Confidence interval |
| OR | Odds ratio |
| IVs | Instrumental variable |
| SNPs | Single-nucleotide polymorphism |
| MR-PRESSO | Mendelian randomization pleiotropy residual sum and outlier |
Appendix A
| MR Analysis | ||||||
|---|---|---|---|---|---|---|
| SNP | β | eaf | se | N | R2 | F |
| rs11184341 | −0.568 | 0.553 | 0.0237 | 16976 | 0.0329 | 576.560 |
| rs11729256 | −0.636 | 0.432 | 0.0241 | 16976 | 0.0393 | 693.680 |
| rs12908520 | −0.636 | 0.420 | 0.0238 | 16976 | 0.0404 | 714.496 |
| rs2602429 | 0.144 | 0.520 | 0.0238 | 16976 | 0.00216 | 36.739 |
| rs4242783 | −0.387 | 0.250 | 0.0274 | 16976 | 0.0116 | 199.280 |
| rs4936098 | −1.012 | 0.076 | 0.0446 | 16976 | 0.0295 | 515.402 |
| rs9349825 | −0.466 | 0.256 | 0.0270 | 16976 | 0.0173 | 298.897 |
| rs12118202 | −0.759 | 0.116 | 0.0366 | 16976 | 0.0247 | 429.479 |
| rs2206482 | 0.268 | 0.312 | 0.0257 | 16976 | 0.00637 | 108.794 |
| rs3860225 | −0.543 | 0.303 | 0.0255 | 16976 | 0.0259 | 452.176 |
| rs4493272 | −0.760 | 0.445 | 0.0236 | 16976 | 0.0577 | 1038.556 |
| rs4685827 | −0.387 | 0.182 | 0.0305 | 16976 | 0.00938 | 160.753 |
| rs912860 | −0.300 | 0.097 | 0.0396 | 16976 | 0.00336 | 57.279 |
| rs9586501 | 0.729 | 0.105 | 0.0386 | 16976 | 0.0206 | 357.364 |
| rs9958960 | −0.765 | 0.034 | 0.0654 | 16976 | 0.00800 | 136.924 |
| rs11128180 | 0.203 | 0.280 | 0.0261 | 16976 | 0.00357 | 60.835 |
| rs12673420 | 0.756 | 0.500 | 0.0236 | 16976 | 0.0571 | 1027.253 |
| rs12747809 | 0.269 | 0.706 | 0.0257 | 16976 | 0.00642 | 109.679 |
| rs12894272 | 0.862 | 0.637 | 0.0244 | 16976 | 0.0683 | 1244.123 |
| rs2444793 | 0.450 | 0.483 | 0.0235 | 16976 | 0.0212 | 367.970 |
| rs2882478 | 0.292 | 0.401 | 0.0239 | 16976 | 0.00871 | 149.105 |
| rs35182105 | −0.508 | 0.019 | 0.102 | 16976 | 0.00147 | 25.041 |
| rs10769159 | −0.610 | 0.568 | 0.0237 | 16976 | 0.0377 | 664.624 |
| rs11783695 | −0.199 | 0.093 | 0.0407 | 16976 | 0.00141 | 23.917 |
| rs6493760 | −0.845 | 0.298 | 0.0276 | 16976 | 0.0523 | 937.570 |
| rs7117576 | −0.132 | 0.017 | 0.0897 | 16976 | 0.000127 | 2.153 |
| rs7583465 | −0.145 | 0.328 | 0.0250 | 16976 | 0.00198 | 33.605 |
| Reverse | ||||||
| rs10485101 | −0.172 | 0.162 | 0.0258 | 16976 | 0.00261 | 44.429 |
| rs28698773 | −0.0987 | 0.269 | 0.0250 | 16976 | 0.000916 | 15.557 |
| rs34473363 | −0.135 | 0.0340 | 0.0286 | 16976 | 0.00130 | 22.160 |
| rs972348 | −0.286 | 0.265 | 0.0251 | 16976 | 0.00764 | 130.631 |
| MR Analysis | Cochran’s Q Test | |||
|---|---|---|---|---|
| Method | Q | Q_df | Q_pval | |
| c. Verrucomicrobiae | MR-Egger | 3.506 | 5 | 0.622 |
| Inverse-variance weighted | 4.832 | 6 | 0.566 | |
| o. Verrucomicrobiales | MR-Egger | 3.506 | 5 | 0.622 |
| Inverse-variance weighted | 4.832 | 6 | 0.566 | |
| f. Verrucomicrobiaceae | MR-Egger | 3.536 | 5 | 0.618 |
| Inverse-variance weighted | 4.823 | 6 | 0.567 | |
| g. Akkermansia | MR-Egger | 3.562 | 5 | 0.614 |
| Inverse-variance weighted | 4.820 | 6 | 0.567 | |
| f. Prevotellaceae | MR-Egger | 7.265 | 6 | 0.297 |
| Inverse-variance weighted | 7.267 | 7 | 0.401 | |
| g. LachnospiraceaeUCG004 | MR-Egger | 1.428 | 5 | 0.921 |
| Inverse-variance weighted | 3.249 | 6 | 0.777 | |
| g. Ruminococcus | MR-Egger | 1.412 | 3 | 0.703 |
| Inverse-variance weighted | 1.475 | 4 | 0.831 | |
| Reverse | ||||
| g. Olsenella | MR-Egger | 0.426 | 2 | 0.808 |
| Inverse-variance weighted | 0.552 | 3 | 0.907 | |
References
- Mine, Y.; Yang, M. Recent advances in the understanding of eggallergens: Basic, industrial, and clinical perspectives. J. Agric. Food Chem. 2008, 56, 4874–4900. [Google Scholar] [CrossRef] [PubMed]
- Urisu, A.; Kondo, Y.; Tsuge, I. Hen’s egg allergy. Chem. Immunol. Allergy 2015, 101, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Díaz, M.; Guadamuro, L.; Espinosa-Martos, I.; Mancabelli, L.; Jiménez, S.; Molinos-Norniella, C.; Pérez-Solis, D.; Milani, C.; Rodríguez, J.; Ventura, M.M.; et al. Microbiota and Derived Parameters in Fecal Samples of Infants with Non-IgE Cow’s Milk Protein Allergy under a Restricted Diet. Nutrients 2018, 10, 1481. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Filippis De, F.; Nocerino, R.; Paparo, L.; Scala Di, C.; Cosenza, L.; Della Gatta, G.; Calignano, A.; Caro De, C.; Laiola, M.; et al. Gut microbiota composition and butyrate production in children affected by non-IgE-mediated cow’s milk allergy. Sci. Rep. 2018, 8, 12500. [Google Scholar] [CrossRef]
- Choi, K.W.; Chen, C.Y.; Stein, M.B.; Klimentidis, Y.C.; Wang, M.J.; Koenen, K.C.; Smoller, J.W. Assessment of Bidirectional Relationships Between Physical Activity and Depression Among Adults: A 2-Sample Mendelian Randomization Study. JAMA Psychiat. 2019, 76, 399–408. [Google Scholar] [CrossRef]
- Smith, G.D.; Ebrahim, S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003, 32, 1–22. [Google Scholar] [CrossRef]
- Hill-Burns, E.M.; Debelius, J.W.; Morton, J.T.; Wissemann, W.T.; Lewis, M.R.; Wallen, Z.D.; Peddada, S.D.; Factor, S.A.; Molho, E.; Zabetian, C.P.; et al. Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Movement Disord. 2017, 32, 739–749. [Google Scholar] [CrossRef]
- Kubo, M.; Fujita, T.; Nishimura, S.; Tokunaga, M.; Utsunomiya, N. Seasonal variation in anti-allergic activity of citrus fruits and flavanone glycoside content. Nat. Med. 2004, 58, 284–294. [Google Scholar]
- Funaguchi, M.; Ohno, Y.; Lin, B.; La, B.; Asai, T.; Yuhgetsu, H.; Sawada, M.; Takemura, G.; Minatoguchi, S.; Fujiwara, T.; et al. Narirutin inhibits airway inflammation in an allergic mouse model. Clin. Exp. Pharmacol. Physiol. 2007, 34, 766–770. [Google Scholar] [CrossRef]
- Sakaue, S.; Kanai, M.; Tanigawa, Y.; Karjalainen, J.; Kurki, M.; Koshiba, S.; Narita, A.; Konuma, T.; Yamamoto, K.; Akiyama, M.; et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Gene 2021, 53, 1415–1424. [Google Scholar] [CrossRef]
- Eijsbouts, C.T.; Zheng, N.; Kennedy, A.; Bonfiglio, F.; Anderson, C.A.; Moutsianas, L.; Holliday, J.; Shi, J.C.Z.; Shringarpure, S.; Voda, A.L.; et al. Genome-wide analysis of 53,400 people with irritable bowel syndrome highlights shared genetic pathways with mood and anxiety disorders. Nat. Gene 2021, 53, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.L.; Zhao, L.N.; Niu, L.Y.; Yan, Y.X.; Chen, Q.S.; Jin, Y.R.; Li, X.W. Oral intervention of narirutin ameliorates the allergic response of ovalvumin allergy. J. Agric. Food Chem. 2022, 70, 13313–13326. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.S.; Zhao, D.F.; Ma, W.J.; Guo, Y.D.; Wang, A.J.; Wang, Q.L.; Lee, D.J. Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. Appl. Microbiol. Biot. 2015, 100, 1421–1426. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microb. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, H.; Zhu, M.J. A sensitive GC/MS detection method for analyzing microbial metabolites short chain fatty acids in fecal and serum samples. Talanta 2019, 196, 249–254. [Google Scholar] [CrossRef]
- Hsu, Y.L.; Chen, C.C.; Lin, Y.T.; Wu, W.K.; Chang, L.C.; Lai, C.H.; Wu, M.S.; Kuo, C.H. Evaluation and Optimization of Sample Handling Methods for Quantification of Short-Chain Fatty Acids in Human Fecal Samples by GC–MS. J. Proteome Res. 2019, 18, 1948–1957. [Google Scholar] [CrossRef]
- Larsson, S.C.; Burgess, S. Appraising the causal role of smoking in multiple diseases: A systematic review and meta-analysis of Mendelian randomization studies. eBioMedicine 2022, 82, 104154. [Google Scholar] [CrossRef]
- Burgess, S.; Dudbridge, F.; Thompson, S.G.; Burgess, S.; Dudbridge, F. Combining information on multiple instrumental variables in Mendelian randomization: Comparison of allele score and summarized data methods. Stat. Med. 2015, 35, 1880–1906. [Google Scholar] [CrossRef] [PubMed]
- Palmer, T.M.; Lawlo, D.A.; Harbord, R.M.; Sheehan, N.A.; Tobias, J.H.; Timpson, N.J.; Smith, G.D.; Sterne, J.A.C. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat. Methods Med. Res. 2011, 21, 223–242. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Barrett, T.; Benson, D.A.; Bolton, E.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Edgar, R.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010, 38, D5–D16. [Google Scholar] [CrossRef] [PubMed]
- Yun, L.; Li, W.; Wu, T.; Zhang, M. Effect of sea cucumber peptides on the immune response and gut microbiota composition in ovalbumin-induced allergic mice. Food Func. 2022, 13, 6338–6349. [Google Scholar] [CrossRef]
- Feehley, T.; Plunkett, C.H.; Bao, R.; Choi Hong, S.M.; Culleen, E.; Belda-Ferre, P.; Campbell, E.; Aitoro, R.; Nocerino, R.; Paparo, L.; et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 2019, 25, 448–453. [Google Scholar] [CrossRef]
- Wright, D.P.; Rosendale, D.I.; Roberton, A.M. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol. Lett. 2000, 190, 73–79. [Google Scholar] [CrossRef]


| id | p Val | or | 95% CI | No. of SNP |
|---|---|---|---|---|
| c. Verrucomicrobiae | 0.00258 | 0.651 | 0.492–0.860 | 7 |
| o. Verrucomicrobiales | 0.00258 | 0.651 | 0.492–0.860 | 7 |
| f. Verrucomicrobiaceae | 0.00256 | 0.650 | 0.492–0.860 | 7 |
| g. Akkermansia | 0.00256 | 0.650 | 0.491–0.860 | 7 |
| f. Prevotellaceae | 0.00389 | 0.702 | 0.552–0.893 | 8 |
| g. LachnospiraceaeUCG004 | 0.00990 | 1.541 | 1.109–2.141 | 7 |
| g. Ruminococcus | 0.0411 | 0.631 | 0.405–0.981 | 5 |
| Reverse | ||||
| p. Actinobacteria | 0.0380 | 0.903 | 0.819–0.994 | 4 |
| c. Coriobacteriia | 0.0307 | 0.898 | 0.814–0.990 | 4 |
| o. Coriobacteriales | 0.0307 | 0.898 | 0.814–0.990 | 4 |
| g. Adlercreutzia | 0.0319 | 0.856 | 0.743–0.987 | 4 |
| g. Olsenella | 0.0409 | 0.802 | 0.649–0.991 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, X.; Liu, H.; Zhang, J.; Yu, Y.; Xiao, J.; Matsui, K.; Li, X.; Jin, Y. Gut Microbiota as Targets for Preventing Ovalbumin-Induced Food Allergy. Nutrients 2025, 17, 1611. https://doi.org/10.3390/nu17101611
Shi X, Liu H, Zhang J, Yu Y, Xiao J, Matsui K, Li X, Jin Y. Gut Microbiota as Targets for Preventing Ovalbumin-Induced Food Allergy. Nutrients. 2025; 17(10):1611. https://doi.org/10.3390/nu17101611
Chicago/Turabian StyleShi, Xiaolei, Huimin Liu, Jiayin Zhang, Yawen Yu, Jing Xiao, Katsuhiko Matsui, Xuwen Li, and Yongri Jin. 2025. "Gut Microbiota as Targets for Preventing Ovalbumin-Induced Food Allergy" Nutrients 17, no. 10: 1611. https://doi.org/10.3390/nu17101611
APA StyleShi, X., Liu, H., Zhang, J., Yu, Y., Xiao, J., Matsui, K., Li, X., & Jin, Y. (2025). Gut Microbiota as Targets for Preventing Ovalbumin-Induced Food Allergy. Nutrients, 17(10), 1611. https://doi.org/10.3390/nu17101611






