Gut Microbiota Interacts with Dietary Habits in Screenings for Early Detection of Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Fecal Occult Blood Test Subsection
2.3. Dietary Habits Assessment
2.4. Colonoscopy and Clinicopathological Data
2.5. Intestinal Microbiota Analysis
2.6. Data Modeling
2.7. Statistical Analysis
3. Results
3.1. Characteristic of the Participants in the Study
3.2. Dietary Habits Assessment of the Participants in This Study
3.3. Diversity of the Gut Microbiota According to the Presence of CRC-Related Pathological Findings
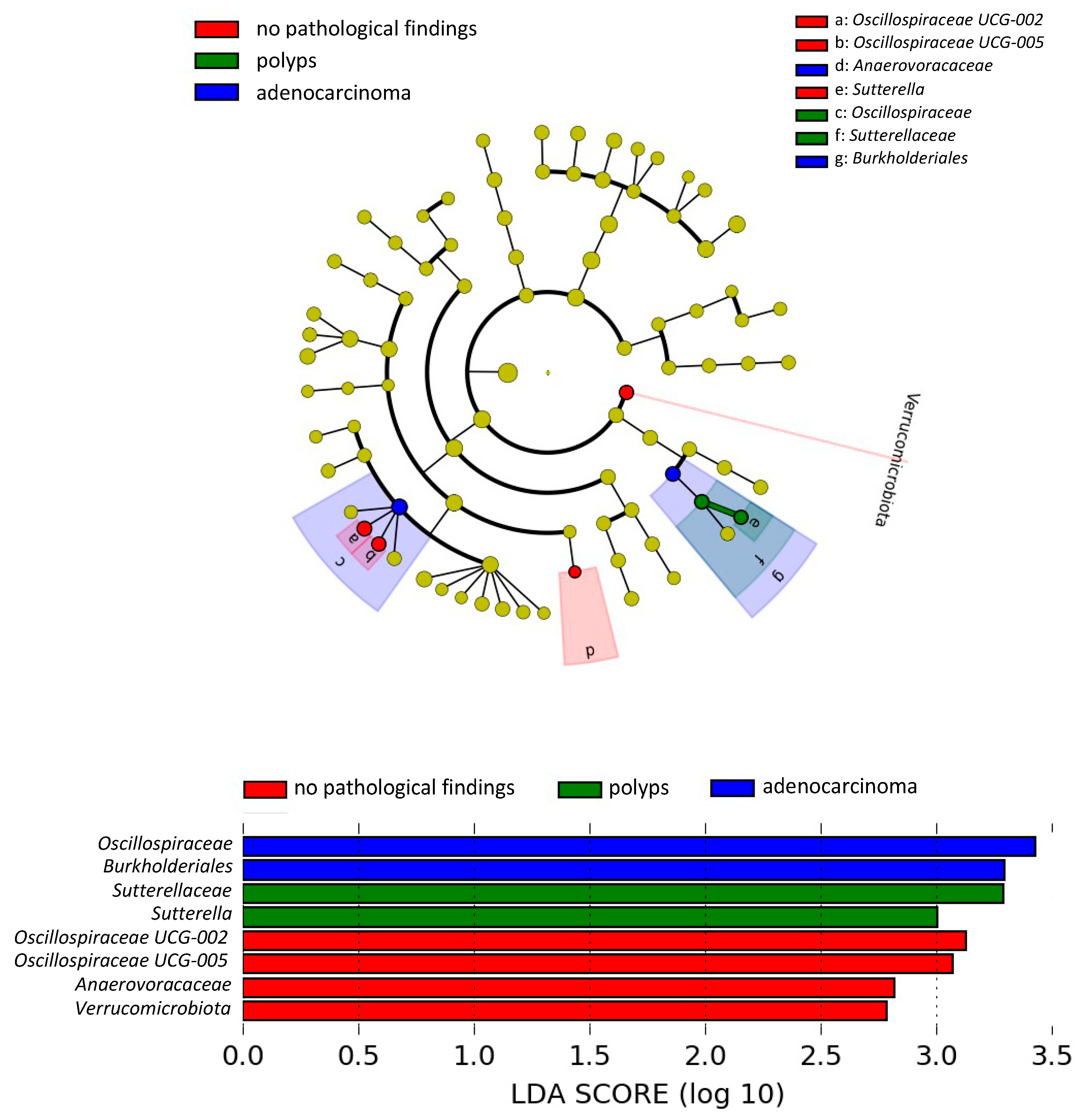
3.4. Differences in the Gut Microbiota According to the Presence of CRC-Related Pathological Findings: LEfSe Analysis
3.5. Random Forest Classifier Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Honari, M.; Shafabakhsh, R.; Reiter, R.J.; Mirzaei, H.; Asemi, Z. Resveratrol is a promising agent for colorectal cancer prevention and treatment: Focus on molecular mechanisms. Cancer Cell Int. 2019, 19, 180. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Levin, T.R.; Corley, D.A.; Jensen, C.D.; Schottinger, J.E.; Quinn, V.P.; Zauber, A.G.; Lee, J.K.; Zhao, W.K.; Udaltsova, N.; Ghai, N.R.; et al. Effects of Organized Colorectal Cancer Screening on Cancer Incidence and Mortality in a Large Community-Based Population. Gastroenterology 2018, 155, 1383–1391.e5. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Liles, E.G.; Bent, S.; Levin, T.R.; Corley, D.A. Accuracy of fecal immunochemical tests for colorectal cancer: Systematic review and meta-analysis. Ann. Intern. Med. 2014, 160, 171. [Google Scholar] [CrossRef]
- Adler, A.; Geiger, S.; Keil, A.; Bias, H.; Schatz, P.; de Vos, T.; Dhein, J.; Zimmermann, M.; Tauber, R.; Wiedenmann, B. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC Gastroenterol. 2014, 14, 183. [Google Scholar] [CrossRef]
- Taber, J.M.; Aspinwall, L.G.; Heichman, K.A.; Kinney, A.Y. Preferences for blood-based colon cancer screening differ by race/ethnicity. Am. J. Health Behav. 2014, 38, 351–361. [Google Scholar] [CrossRef]
- Weiss, K.T.; Zeman, F.; Schreml, S. A randomized trial of early endovenous ablation in venous ulceration: A critical appraisal. Br. J. Dermatol. 2019, 180, 51–55. [Google Scholar]
- Heiss, C.N.; Olofsson, L.E. The role of the gut microbiota in development, function and disorders of the central nervous system and the enteric nervous system. J. Neuroendocrinol. 2019, 31, e12684. [Google Scholar] [CrossRef]
- Qiu, P.; Ishimoto, T.; Fu, L.; Zhang, J.; Zhang, Z.; Liu, Y. The Gut Microbiota in Inflammatory Bowel Disease. Front. Cell Infect. Microbiol. 2022, 12, 733992. [Google Scholar] [CrossRef]
- Zhao, T.; Wei, Y.; Zhu, Y.; Xie, Z.; Hai, Q.; Li, Z.; Qin, D. Gut microbiota and rheumatoid arthritis: From pathogenesis to novel therapeutic opportunities. Front. Immunol. 2022, 13, 1007165. [Google Scholar] [CrossRef]
- Wong, C.C.; Yu, J. Gut microbiota in colorectal cancer development and therapy. Nat. Rev. Clin. Oncol. 2023, 20, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef] [PubMed]
- Roleira, F.M.; Tavares-da-Silva, E.J.; Varela, C.L.; Costa, S.C.; Silva, T.; Garrido, J.; Borges, F. Plant derived and dietary phenolic antioxidants: Anticancer properties. Food Chem. 2015, 183, 235–258. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Ussar, S.; Griffin, N.W.; Bezy, O.; Fujisaka, S.; Vienberg, S.; Softic, S.; Deng, L.; Bry, L.; Gordon, J.I.; Kahn, C.R. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metab. 2015, 22, 516–530. [Google Scholar] [CrossRef]
- Martinez-Gonzalez, M.A.; Fernandez-Jarne, E.; Serrano-Martinez, M.; Wright, M.; Gomez-Gracia, E. Development of a short dietary intake questionnaire for the quantitative estimation of adherence to a cardioprotective Mediterranean diet. Eur. J. Clin. Nutr. 2004, 58, 1550–1552. [Google Scholar] [CrossRef]
- Santos-Marcos, J.A.; Barroso, A.; Rangel-Zuniga, O.A.; Perdices-Lopez, C.; Haro, C.; Sanchez-Garrido, M.A.; Molina-Abril, H.; Ohlsson, C.; Perez-Martinez, P.; Poutanen, M.; et al. Interplay between gonadal hormones and postnatal overfeeding in defining sex-dependent differences in gut microbiota architecture. Aging 2020, 12, 19979–20000. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Schutte, S.; Esser, D.; Hoevenaars, F.P.M.; Hooiveld, G.; Priebe, M.G.; Vonk, R.J.; Wopereis, S.; Afman, L.A. A 12-wk whole-grain wheat intervention protects against hepatic fat: The Graandioos study, a randomized trial in overweight subjects. Am. J. Clin. Nutr. 2018, 108, 1264–1274. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 27. [Google Scholar]
- Garrett, W.S. The gut microbiota and colon cancer. Science 2019, 364, 1133–1135. [Google Scholar] [CrossRef]
- Lu, F.; Lei, T.; Zhou, J.; Liang, H.; Cui, P.; Zuo, T.; Ye, L.; Chen, H.; Huang, J. Using gut microbiota as a diagnostic tool for colorectal cancer: Machine learning techniques reveal promising results. J. Med. Microbiol. 2023, 72, 001699. [Google Scholar] [CrossRef]
- Dai, Z.; Coker, O.O.; Nakatsu, G.; Wu, W.K.K.; Zhao, L.; Chen, Z.; Chan, F.K.L.; Kristiansen, K.; Sung, J.J.Y.; Wong, S.H.; et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome 2018, 6, 70. [Google Scholar] [CrossRef]
- Zhang, M.; Lv, Y.; Hou, S.; Liu, Y.; Wang, Y.; Wan, X. Differential Mucosal Microbiome Profiles across Stages of Human Colorectal Cancer. Life 2021, 11, 831. [Google Scholar] [CrossRef]
- Yang, T.W.; Lee, W.H.; Tu, S.J.; Huang, W.C.; Chen, H.M.; Sun, T.H.; Tsai, M.C.; Wang, C.C.; Chen, H.Y.; Huang, C.C.; et al. Enterotype-based Analysis of Gut Microbiota along the Conventional Adenoma-Carcinoma Colorectal Cancer Pathway. Sci. Rep. 2019, 9, 10923. [Google Scholar] [CrossRef] [PubMed]
- LaCourse, K.D.; Johnston, C.D.; Bullman, S. The relationship between gastrointestinal cancers and the microbiota. Lancet Gastroenterol. Hepatol. 2021, 6, 498–509. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Lee, D.Y.; Kang, J.H.; Kim, J.H.; Jeong, J.W.; Kim, H.W.; Oh, D.H.; Yoon, S.H.; Hur, S.J. Relationship between gut microbiota and colorectal cancer: Probiotics as a potential strategy for prevention. Food Res. Int. 2022, 156, 111327. [Google Scholar] [CrossRef] [PubMed]
- Carmody, R.N.; Gerber, G.K.; Luevano, J.M., Jr.; Gatti, D.M.; Somes, L.; Svenson, K.L.; Turnbaugh, P.J. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015, 17, 72–84. [Google Scholar] [CrossRef]
- Albenberg, L.G.; Wu, G.D. Diet and the intestinal microbiome: Associations, functions, and implications for health and disease. Gastroenterology 2014, 146, 1564–1572. [Google Scholar] [CrossRef]
- Bradbury, K.E.; Appleby, P.N.; Key, T.J. Fruit, vegetable, and fiber intake in relation to cancer risk: Findings from the European Prospective Investigation into Cancer and Nutrition (EPIC). Am. J. Clin. Nutr. 2014, 100 (Suppl. S1), 394S–398S. [Google Scholar] [CrossRef]
- van Duijnhoven, F.J.; Bueno-De-Mesquita, H.B.; Ferrari, P.; Jenab, M.; Boshuizen, H.C.; Ros, M.M.; Casagrande, C.; Tjonneland, A.; Olsen, A.; Overvad, K.; et al. Fruit, vegetables, and colorectal cancer risk: The European Prospective Investigation into Cancer and Nutrition. Am. J. Clin. Nutr. 2009, 89, 1441–1452. [Google Scholar] [CrossRef]
- Vogtmann, E.; Xiang, Y.B.; Li, H.L.; Levitan, E.B.; Yang, G.; Waterbor, J.W.; Gao, J.; Cai, H.; Xie, L.; Wu, Q.J.; et al. Fruit and vegetable intake and the risk of colorectal cancer: Results from the Shanghai Men’s Health Study. Cancer Causes Control 2013, 24, 1935–1945. [Google Scholar] [CrossRef]
- Alzate-Yepes, T.; Perez-Palacio, L.; Martinez, E.; Osorio, M. Mechanisms of Action of Fruit and Vegetable Phytochemicals in Colorectal Cancer Prevention. Molecules 2023, 28, 4322. [Google Scholar] [CrossRef]
- Chazelas, E.; Srour, B.; Desmetz, E.; Kesse-Guyot, E.; Julia, C.; Deschamps, V.; Druesne-Pecollo, N.; Galan, P.; Hercberg, S.; Latino-Martel, P.; et al. Sugary drink consumption and risk of cancer: Results from NutriNet-Sante prospective cohort. BMJ 2019, 366, l2408. [Google Scholar] [CrossRef]
- Boyle, P.; Koechlin, A.; Autier, P. Sweetened carbonated beverage consumption and cancer risk: Meta-analysis and review. Eur. J. Cancer Prev. 2014, 23, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Eshaghian, N.; Zare, M.J.; Mohammadian, M.K.; Gozidehkar, Z.; Ahansaz, A.; Askari, G.; Asadi, M.; Milajerdi, A.; Sadeghi, O. Sugar sweetened beverages, natural fruit juices, and cancer: What we know and what still needs to be assessed. Front. Nutr. 2023, 10, 1301335. [Google Scholar] [CrossRef] [PubMed]
- Llaha, F.; Gil-Lespinard, M.; Unal, P.; de Villasante, I.; Castaneda, J.; Zamora-Ros, R. Consumption of Sweet Beverages and Cancer Risk. A Systematic Review and Meta-Analysis of Observational Studies. Nutrients 2021, 13, 516. [Google Scholar] [CrossRef]
- Zoltick, E.S.; Smith-Warner, S.A.; Yuan, C.; Wang, M.; Fuchs, C.S.; Meyerhardt, J.A.; Chan, A.T.; Ng, K.; Ogino, S.; Stampfer, M.J.; et al. Sugar-sweetened beverage, artificially sweetened beverage and sugar intake and colorectal cancer survival. Br. J. Cancer 2021, 125, 1016–1024. [Google Scholar] [CrossRef]
- Rehman, O.U.; Fatima, E.; Arabpour, J. The association of excessive soft drinks consumption with early-onset colorectal cancer. Eur. J. Cancer 2023, 192, 113242. [Google Scholar] [CrossRef]

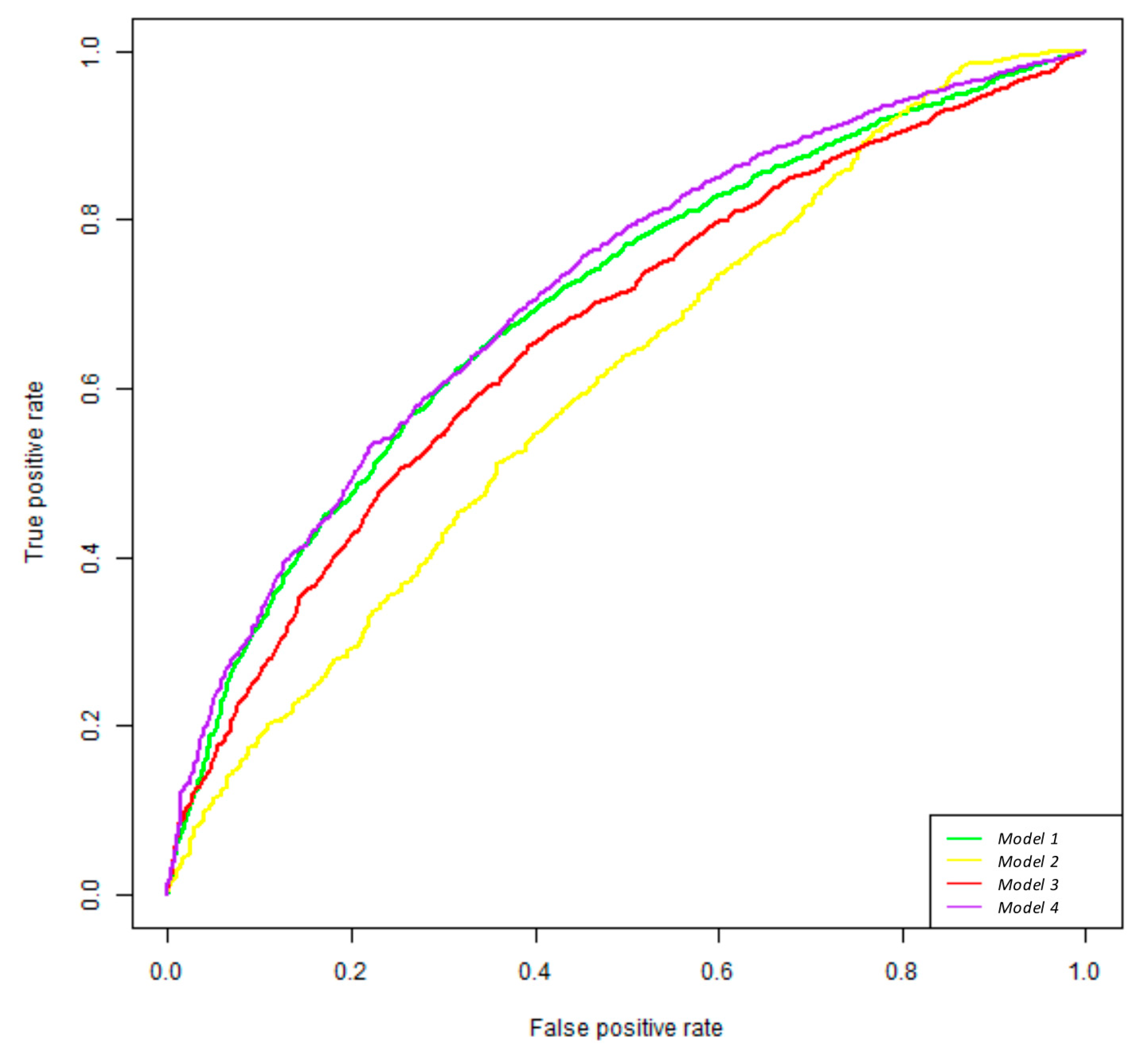
| 20 More Important Variables in the Model 1 | Importance |
|---|---|
| Ruminococcaceae family | 100.00 |
| Collinsella genus | 58.21 |
| Faecalibacterium genus | 54.12 |
| Anaerovoracaceae family | 52.08 |
| Streptococcus genus | 47.10 |
| Negativibacillus genus | 42.81 |
| Barnesiella genus | 41.27 |
| Streptococcaceae family | 35.99 |
| Veillonellaceae family | 35.59 |
| Verrucomicrobiota phylum | 31.90 |
| Gammaproteobacteria class | 31.77 |
| Burkholderiales order | 24.83 |
| Oscillospirales order | 24.16 |
| unknown genus from Lachnospiraceae family | 20.54 |
| NK4A214 group (Oscillospiraceae family) | 20.21 |
| Coriobacteriales order | 20.12 |
| UCG-005 genus (Oscillospiraceae family) | 19.82 |
| Proteobacteria phylum | 19.42 |
| Coriobacteriia class | 19.22 |
| Bacilli order | 18.57 |
| 20 More Important Variables in the Model 3 | Importance |
|---|---|
| Mediterranean diet item 3 | 100.00 |
| Mediterranean diet item 7 | 54.90 |
| Sutterella genus | 26.30 |
| Oscillospirales order | 20.41 |
| Monoglobus genus | 15.77 |
| UCG-002 genus (Oscillospiraceae family) | 15.76 |
| Proteobacteria phylum | 15.01 |
| Ruminococcaceae family | 14.64 |
| Monoglobales order | 13.65 |
| Streptococcaceae family | 13.64 |
| Streptococcus genus | 13.46 |
| Mediterranean diet item 4 | 12.83 |
| Parasutterella genus | 12.49 |
| Eubacterium eligens group | 12.00 |
| Parabacteroides genus | 11.91 |
| Subdoligranulum genus | 11.24 |
| Burkholderiales order | 10.47 |
| Bacilli class | 10.35 |
| Monoglobaceae family | 10.30 |
| UCG-005 genus (Oscillospiraceae family) | 10.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vega-Rojas, A.; Haro, C.; Molina-Abril, H.; Guil-Luna, S.; Santos-Marcos, J.A.; Gutierrez-Mariscal, F.M.; Garcia-Fernandez, H.; Caballero-Villarraso, J.; Rodriguez-Ariza, A.; Lopez-Miranda, J.; et al. Gut Microbiota Interacts with Dietary Habits in Screenings for Early Detection of Colorectal Cancer. Nutrients 2025, 17, 84. https://doi.org/10.3390/nu17010084
Vega-Rojas A, Haro C, Molina-Abril H, Guil-Luna S, Santos-Marcos JA, Gutierrez-Mariscal FM, Garcia-Fernandez H, Caballero-Villarraso J, Rodriguez-Ariza A, Lopez-Miranda J, et al. Gut Microbiota Interacts with Dietary Habits in Screenings for Early Detection of Colorectal Cancer. Nutrients. 2025; 17(1):84. https://doi.org/10.3390/nu17010084
Chicago/Turabian StyleVega-Rojas, Ana, Carmen Haro, Helena Molina-Abril, Silvia Guil-Luna, Jose Antonio Santos-Marcos, Francisco Miguel Gutierrez-Mariscal, Helena Garcia-Fernandez, Javier Caballero-Villarraso, Antonio Rodriguez-Ariza, Jose Lopez-Miranda, and et al. 2025. "Gut Microbiota Interacts with Dietary Habits in Screenings for Early Detection of Colorectal Cancer" Nutrients 17, no. 1: 84. https://doi.org/10.3390/nu17010084
APA StyleVega-Rojas, A., Haro, C., Molina-Abril, H., Guil-Luna, S., Santos-Marcos, J. A., Gutierrez-Mariscal, F. M., Garcia-Fernandez, H., Caballero-Villarraso, J., Rodriguez-Ariza, A., Lopez-Miranda, J., Perez-Martinez, P., Hervas, A., & Camargo, A. (2025). Gut Microbiota Interacts with Dietary Habits in Screenings for Early Detection of Colorectal Cancer. Nutrients, 17(1), 84. https://doi.org/10.3390/nu17010084






