Bifidobacterium longum K5 Prevents Enterohaemorrhagic Escherichia coli O157:H7 Infection in Mice through the Modulation of the Gut Microbiota
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Animals and Experimental Design
2.3. Collection of Samples
2.4. Disease Activity Index (DAI) Score
2.5. Determination of MPO Activity
2.6. H&E and AB-PAS Staining
2.7. Quantitative RT-PCR
2.8. Detection of Gut Microbiota
2.9. Detection of SCFAs
2.10. Statistical Analysis
3. Results
3.1. Effects of B. longum K5 in Alleviating Clinical Symptoms
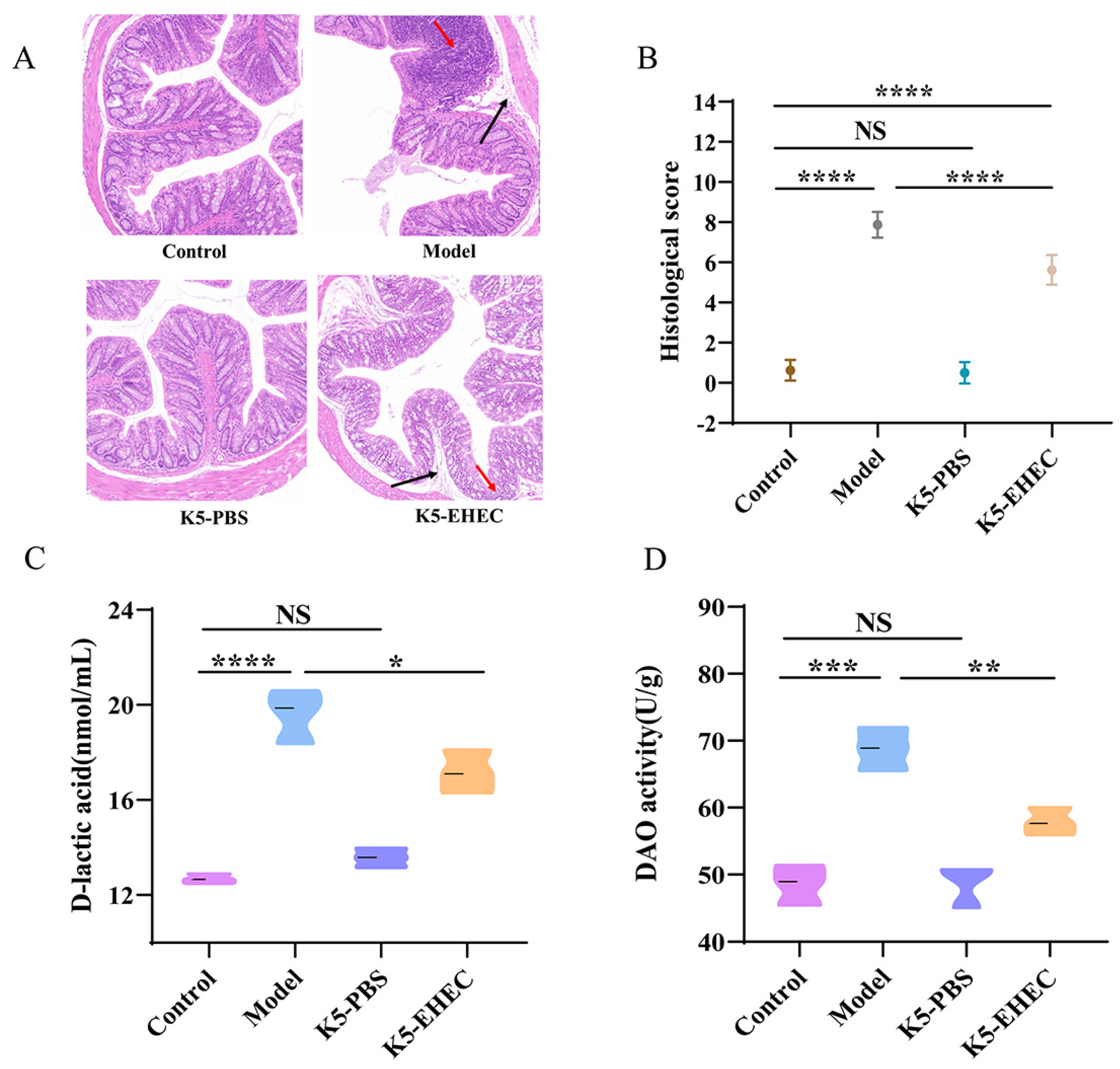
3.2. Effects of B. longum K5 on Intestinal Pathological Changes
3.3. Effect of B. longum K5 on Intestinal Permeability
3.4. Effect of B. longum K5 on Gut Barrier Function
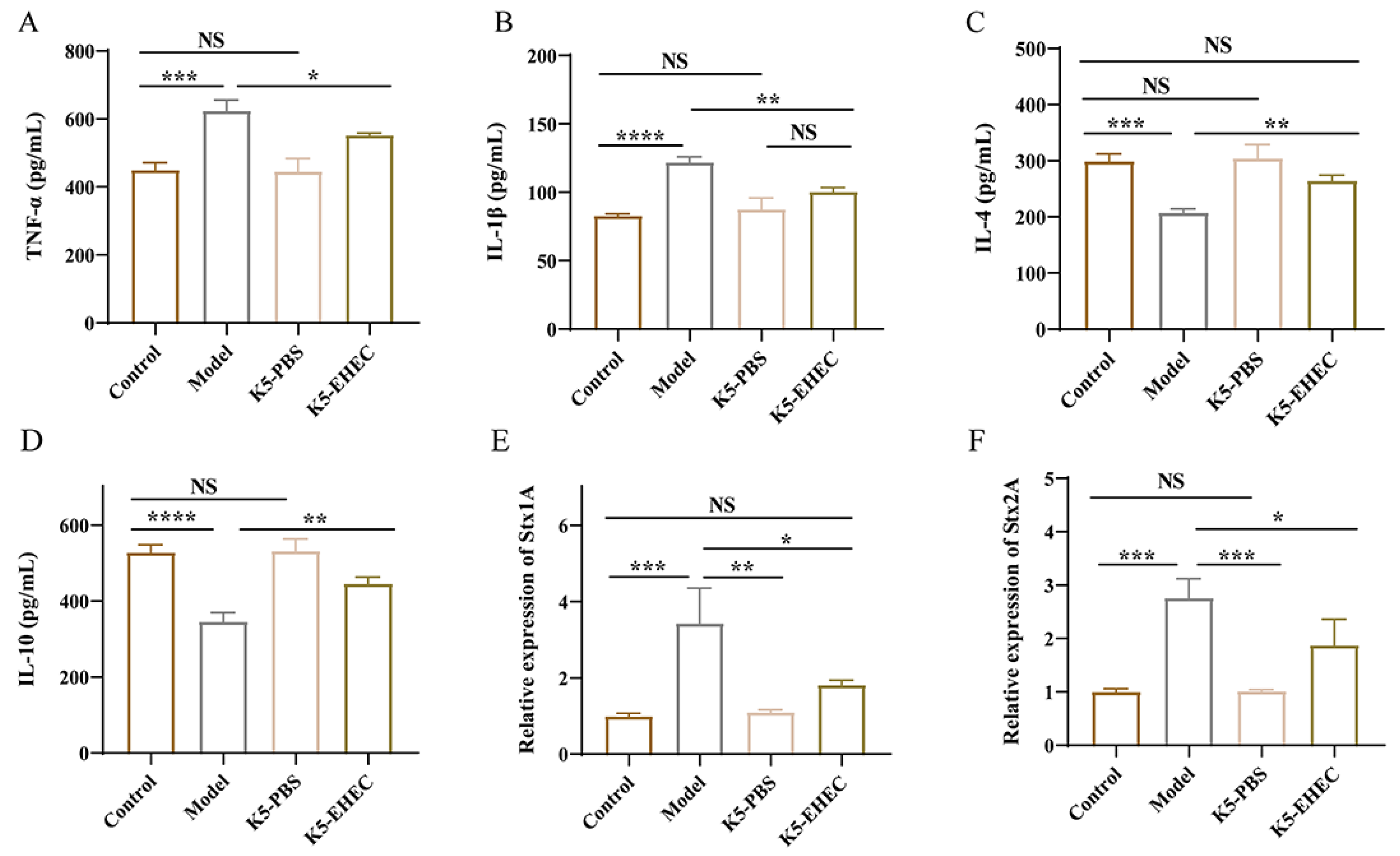
3.5. Effect of B. longum K5 on Gut Inflammation
3.6. Effect of B. longum K5 on the Expression of Virulence Factors
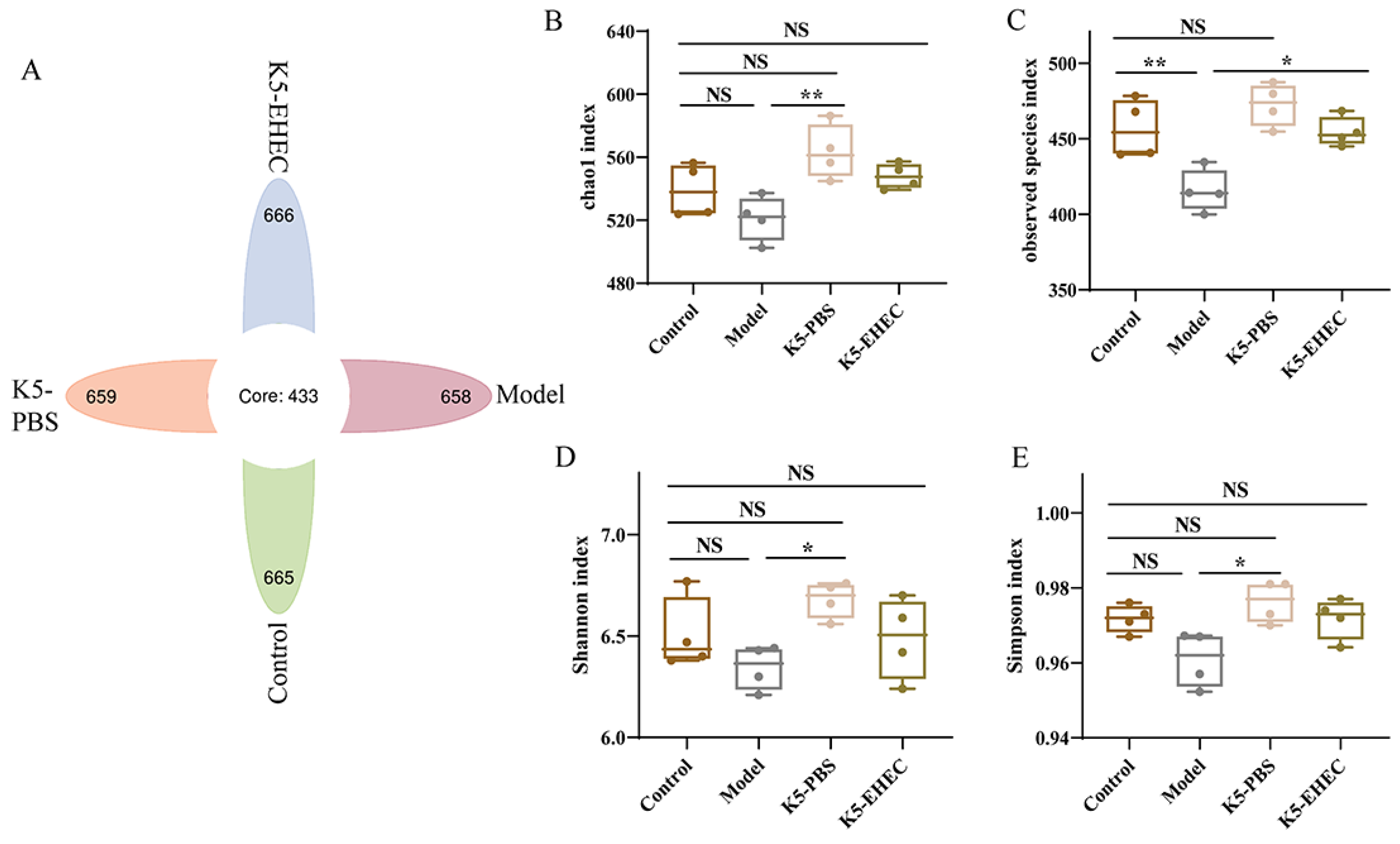
3.7. Effect of B. longum K5 on the Diversity of the Gut Microbiota
3.8. Effect of B. longum K5 on the Structure of the Gut Microbiota
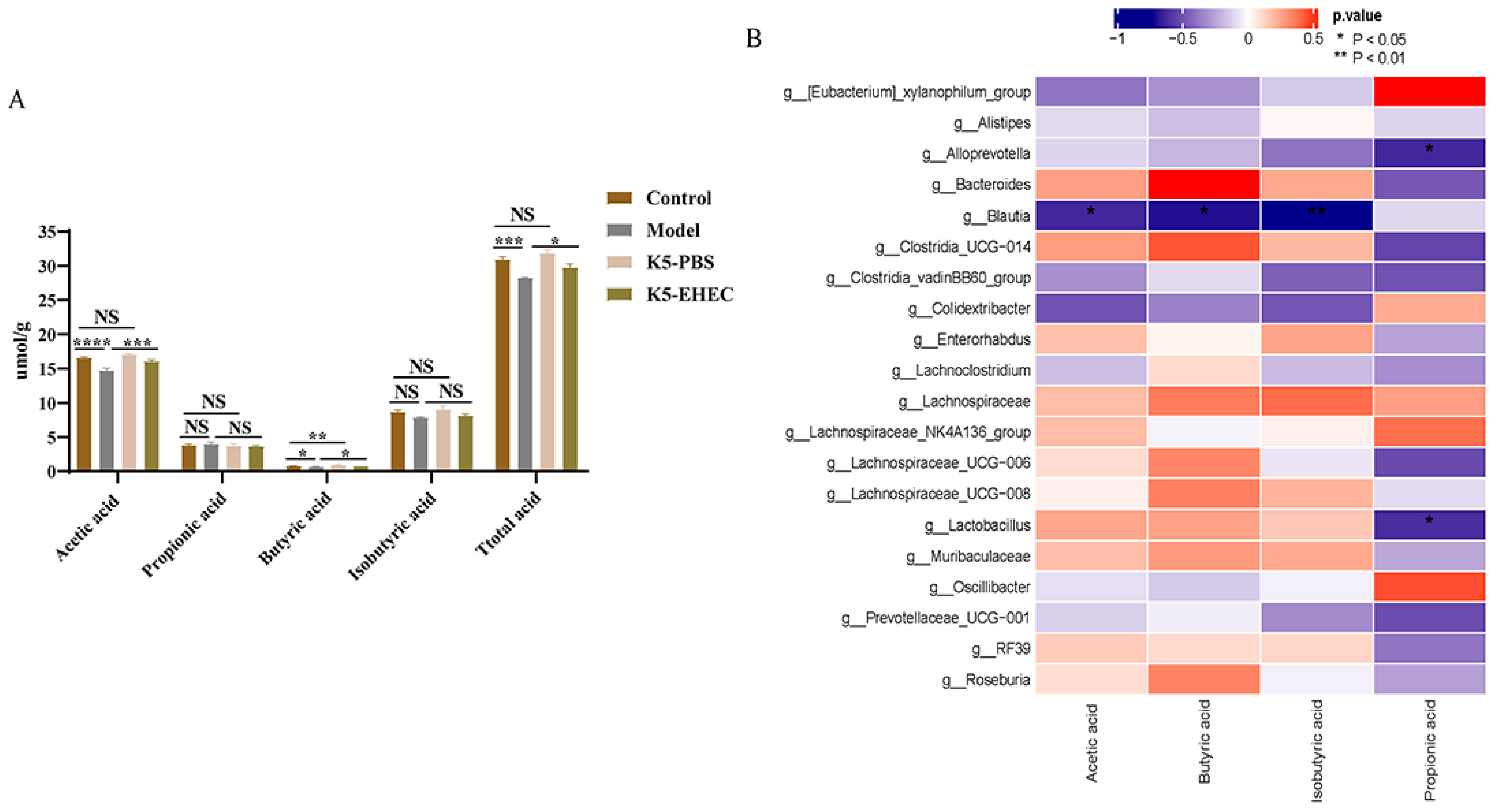
3.9. Production of SCFAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed]
- Srutkova, D.; Schwarzer, M.; Hudcovic, T.; Zakostelska, Z.; Drab, V.; Spanova, A.; Rittich, B.; Kozakova, H.; Schabussova, I. Bifidobacterium longum CCM 7952 promotes epithelial barrier function and prevents acute DSS-induced colitis in strictly strain-specific manner. PLoS ONE 2015, 10, e0134050. [Google Scholar] [CrossRef]
- Nguyen, Y.; Sperandio, V. Enterohemorrhagic E. coli (EHEC) pathogenesis. Front. Cell. Infect. Microbiol. 2012, 2, 90. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.; Yoon, J.W.; Hovde, C.J. A brief overview of Escherichia coli O157: H7 and its plasmid O157. J. Microbiol. Biotechnol. 2010, 20, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Wasan, A.; Sharma, R.K. Recent developments in probiotics: An emphasis on Bifidobacterium. Food Biosci. 2021, 41, 100993. [Google Scholar] [CrossRef]
- Jung, H.L.; Soon, H.S. Antimicrobial and Immunomodulatory Effects of Bifidobacterium Strains: A Review. J. Microbiol. Biotechnol. 2020, 30, 1793. [Google Scholar]
- Delcaru, C.; Alexandru, I.; Podgoreanu, P.; Cristea, V.C.; Bleotu, C.; Chifiriuc, M.C.; Bezirtzoglou, E.; Lazar, V. Antagonistic activities of some Bifidobacterium sp. strains isolated from resident infant gastrointestinal microbiota on Gram-negative enteric pathogens. Anaerobe 2016, 39, 39–44. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Westermann, C.; Gleinser, M.; Corr, S.C.; Riedel, C.U. A critical evaluation of bifidobacterial adhesion to the host tissue. Front. Microbiol. 2016, 7, 1220. [Google Scholar] [CrossRef]
- Zhang, C.; Yu, Z.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Colonization and probiotic function of Bifidobacterium longum. J. Funct. Foods 2019, 53, 157–165. [Google Scholar] [CrossRef]
- Yoshimura, K.; Matsui, T.; Itoh, K. Prevention of Escherichia coli O157: H7 infection in gnotobiotic mice associated with Bifidobacterium strains. Antonie Van Leeuwenhoek 2010, 97, 107–117. [Google Scholar] [CrossRef]
- Juge, N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 2012, 20, 30–39. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019, 103, 6463–6472. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, S.; Dong, J.; Shi, J. Identification, Characterization, and Antioxidant Potential of Bifidobacterium longum subsp longum Strains Isolated From Feces of Healthy Infants. Front. Microbiol. 2021, 12, 3154. [Google Scholar]
- Zhao, L.; Xie, Q.; Evivie, S.E.; Yue, Y.; Yang, H.; Lv, X.; Liu, F.; Li, B.; Huo, G. Bifidobacterium longum subsp longum K5 alleviates inflammatory response and prevents intestinal barrier injury induced by LPS in vitro based on comparative genomics. J. Funct. Foods 2022, 92, 105030. [Google Scholar]
- Dong, J.; Ping, L.; Xie, Q.; Liu, D.; Zhao, L.; Evivie, S.E.; Wang, Z.; Li, B.; Huo, G. Lactobacillus plantarum KLDS1.0386 with antioxidant capacity ameliorates the lipopolysaccharide-induced acute liver injury in mice by NF-κB and Nrf2 pathway. Food Biosci. 2022, 47, 101589. [Google Scholar] [CrossRef]
- Kwon, J.; Lee, C.; Heo, S.; Kim, B.; Hyun, C.-K. DSS-induced colitis is associated with adipose tissue dysfunction and disrupted hepatic lipid metabolism leading to hepatosteatosis and dyslipidemia in mice. Sci. Rep. 2021, 11, 5283. [Google Scholar] [CrossRef] [PubMed]
- Jiaqi, G.; Fei, L.; Sijia, Z.; Etareri, E.S.; Jialu, S.; Na, L.; Li, Z.; Yingxue, Y.; Qinggang, X.; Guicheng, H.; et al. Effect of Bifidobacterium longum subsp. longum on the proliferative and tight-junction activities of Human Fetal Colon Epithelial Cells. J. Funct. Foods 2021, 86, 104715. [Google Scholar]
- Na, L.; Shengnan, L.; Qingxue, C.; Lina, Z.; Bailiang, L.; Guicheng, H. Distinct gut microbiota and metabolite profiles induced by delivery mode in healthy Chinese infants. J. Proteom. 2021, 232, 104071. [Google Scholar]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Fenfen, Y.; Na, L.; Jialu, S.; Huizhen, L.; Yingxue, Y.; Wenshu, J.; Nana, W.; Yue, S.; Guicheng, H.; Bailiang, L. Lactobacillus acidophilus alleviates type 2 diabetes by regulating hepatic glucose, lipid metabolism and gut microbiota in mice. Food Funct. 2019, 10, 5804–5815. [Google Scholar]
- Cookson, A.L.; Woodward, M.J. The role of intimin in the adherence of enterohaemorrhagic Escherichia coli (EHEC) O157: H7 to HEp-2 tissue culture cells and to bovine gut explant tissues. Int. J. Med. Microbiol. 2003, 292, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Moxley, R.A. Escherichia coli 0157:H7: An update on intestinal colonization and virulence mechanisms. Anim. Health Res. Rev. 2004, 5, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Lebeis, S.L.; Sherman, M.A.; Kalman, D. Protective and destructive innate immune responses to enteropathogenic Escherichia coli and related A/E pathogens. Future Microbiol. 2008, 3, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Obrig, T.G. Shiga toxin mode of action in E. coli O157:H7 disease. Front. Biosci. 1997, 2, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, Y.; Song, X.; Xia, Y.; Lai, P.F.H.; Ai, L. Lactobacillus casei LC2W can inhibit the colonization of Escherichia coli O157: H7 in vivo and reduce the severity of colitis. Food Funct. 2019, 10, 5843–5852. [Google Scholar] [CrossRef]
- Bonvicini, F.; Pagnotta, E.; Punzo, A.; Calabria, D.; Simoni, P.; Mirasoli, M.; Passerini, N.; Bertoni, S.; Ugolini, L.; Lazzeri, L.; et al. Effect of Lactobacillus acidophilus fermented broths enriched with Eruca sativa seed extracts on intestinal barrier and inflammation in a co-culture system of an enterohemorrhagic Escherichia coli and human intestinal cells. Nutrients 2020, 12, 3064. [Google Scholar] [CrossRef]
- Yun, B.; Song, M.; Park, D.J.; Oh, S. Beneficial effect of Bifidobacterium longum ATCC 15707 on survival rate of Clostridium difficile infection in mice. Korean J. Food Sci. Anim. Resour. 2017, 37, 368. [Google Scholar] [CrossRef]
- Jiang, J.; Wu, C.; Zhang, C.; Zhang, Q.; Yu, L.; Zhao, J.; Zhang, H.; Narbad, A.; Chen, W.; Zhai, Q. Strain-specific effects of Bifidobacterium longum on hypercholesterolemic rats and potential mechanisms. Int. J. Mol. Sci. 2021, 22, 1305. [Google Scholar] [CrossRef]
- Finamore, A.; Roselli, M.; Donini, L.; Brasili, E.; Rami, R.; Carnevali, P.; Mistura, L.; Pinto, A.; Giusti, A.; Mengheri, E. Supplementation with Bifidobacterium longum Bar33 and Lactobacillus helveticus Bar13 mixture improves immunity in elderly humans (over 75 years) and aged mice. Nutrition 2019, 63, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.A.C.; Balciunas, E.M.; Converti, A.; Cotter, P.D.; de Souza Oliveira, R.P. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol. Adv. 2013, 31, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Makras, L.; De Vuyst, L. The in vitro inhibition of Gram-negative pathogenic bacteria by Bifidobacteria is caused by the production of organic acids. Int. Dairy J. 2006, 16, 1049–1057. [Google Scholar] [CrossRef]
- Nair, M.S.; Amalaradjou, M.A.; Venkitanarayanan, K. Antivirulence properties of probiotics in combating microbial pathogenesis. Adv. Appl. Microbiol. 2017, 98, 1–29. [Google Scholar]
- Bian, X.; Wang, T.T.; Xu, M.; Evivie, S.E.; Luo, G.W.; Liang, H.Z.; Yu, S.F.; Huo, G.C. Effect of Lactobacillus Strains on Intestinal Microflora and Mucosa Immunity in Escherichia coli O157:H7-Induced Diarrhea in Mice. Curr. Microbiol. 2016, 73, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Mélanie, G.; Ehab, E.K.; Nassra, D.; Denis, R. Effect of Bifidobacterium thermacidophilum probiotic feeding on enterohemorrhagic Escherichia coli O157:H7 infection in BALB/c mice. Int. J. Food Microbiol. 2006, 111, 26–33. [Google Scholar]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef] [PubMed]
- König, J.; Wells, J.; Cani, P.D.; García-Ródenas, C.L.; MacDonald, T.; Mercenier, A.; Whyte, J.; Troost, F.; Brummer, R.-J. Human intestinal barrier function in health and disease. Clin. Transl. Gastroenterol. 2016, 7, e196. [Google Scholar] [CrossRef]
- Groschwitz, K.R.; Hogan, S.P. Intestinal barrier function: Molecular regulation and disease pathogenesis. J. Allergy Clin. Immunol. 2009, 124, 21–22. [Google Scholar] [CrossRef]
- Bao, C.L.; Liu, S.Z.; Shang, Z.D.; Liu, Y.J.; Wang, J.; Zhang, W.X.; Dong, B.; Cao, Y.H. Bacillus amyloliquefaciens TL106 protects mice against enterohaemorrhagic Escherichia coli O157: H7-induced intestinal disease through improving immune response, intestinal barrier function and gut microbiota. J. Appl. Microbiol. 2021, 131, 470–484. [Google Scholar] [CrossRef]
- Kim, J.J.; Khan, W.I. Goblet cells and mucins: Role in innate defense in enteric infections. Pathogens 2013, 2, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Gundamaraju, R.; Chong, W.C. Consequence of distinctive expression of MUC2 in colorectal cancers: How much is actually bad? Biochim. Biophys. Acta BBA Rev. Cancer 2021, 1876, 188579. [Google Scholar] [CrossRef] [PubMed]
- Martens, E.C.; Neumann, M.; Desai, M.S. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Microbiol. 2018, 16, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yu, X.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. The role of MUC2 mucin in intestinal homeostasis and the impact of dietary components on MUC2 expression. Int. J. Biol. Macromol. 2020, 164, 884–891. [Google Scholar] [CrossRef]
- Wang, G.; Tang, H.; Zhang, Y.; Xiao, X.; Xia, Y.; Ai, L. The intervention effects of Lactobacillus casei LC2W on Escherichia coli O157: H7-induced mouse colitis. Food Sci. Hum. Wellness 2020, 9, 289–294. [Google Scholar] [CrossRef]
- Gobert, A.P.; Coste, A.; Guzman, C.A.; Vareille, M.; Hindré, T.; de Sablet, T.; Girardeau, J.-P.; Martin, C. Modulation of chemokine gene expression by Shiga-toxin producing Escherichia coli belonging to various origins and serotypes. Microbes Infect. 2008, 10, 159–165. [Google Scholar] [CrossRef]
- Shi, Y.F. The role of inflammatory cytokines in bacterial infection. Int. J. Intern. Med. 2009, 36, 112–115. [Google Scholar]
- Zhao, X.; Wang, L.; Zhu, C.; Xia, X.; Zhang, S.; Wang, Y.; Zhang, H.; Xu, Y.; Chen, S.; Jiang, J. The antimicrobial peptide mastoparan X protects against enterohemorrhagic Escherichia coli O157: H7 infection, inhibits inflammation, and enhances the intestinal epithelial barrier. Front. Microbiol. 2021, 12, 644887. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Suzuki, R.; Koyanagi, Y.; Isogai, H.; Yoneyama, H.; Isogai, E. Inhibition of enterohemorrhagic Escherichia coli O157: H7 infection in a gnotobiotic mouse model with pre-colonization by Bacteroides strains. Biomed. Rep. 2019, 10, 175–182. [Google Scholar] [CrossRef]
- Lawley, T.D.; Walker, A.W. Intestinal colonization resistance. Immunology 2013, 138, 1–11. [Google Scholar] [CrossRef]
- Kelly, D.; Conway, S.; Aminov, R. Commensal gut bacteria: Mechanisms of immune modulation. Trends Immunol. 2005, 26, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhao, M.; Lu, Z.; Lv, F.; Zhao, H.; Bie, X.L. L. johnsonii, L. plantarum, and L. rhamnosus alleviated Enterohaemorrhagic Escherichia coli-induced diarrhoea in mice by regulating gut microbiota. Microb. Pathog. 2021, 154, 104856. [Google Scholar] [CrossRef] [PubMed]
- Den Besten, G.; Van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.J. The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Biswas, D. Short chain and polyunsaturated fatty acids in host gut health and foodborne bacterial pathogen inhibition. Crit. Rev. Food Sci. Nutr. 2017, 57, 3987–4002. [Google Scholar] [CrossRef]
- Wang, L.; Hu, L.; Xu, Q.; Jiang, T.; Fang, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Bifidobacteria exert species-specific effects on constipation in BALB/c mice. Food Funct. 2017, 8, 3587–3600. [Google Scholar] [CrossRef]



| Group | −6–0 d | 1–7 d | 7–14 d |
|---|---|---|---|
| Control | Diet ad libitum | 0.2 mL PBS | 0.2 mL PBS |
| Model | Diet ad libitum | 0.2 mL PBS | 0.2 mL EHEC O157:H7 |
| K5-PBS | Diet ad libitum | 0.2 mL B. longum K5 | 0.2 mL PBS |
| K5-EHEC | Diet ad libitum | 0.2 mL B. longum K5 | 0.2 mL EHEC O157:H7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Li, C.; Cao, T.; Lv, X.; Yue, Y.; Li, S.; Cheng, Y.; Liu, F.; Huo, G.; Li, B. Bifidobacterium longum K5 Prevents Enterohaemorrhagic Escherichia coli O157:H7 Infection in Mice through the Modulation of the Gut Microbiota. Nutrients 2024, 16, 1164. https://doi.org/10.3390/nu16081164
Liu D, Li C, Cao T, Lv X, Yue Y, Li S, Cheng Y, Liu F, Huo G, Li B. Bifidobacterium longum K5 Prevents Enterohaemorrhagic Escherichia coli O157:H7 Infection in Mice through the Modulation of the Gut Microbiota. Nutrients. 2024; 16(8):1164. https://doi.org/10.3390/nu16081164
Chicago/Turabian StyleLiu, Deyu, Chunyan Li, Ting Cao, Xiuli Lv, Yingxue Yue, Shuang Li, Yang Cheng, Fei Liu, Guicheng Huo, and Bailiang Li. 2024. "Bifidobacterium longum K5 Prevents Enterohaemorrhagic Escherichia coli O157:H7 Infection in Mice through the Modulation of the Gut Microbiota" Nutrients 16, no. 8: 1164. https://doi.org/10.3390/nu16081164
APA StyleLiu, D., Li, C., Cao, T., Lv, X., Yue, Y., Li, S., Cheng, Y., Liu, F., Huo, G., & Li, B. (2024). Bifidobacterium longum K5 Prevents Enterohaemorrhagic Escherichia coli O157:H7 Infection in Mice through the Modulation of the Gut Microbiota. Nutrients, 16(8), 1164. https://doi.org/10.3390/nu16081164







