Bifidobacterium longum Subsp. infantis Promotes IgA Level of Growing Mice in a Strain-Specific and Intestinal Niche-Dependent Manner
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria
2.2. Animal Experiments
2.3. Biochemical Indicator Measurement
2.4. qRT-PCR Analysis
2.5. Separation of sIgA-Coated Bacteria
2.6. DNA Extraction and 16S rRNA Gene Sequencing
2.7. Statistical Analyses
3. Results
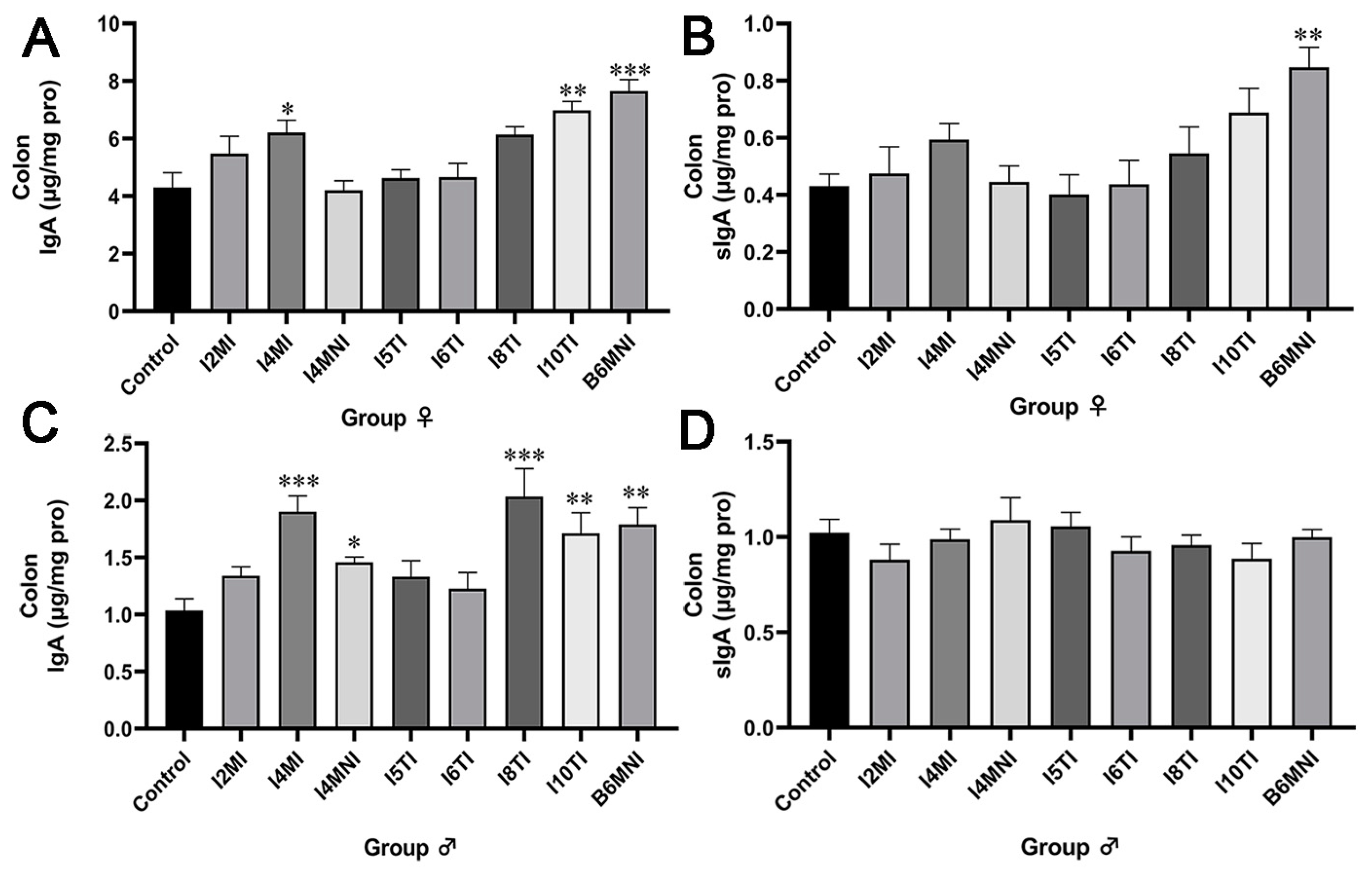
3.1. B. longum Subsp. infantis Influenced IgA and sIgA Levels in the Colon of Mice
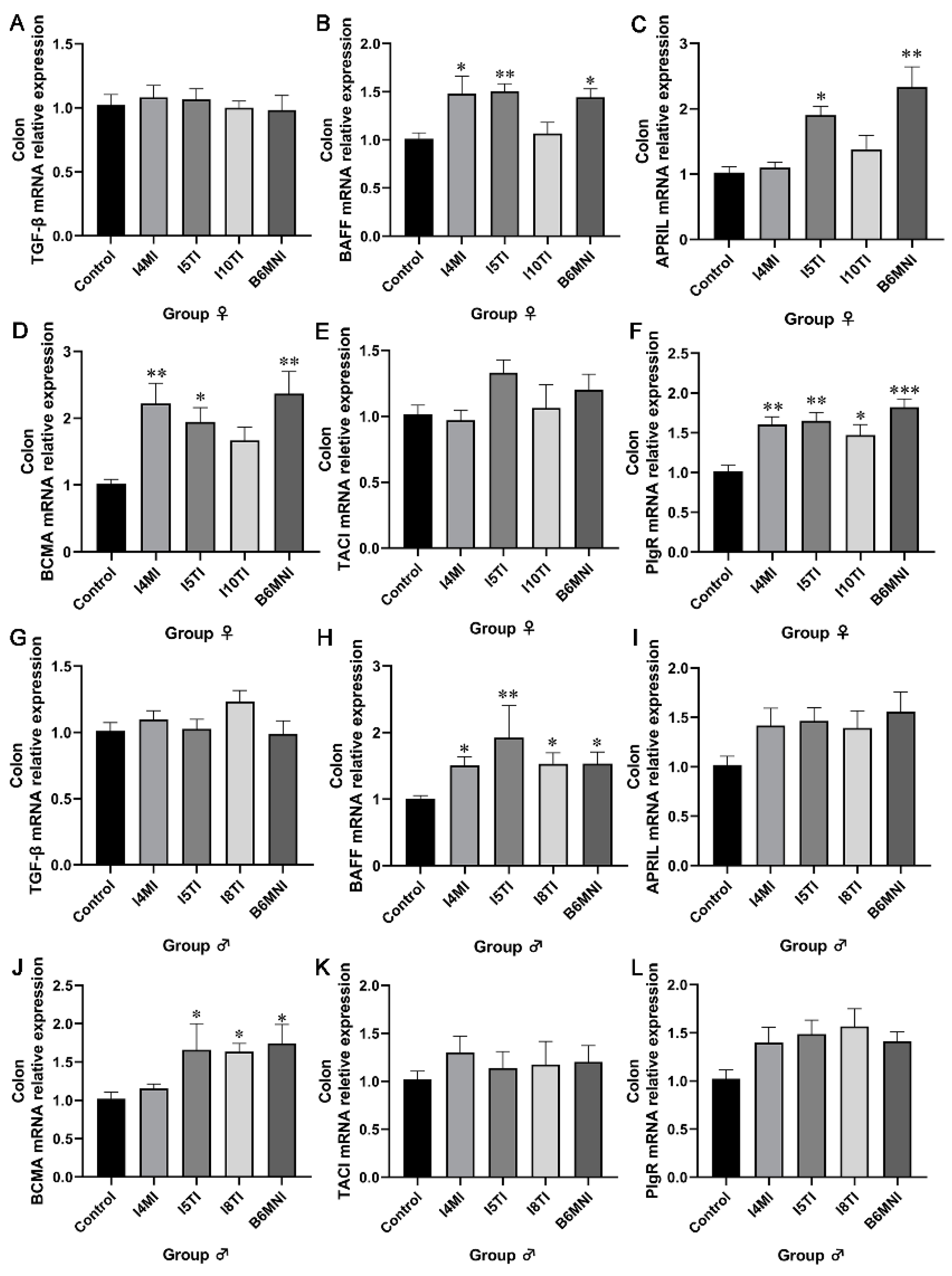
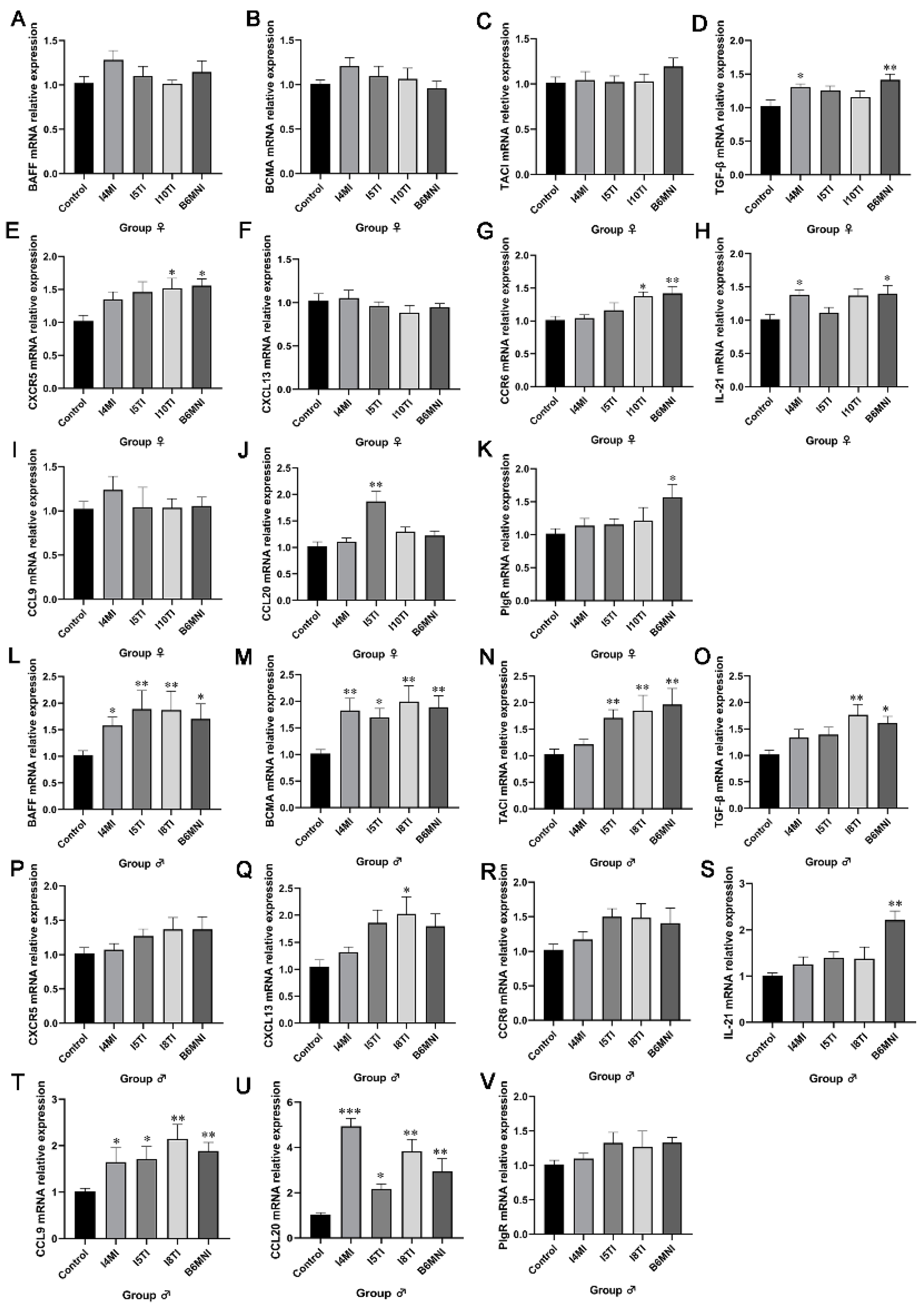
3.2. B. longum Subsp. infantis Influenced IgA Synthesis Genes
3.3. B. longum Subsp. infantis Influenced IgA+ Plasmocyte Synthetic-Related Genes in PPs
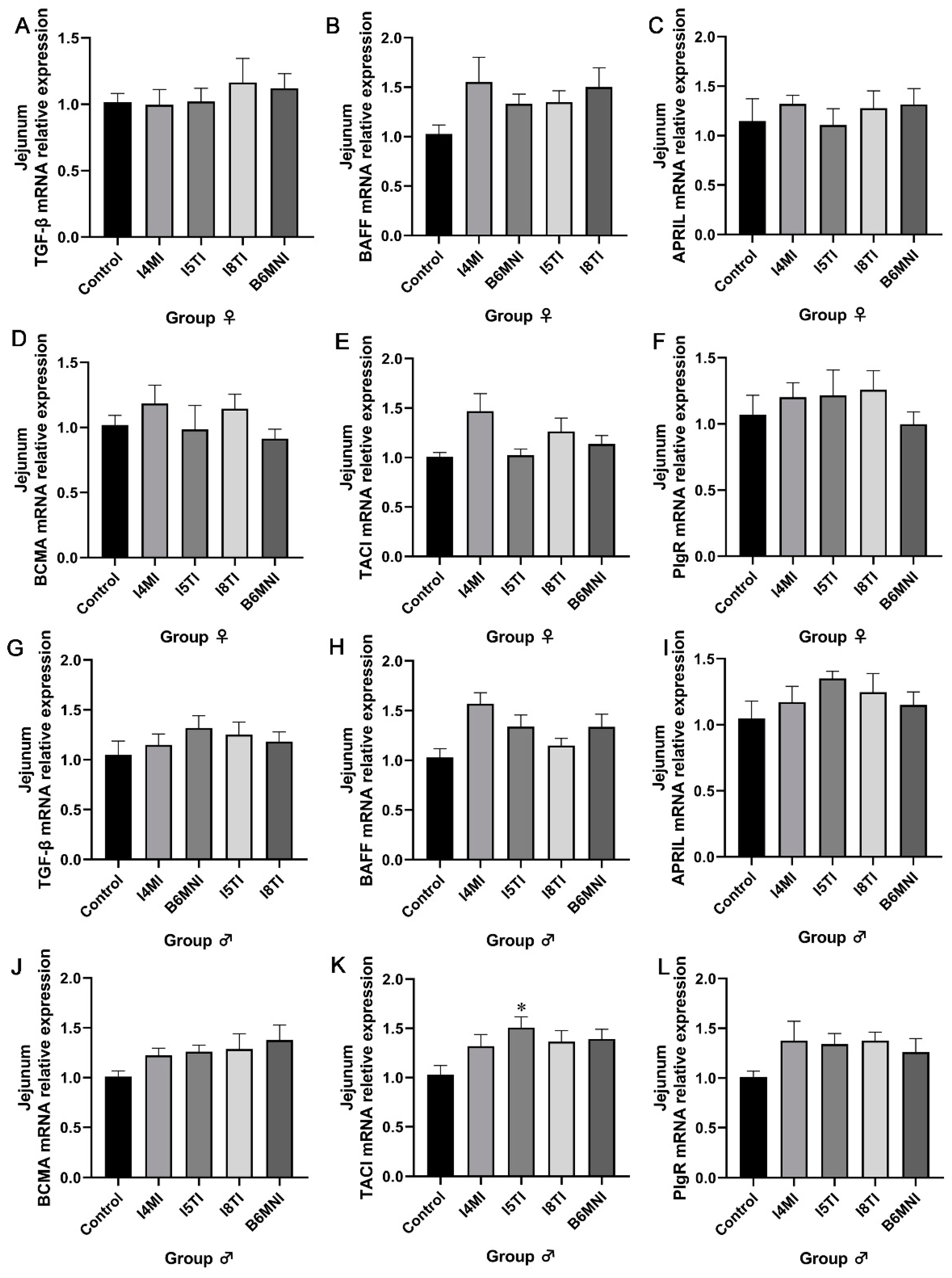
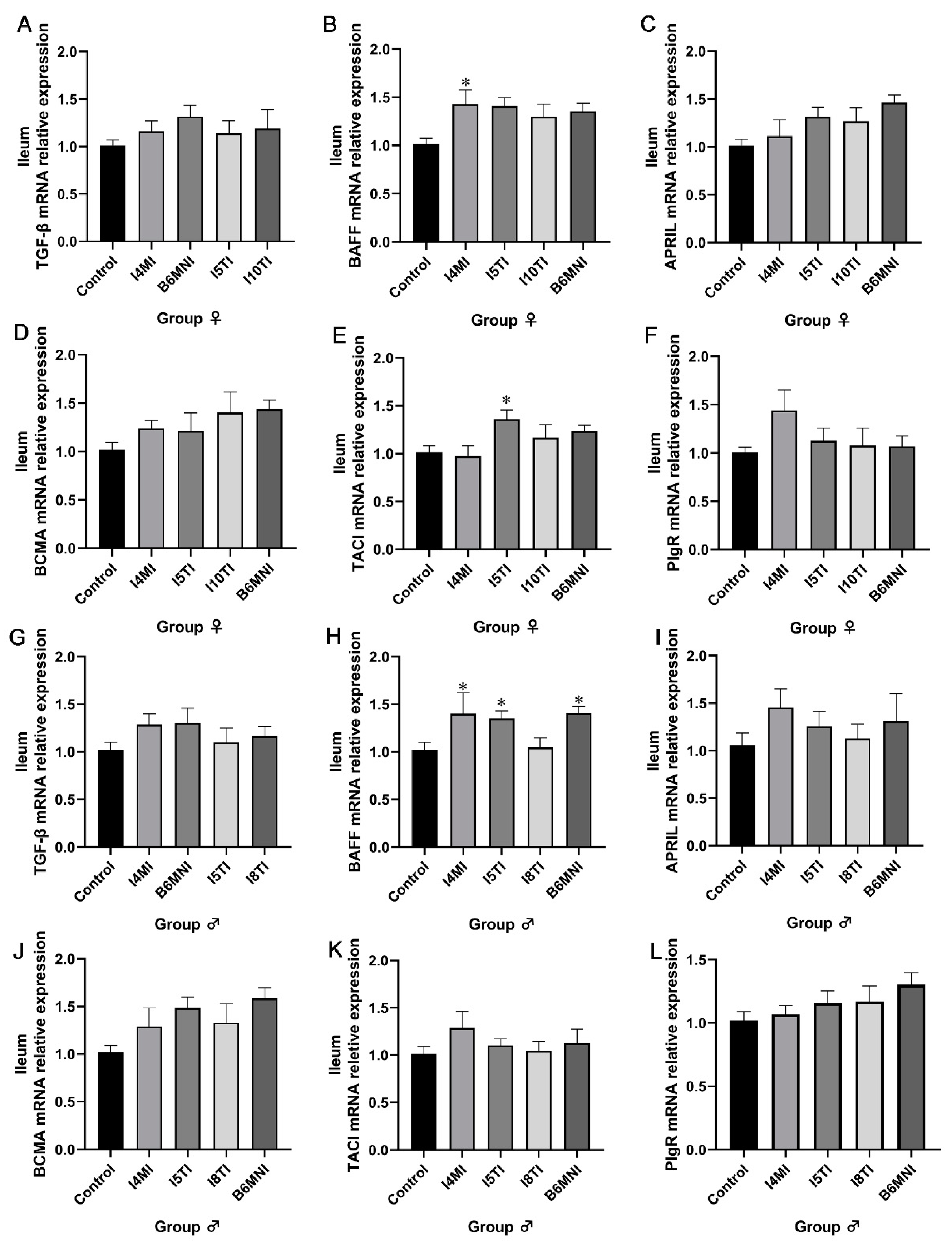
3.4. B. longum Subsp. infantis Influenced IgA Synthesis-Related Genes in Jejunum and Ileum
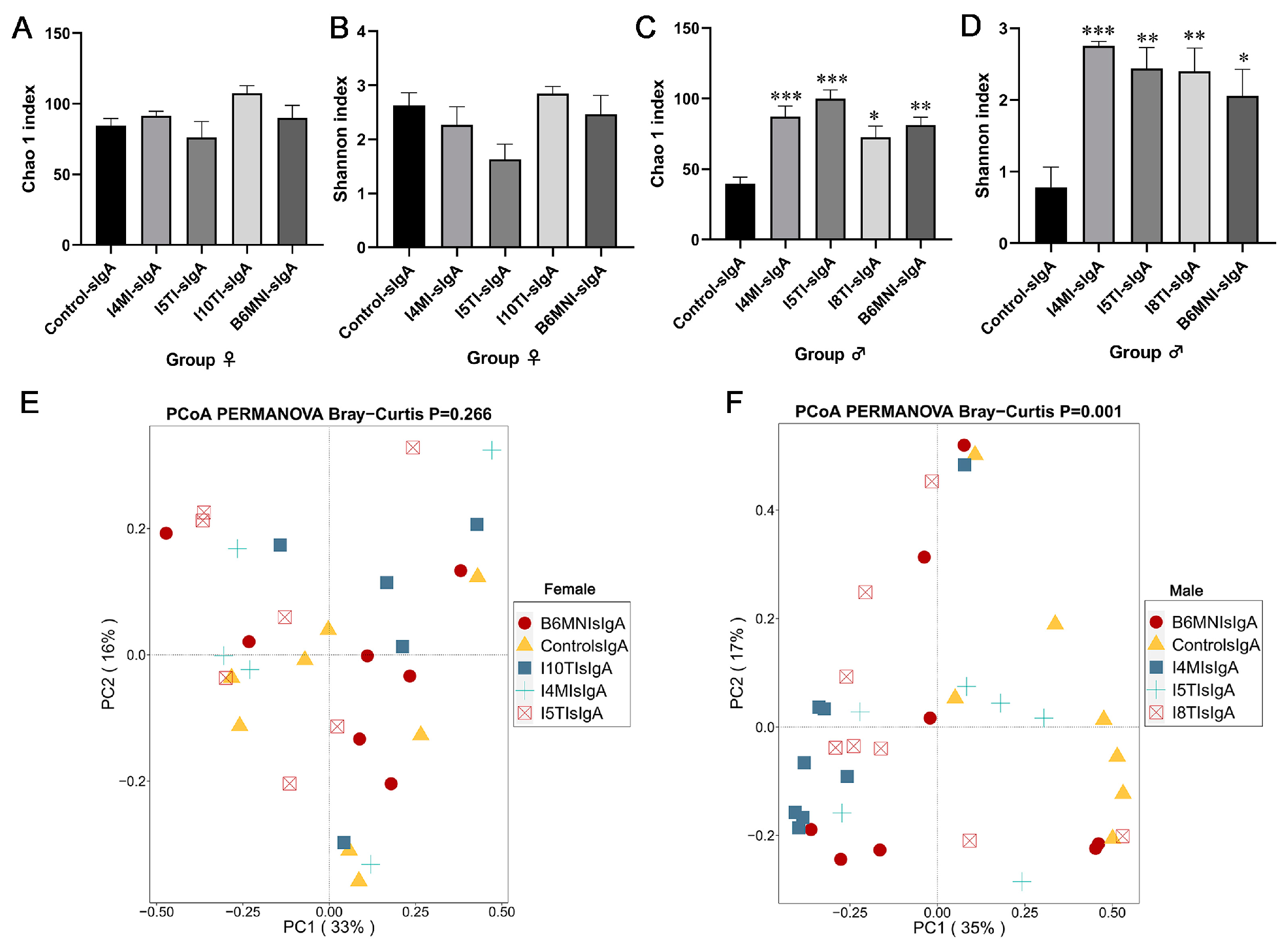
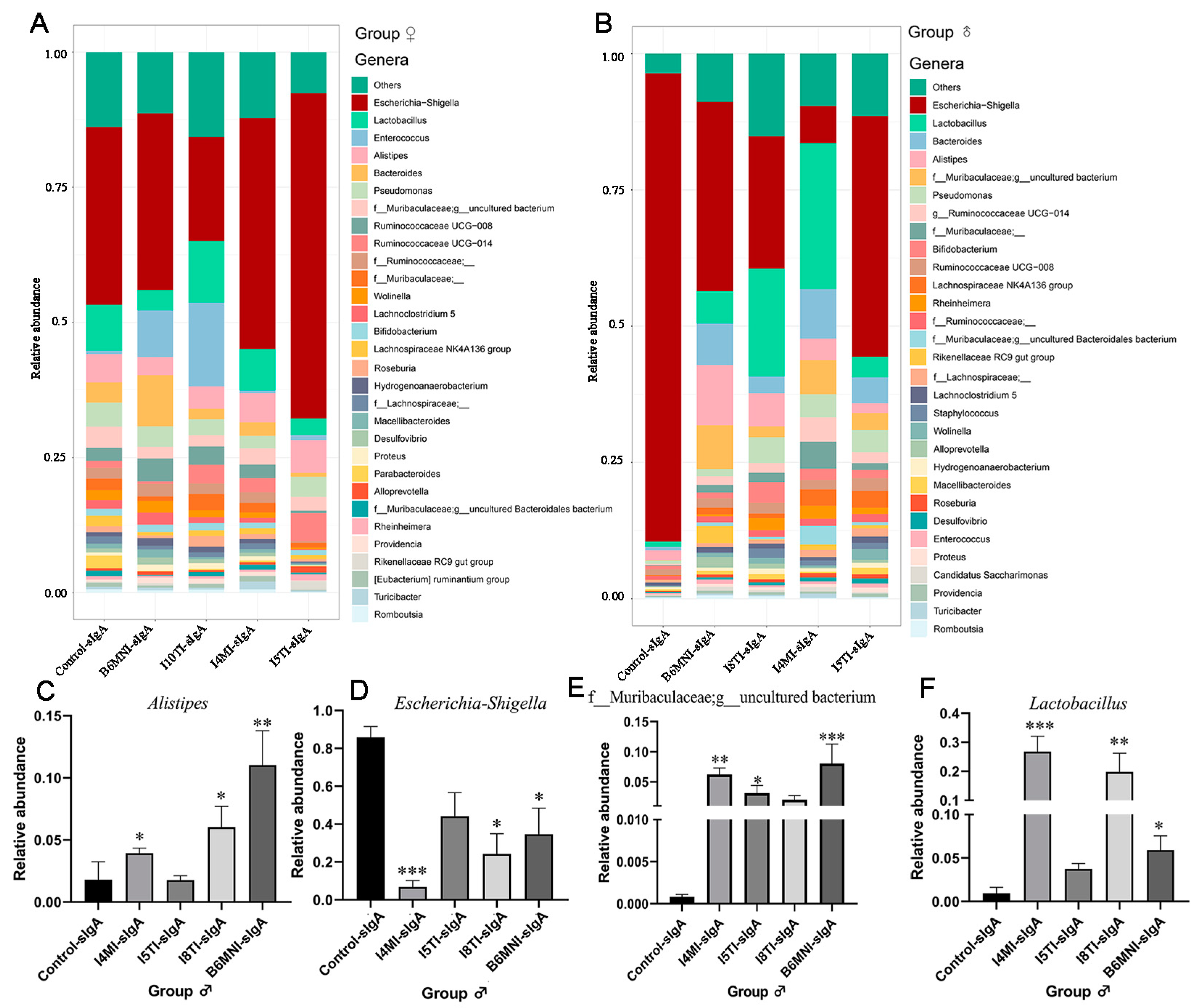
3.5. Variety of sIgA-Coated Bacterial Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ding, M.; Yang, B.; Ross, R.P.; Stanton, C.; Zhao, J.; Zhang, H.; Chen, W. Crosstalk between sIgA-coated bacteria in infant gut and early-life health. Trends Microbiol. 2021, 29, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Zhao, W.; Li, Y.; Zou, X.; Dong, X. Intestinal immune development is accompanied by temporal deviation in microbiota composition of newly hatched pigeon squabs. Microbiol. Spectr. 2022, 10, 1892–1921. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Godoy-Vitorino, F.; Knight, R.; Blaser, M.J. Role of the microbiome in human development. Gut 2019, 68, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, E.D.; Sonnenburg, J.L. The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 2019, 17, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.-C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.-C.; Arévalo, A.; Stiemsma, L.; Dimitriu, P.; Chico, M.E.; Loor, S.; Vaca, M.; Boutin, R.C.T.; Morien, E.; Jin, M.; et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J. Allergy Clin. Immunol. 2018, 142, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Fagarasan, S.; Kawamoto, S.; Kanagawa, O.; Suzuki, K. Adaptive Immune Regulation in the Gut: T Cell–Dependent and T Cell–Independent IgA Synthesis. Annu. Rev. Immunol. 2010, 28, 243–273. [Google Scholar] [CrossRef] [PubMed]
- Huus, K.E.; Bauer, K.C.; Brown, E.M.; Bozorgmehr, T.; Woodward, S.E.; Serapio-Palacios, A.; Boutin, R.C.T.; Petersen, C.; Finlay, B.B. Commensal Bacteria Modulate Immunoglobulin A Binding in Response to Host Nutrition. Cell Host Microbe 2020, 27, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Catanzaro, J.R.; Strauss, J.D.; Bielecka, A.; Porto, A.F.; Lobo, F.M.; Urban, A.; Schofield, W.B.; Palm, N.W. IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci. Rep. 2019, 9, 13574. [Google Scholar] [CrossRef] [PubMed]
- Mirpuri, J.; Raetz, M.; Sturge, C.R.; Wilhelm, C.L.; Benson, A.; Savani, R.C.; Hooper, L.V.; Yarovinsky, F. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 2014, 5, 28–39. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Ladinsky, M.S.; Yu, K.B.; Sanders, J.G.; Yoo, B.B.; Chou, W.C.; Conner, M.E.; Earl, A.M.; Knight, R.; Bjorkman, P.J.; et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 2018, 300, 795–800. [Google Scholar] [CrossRef]
- Zhong, Z.; Zhang, H.; Nan, K.; Zhong, J.; Wu, Q.; Lu, L.; Yue, Y.; Zhang, Z.; Guo, M.; Wang, Z.; et al. Fasting-mimicking diet drives antitumor immunity against colorectal cancer by reducing IgA-producing cells. Cancer Res. 2023, 83, 3529–3543. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Xun, M.; Ren, S.; Wang, J. Effects of dietary organic acids and probiotics on laying performance, egg quality, serum antioxidants and expressions of reproductive genes of laying ducks in the late phase of production. Poult. Sci. 2022, 101, 102189. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.A.; Marette, A. Potential health benefits of combining yogurt and fruits based on their probiotic and prebiotic properties. Adv. Nutr. 2017, 8, 155S–164S. [Google Scholar] [CrossRef] [PubMed]
- Slavin, J. Fiber and prebiotics: Mechanisms and health benefits. Nutrients 2013, 5, 1417–1435. [Google Scholar] [CrossRef] [PubMed]
- Robertson, R.C.; Manges, A.R.; Finlay, B.B.; Prendergast, A.J. The human microbiome and child growth First 1000 days and beyond. Trends Microbiol. 2019, 27, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Le Doare, K.; Holder, B.; Bassett, A.; Pannaraj, P.S. Mother’s milk: A purposeful contribution to the development of the infant microbiota and immunity. Front. Immunol. 2018, 9, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Eidelman, A.I. Impact of the Infant Formula Shortage on Breastfeeding Rates. J. Pediatr. Health Care 2022, 37, 279–286. [Google Scholar]
- Terahara, M.; Nakamura, Y.; Tsuboi, M.; Jinno, S.; Tsukahara, T.; Miyake, T.; Shimojo, N. Effects of the intake of non-live Bifidobacterium bifidum on the faecal IgA of full-term infants: A double-blind, randomised, placebo-controlled study. Biosci. Microbiota Food Health 2021, 40, 196–203. [Google Scholar] [CrossRef]
- Hiraku, A.; Nakata, S.; Murata, M.; Xu, C.; Mutoh, N.; Arai, S.; Odamaki, T.; Iwabuchi, N.; Tanaka, M.; Tsuno, T.; et al. Early Probiotic Supplementation of Healthy Term Infants with Bifidobacterium longum subsp. infantis M-63 Is Safe and Leads to the Development of Bifidobacterium-Predominant Gut Microbiota: A Double-Blind, Placebo-Controlled Trial. Nutrients 2023, 15, 1402. [Google Scholar] [CrossRef]
- Kim, J.Y.; Bang, S.-J.; Kim, J.-Y.; Choi, E.J.; Heo, K.; Shim, J.-J.; Lee, J.-L. The probiotic Bifidobacterium animalis strain ssp. HY8002 potentially iImproves the mucosal integrity of an altered intestinal microbial environment. Front. Immunol. 2022, 13, 817591. [Google Scholar]
- Nocerino, R.; De Filippis, F.; Cecere, G.; Marino, A.; Micillo, M.; Di Scala, C.; de Caro, C.; Calignano, A.; Bruno, C.; Paparo, L.; et al. The therapeutic efficacy of Bifidobacterium animalis subsp. lactis BB-12® in infant colic: A randomised, double blind, placebo-controlled trial. Aliment. Pharmacol. Ther. 2020, 51, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-H.; Um, J.-I.; Lee, B.-J.; Goh, J.-S.; Park, S.-Y.; Kim, W.-S.; Kim, P.-H. Encapsulated Bifidobacterium bifidum potentiates intestinal IgA production. Cell. Immunol. 2002, 219, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K. Diversified IgA-Bacteria Interaction in Gut Homeostasis. Adv. Exp. Med. Biol. 2020, 1254, 105–166. [Google Scholar] [PubMed]
- Wu, B.-B.; Yang, Y.; Xu, X.; Wang, W.-P. Effects of Bifidobacterium supplementation on intestinal microbiota composition and the immune response in healthy infants. World J. Pediatr. 2016, 12, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, A.J.; Slack, E. The functional interactions of commensal bacteria with intestinal secretory IgA. Curr. Opin. Gastroenterol. 2007, 23, 673–678. [Google Scholar] [CrossRef]
- Mikulic, J.; Longet, S.; Favre, L.; Benyacoub, J.; Corthesy, B. Secretory IgA in complex with Lactobacillus rhamnosus potentiates mucosal dendritic cell-mediated Treg cell differentiation via TLR regulatory proteins, RALDH2 and secretion of IL-10 and TGF-β. Cell Mol. Immunol. 2017, 14, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Fransen, F.; van Beek, A.A.; Borghuis, T.; Meijer, B.; Hugenholtz, F.; van der Gaast-de Jongh, C.; Savelkoul, H.F.; de Jonge, M.I.; Faas, M.M.; Boekschoten, M.V.; et al. The impact of gut microbiota on gender-specific differences in immunity. Front. Immunol. 2017, 8, 754. [Google Scholar] [CrossRef]
- Underwood, M.A.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Bifidobacterium longum subspecies infantis: Champion colonizer of the infant gut. Pediatr. Res. 2015, 77, 229–235. [Google Scholar] [CrossRef]
- Ding, M.; Zheng, Y.; Liu, F.; Tian, F.; Ross, R.P.; Stanton, C.; Yu, R.; Zhao, J.; Zhang, H.; Yang, B.; et al. Lactation time influences the composition of Bifidobacterium and lactobacillus at species level in human breast milk. Benef. Microbes 2022, 13, 319–330. [Google Scholar] [CrossRef]
- Ding, M.; Yang, B.; Khine, W.W.T.; Lee, Y.-K.; Rahayu, E.S.; Ross, R.P.; Stanton, C.; Zhao, J.; Zhang, H.; Chen, W. The species-level composition of the fecal Bifidobacterium and Lactobacillus genera in Indonesian children differs from that of their mothers. Microorganisms 2021, 9, 1995. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Chen, H.; Yu, R.; Ross, R.P.; Stanton, C.; Zhang, H.; Yang, B.; Chen, W. Shared and non-shared sIgA-coated and -uncoated bacteria in intestine of mother and infant pairs. Int. J. Mol. Sci. 2022, 23, 9873. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Amano, H.; Kawano, S.; Minowa, K.; Ando, S.; Watanabe, T.; Nakano, S.; Suzuki, J.; Morimoto, S.; Tokano, Y.; et al. Increased serum concentration of BAFF/APRIL and IgA2 subclass in patients with mixed connective tissue disease complicated by interstitial lung disease. Mod. Rheumatol. 2014, 24, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Michael, M.R.; Wieske, L.; Koel-Simmelink, M.J.; van Schaik, I.N.; Teunissen, C.E.; Eftimov, F. Serum B-cell activating factor is not a potential biomarker for disease activity in chronic inflammatory demyelinating polyneuropathy. J. Neuroimmunol. 2023, 382, 578169. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, R.; Rodríguez-Aguilar, L.; Llorente, S.; Jimenez-Coll, V.; Martínez-Banaclocha, H.; Galián, J.A.; Botella, C.; Moya-Quiles, M.R.; Muro-Perez, M.; Minguela, A.; et al. Early cytomegalovirus reactivation in renal recipients is associated with high levels of B cell maturation antigen transcript expression prior to transplantation. Int. J. Mol. Sci. 2023, 24, 10491. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Carmona, Y.; Fribourg, M.; Sowa, A.; Cerutti, A.; Cunningham-Rundles, C. TACI and endogenous APRIL in B cell maturation. Clin. Immunol. 2023, 253, 109689. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, A. Location, location, location: B-cell differentiation in the gut lamina propria. Mucosal Immunol. 2008, 1, 8–10. [Google Scholar] [CrossRef] [PubMed]
- Date, Y.; Ebisawa, M.; Fukuda, S.; Shima, H.; Obata, Y.; Takahashi, D.; Kato, T.; Hanazato, M.; Nakato, G.; Williams, I.R.; et al. NALT M cells are important for immune induction for the common mucosal immune system. Int. Immunol. 2017, 29, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Ladjemi, M.Z.; Martin, C.; Lecocq, M.; Detry, B.; Nana, F.A.; Moulin, C.; Weynand, B.; Fregimilicka, C.; Bouzin, C.; Thurion, P.; et al. Increased IgA expression in lung lymphoid follicles in severe chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2019, 199, 592–609. [Google Scholar] [CrossRef]
- Enani, S.; Przemska-Kosicka, A.; Childs, C.E.; Maidens, C.; Dong, H.; Conterno, L.; Tuohy, K.; Todd, S.; Gosney, M.; Yaqoob, P. Impact of ageing and a synbiotic on the immune response to seasonal influenza vaccination; a randomised controlled trial. Clin. Nutr. 2018, 37, 443–451. [Google Scholar] [CrossRef]
- Manneck, D.; Braun, H.-S.; Schrapers, K.T.; Stumpff, F. TRPV3 and TRPV4 as candidate proteins for intestinal ammonium absorption. Acta Physiol. 2021, 233, e13694. [Google Scholar] [CrossRef] [PubMed]
- Lagos, L.; Bekkelund, A.K.; Skugor, A.; Ånestad, R.; Åkesson, C.P.; Press, C.M.; Øverland, M. Cyberlindnera jadinii yeast as a protein source for weaned piglets—Impact on immune response and gut microbiota. Front. Immunol. 2020, 11, 1924. [Google Scholar] [CrossRef] [PubMed]
- Martinsson, K.; Kling, L.L.; Roos-Ljungberg, K.; Griazeva, I.; Samoylovich, M.; Paul, S.; Rönnelid, J.; Weitoft, T.; Wetterö, J.; Kastbom, A. Extramucosal formation and prognostic value of secretory antibodies in rheumatoid arthritis. Arthritis Rheumatol. 2022, 74, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Chintalacharuvu, K.R.; Tavill, A.S.; Louis, L.N.; Vaerman, J.P.; Lamm, M.E.; Kaetzel, C.S. Disulfide bond formation between dimeric immunoglobulin A and the polymeric immunoglobulin receptor during hepatic transcytosis. Hepatology 1994, 9, 162–173. [Google Scholar]
- Blackburn, J.B.; Schaff, J.A.; Gutor, S.; Du, R.-H.; Nichols, D.; Sherrill, T.; Gutierrez, A.J.; Xin, M.K.; Wickersham, N.; Zhang, Y.; et al. Secretory cells are the primary source of pIgR in small airways. Am. J. Respir. Cell Mol. Biol. 2022, 67, 334–345. [Google Scholar] [CrossRef]
- Kato, L.M.; Kawamoto, S.; Maruya, M.; Fagarasan, S. The role of the adaptive immune system in regulation of gut microbiota. Immunol. Rev. 2014, 260, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Juan, Z.; Qing, Z.; Yongping, L.; Qian, L.; Wu, W.; Wen, Y.; Tong, J.; Ding, B. Probiotics for the treatment of docetaxel-related weight gain of breast cancer patients—A single-center, randomized, double-blind, and placebo-controlled trial. Front. Nutr. 2021, 8, 762929. [Google Scholar] [CrossRef]
- Gopalakrishna, K.P.; Macadangdang, B.R.; Rogers, M.B.; Tometich, J.T.; Firek, B.A.; Baker, R.; Ji, J.; Burr, A.H.P.; Ma, C.; Good, M.; et al. Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat. Med. 2019, 25, 1110–1115. [Google Scholar] [CrossRef]
- Qi, C.; Ding, M.; Li, S.; Zhou, Q.; Li, D.; Yu, R.; Sun, J. Sex-dependent modulation of immune development in mice by secretory IgA–coated Lactobacillus reuteri isolated from breast milk. J. Dairy Sci. 2021, 104, 3863–3875. [Google Scholar] [CrossRef]
- Org, E.; Mehrabian, M.; Parks, B.W.; Shipkova, P.; Liu, X.; Drake, T.A.; Lusis, A.J. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes 2016, 7, 313–322. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Burrows, M.; Khan, A.A.; Graham, L.; Volchkov, P.; Becker, L.; Antonopoulos, D.; Umesaki, Y.; Chervonsky, A.V. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013, 39, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Borgo, F.; Garbossa, S.; Riva, A.; Severgnini, M.; Luigiano, C.; Benetti, A.; Pontiroli, A.E.; Morace, G.; Borghi, E. Body mass index and sex affect diverse microbial niches within the gut. Front. Microbiol. 2018, 9, 213–225. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| TGF-β | CTCCCGTGGCTTCTAGTGC | GCCTTAGTTTGGACAGGATCTG |
| BAFF | GGAACAGACGCGCTTTCCA | GGCCGGTCATTACCTTTTCGT |
| APRIL | GGAACAGACGCGCTTTCCA | GGCCGGTCATTACCTTTTCGT |
| BCMA | GCGCAACAGTGTTTCCACAG | CGCTTGGATCACAGTAAGGCT |
| TACI | ATGGCATTCTGCCCCAAAGAT | ATGGTCGTAGTACCTGCCTTG |
| pIgR | ATGAGGCTCTACTTGTTCACGC | CGCCTTCTATACTACTCACCTCC |
| CXCR5 | ATGAACTACCCACTAACCCTGG | TGTAGGGGAATCTCCGTGCT |
| CXCL13 | GGCCACGGTATTCTGGAAGC | GGGCGTAACTTGAATCCGATCTA |
| CCR6 | CCTGGGCAACATTATGGTGGT | CAGAACGGTAGGGTGAGGACA |
| IL-21 | GGACCCTTGTCTGTCTGGTAG | TGTGGAGCTGATAGAAGTTCAGG |
| CCL9 | CCCTCTCCTTCCTCATTCTTACA | AGTCTTGAAAGCCCATGTGAAA |
| CCL20 | GCCTCTCGTACATACAGACGC | GCCTCTCGTACATACAGACGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, M.; Li, B.; Chen, H.; Ross, R.P.; Stanton, C.; Zhao, J.; Chen, W.; Yang, B. Bifidobacterium longum Subsp. infantis Promotes IgA Level of Growing Mice in a Strain-Specific and Intestinal Niche-Dependent Manner. Nutrients 2024, 16, 1148. https://doi.org/10.3390/nu16081148
Ding M, Li B, Chen H, Ross RP, Stanton C, Zhao J, Chen W, Yang B. Bifidobacterium longum Subsp. infantis Promotes IgA Level of Growing Mice in a Strain-Specific and Intestinal Niche-Dependent Manner. Nutrients. 2024; 16(8):1148. https://doi.org/10.3390/nu16081148
Chicago/Turabian StyleDing, Mengfan, Bowen Li, Haiqin Chen, Reynolds Paul Ross, Catherine Stanton, Jianxin Zhao, Wei Chen, and Bo Yang. 2024. "Bifidobacterium longum Subsp. infantis Promotes IgA Level of Growing Mice in a Strain-Specific and Intestinal Niche-Dependent Manner" Nutrients 16, no. 8: 1148. https://doi.org/10.3390/nu16081148
APA StyleDing, M., Li, B., Chen, H., Ross, R. P., Stanton, C., Zhao, J., Chen, W., & Yang, B. (2024). Bifidobacterium longum Subsp. infantis Promotes IgA Level of Growing Mice in a Strain-Specific and Intestinal Niche-Dependent Manner. Nutrients, 16(8), 1148. https://doi.org/10.3390/nu16081148