S/O/W Emulsion with CAPE Ameliorates DSS-Induced Colitis by Regulating NF-κB Pathway, Gut Microbiota and Fecal Metabolome in C57BL/6 Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Preparation of S/O/W Emulsion with CAPE
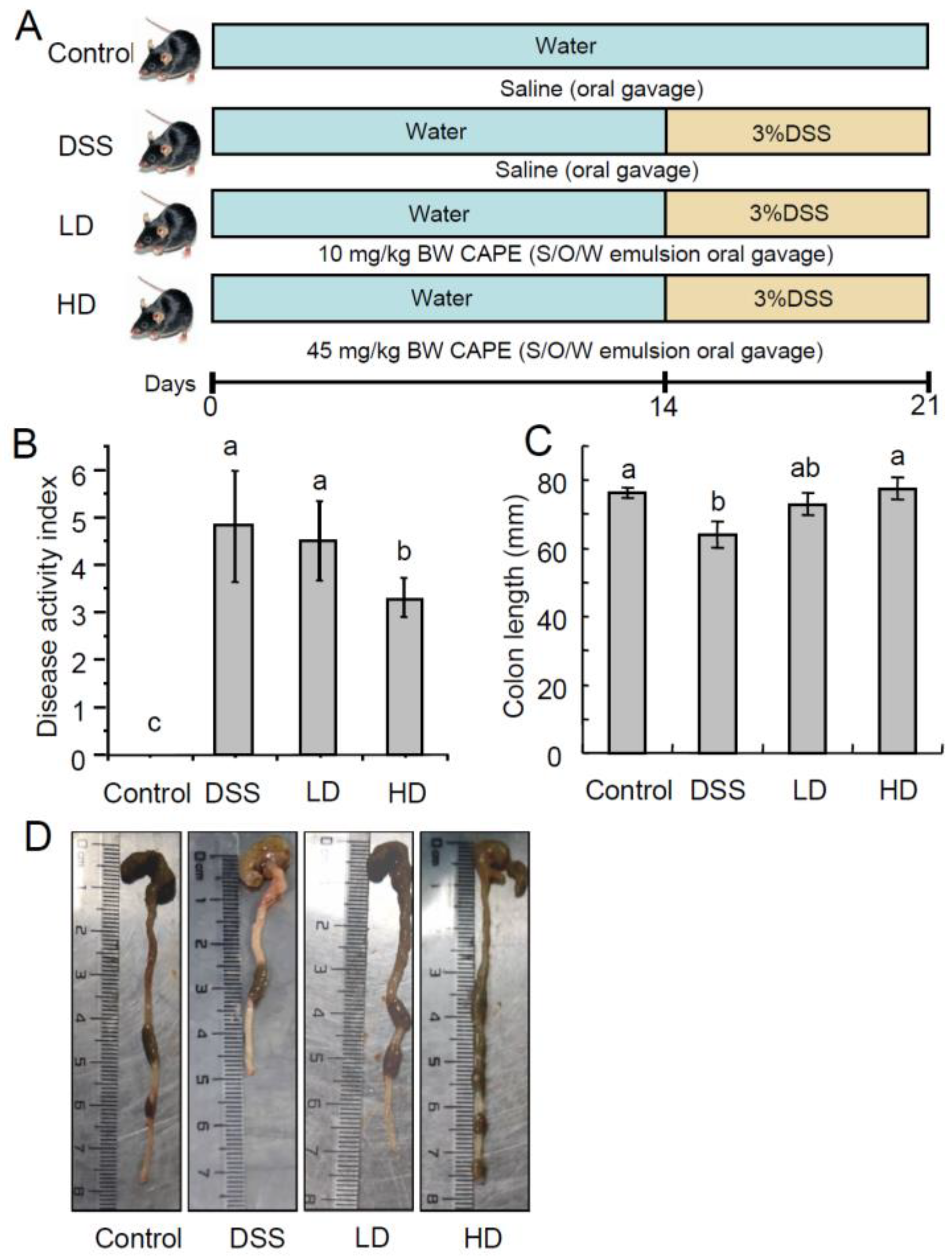
2.3. Animal Experiment
2.4. Disease Activity Index (DAI)
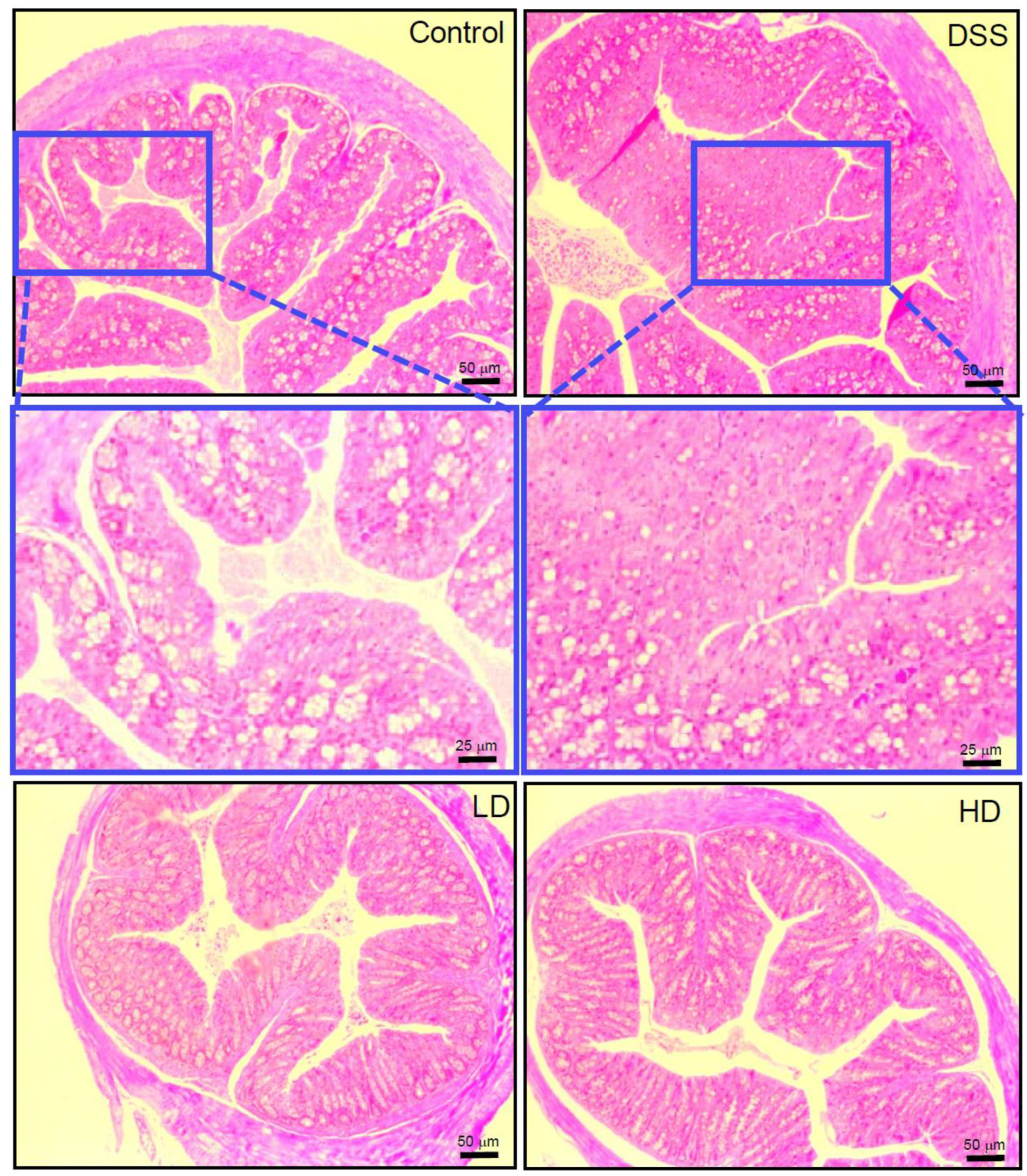
2.5. Histopathology of Colitis
2.6. Analysis of Inflammatory Cytokines in Colonic Tissue
2.7. Western Blot Analysis
2.8. Gut Microbiota Analysis
2.9. Quantification of Short-Chain Fatty Acids (SCFAs)
2.10. Fecal Metabolite Analysis
2.11. Statistical Analysis
3. Results
3.1. The CAPE-Emulsion Alleviated Symptoms of DSS-Induced Colitis in Mice
3.2. The CAPE-Emulsion Reduced the Overproduction of Inflammatory Cytokines by Inhabiting NF-κB Signal Path in DSS-Induced Colitis Mice
3.3. The CAPE-Emulsion Recovered the SCFAs in DSS-Induced Colitis Mice
3.4. The CAPE-Emulsion Regulated the Gut Microbiota in DSS-Induced Colitis Mice
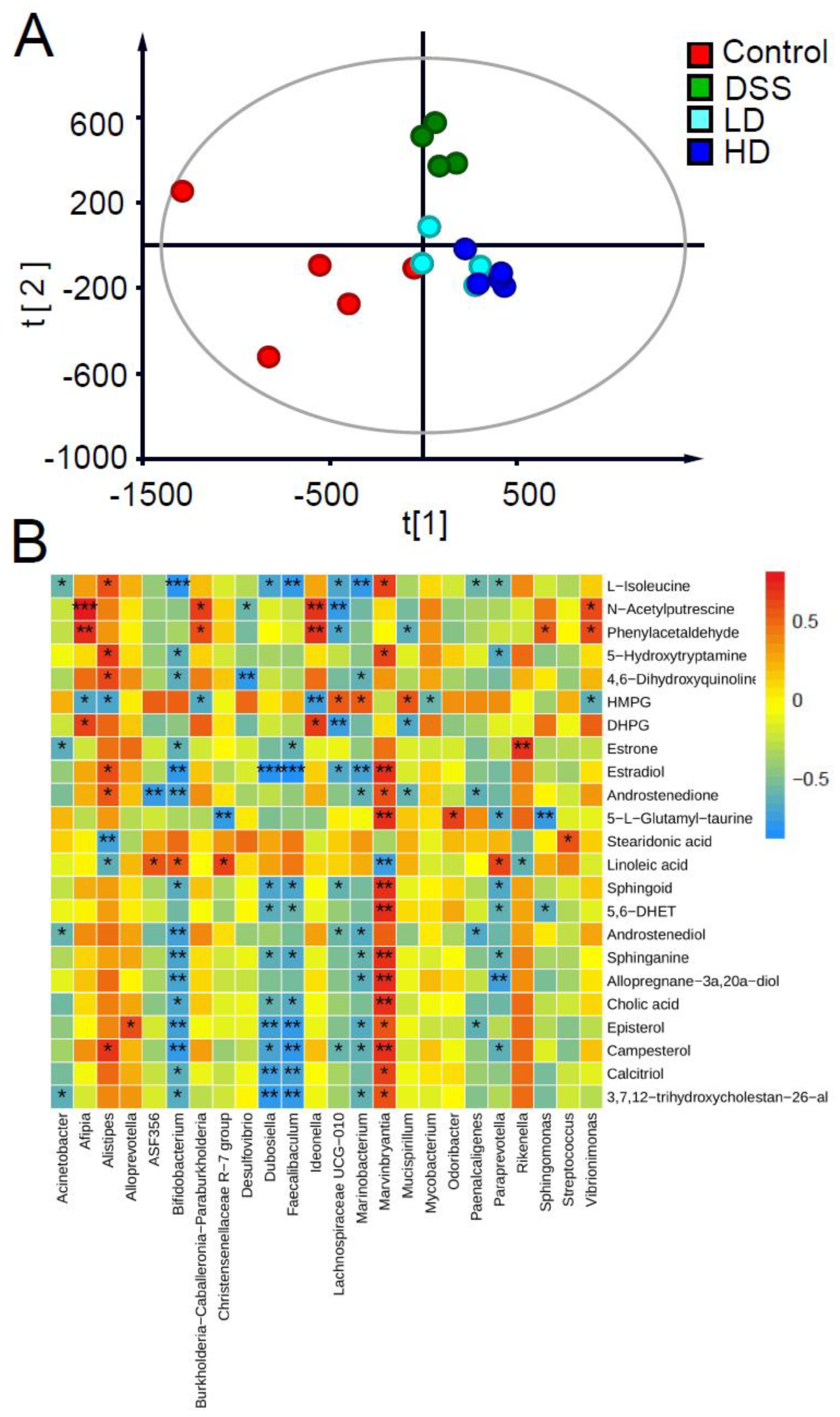
3.5. Effect of the CAPE-Emulsion on Fecal Metabolome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lin, W.S.; Lai, Y.J.; Kalyanam, N.; Ho, C.T.; Pan, M.H. S-Allylcysteine Inhibits PhIP/DSS-Induced Colon Carcinogenesis through Mitigating Inflammation, Targeting Keap1, and Modulating Microbiota Composition in Mice. Mol. Nutr. Food Res. 2020, 64, 2000576. [Google Scholar] [CrossRef]
- Manichanh, C.; Borruel, N.; Casellas, F.; Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 599–608. [Google Scholar] [CrossRef]
- Nagalingam NA, L.S. Role of the microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 2012, 18, 968–984. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Roy, B.C.; Khan, S.A.; Septer, S.; Umar, S. Microbiome, Metabolome and Inflammatory Bowel Disease. Microorganisms 2016, 4, 20. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef]
- Hu, Q.; Yuan, B.; Wu, X.; Du, H.; Gu, M.; Han, Y.; Yang, W.; Song, M.; Xiao, H. Dietary Intake of Pleurotus eryngii Ameliorated Dextran-Sodium-Sulfate-Induced Colitis in Mice. Mol. Nutr. Food Res. 2019, 63, e1801265. [Google Scholar] [CrossRef]
- Shao, X.; Sun, C.; Tang, X.; Zhang, X.; Han, D.; Liang, S.; Qu, R.; Hui, X.; Shan, Y.; Hu, L.; et al. Anti-Inflammatory and Intestinal Microbiota Modulation Properties of Jinxiang Garlic (Allium sativum L.) Polysaccharides toward Dextran Sodium Sulfate-Induced Colitis. J. Agric. Food Chem. 2020, 68, 12295–12309. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Xiao, N.; Zeng, L.; Xiao, J.; Huang, J.; Xu, Y.; Chen, Y.; Ren, Y.; Du, B. Structural characteristics of a mannoglucan isolated from Chinese yam and its treatment effects against gut microbiota dysbiosis and DSS-induced colitis in mice. Carbohydr. Polym. 2020, 250, 116958. [Google Scholar] [CrossRef]
- Zhao, B.; Wu, J.; Li, J.; Bai, Y.; Luo, Y.; Ji, B.; Xia, B.; Liu, Z.; Tan, X.; Lv, J.; et al. Lycopene Alleviates DSS-Induced Colitis and Behavioral Disorders via Mediating Microbes-Gut-Brain Axis Balance. J. Agric. Food Chem. 2020, 68, 3963–3975. [Google Scholar] [CrossRef]
- Xuan, H.; Ou, A.; Hao, S.; Shi, J.; Jin, X. Galangin Protects against Symptoms of Dextran Sodium Sulfate-induced Acute Colitis by Activating Autophagy and Modulating the Gut Microbiota. Nutrients 2020, 12, 347. [Google Scholar] [CrossRef]
- Feng, K.; Wei, Y.-S.; Hu, T.-G.; Linhardt, R.J.; Zong, M.-H.; Wu, H. Colon-targeted delivery systems for nutraceuticals: A review of current vehicles, evaluation methods and future prospects. Trends Food Sci. Technol. 2020, 102, 203–222. [Google Scholar] [CrossRef]
- Wlodarska, M.; Kostic, A.D.; Xavier, R.J. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe 2015, 17, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Luo, J.; Han, Y.; Du, H.; Liu, J.; He, W.; Zhu, J.; Xiao, J.; Wang, J.; Cao, Y.; et al. Dietary Tangeretin Alleviated Dextran Sulfate Sodium-Induced Colitis in Mice via Inhibiting Inflammatory Response, Restoring Intestinal Barrier Function, and Modulating Gut Microbiota. J. Agric. Food Chem. 2021, 69, 7663–7674. [Google Scholar] [CrossRef]
- Tolba, M.F.; Omar, H.A.; Azab, S.S.; Khalifa, A.E.; Abdel-Naim, A.B.; Abdel-Rahman, S.Z.-A. Caffeic acid phenethyl ester: A review of its antioxidant activity, protective effects against ischemia-reperfusion injury and drug adverse reactions. Crit. Rev. Food Sci. Nutr. 2016, 56, 2183–2190. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Cui, H.; Ma, Z.; Liu, X.; Yang, L. Recent progresses in the pharmacological activities of caffeic acid phenethyl ester. Naunyn-Schmiedeberg Arch. Pharmacol. 2021, 394, 1327–1339. [Google Scholar] [CrossRef]
- Natarajan, K.; Singh, S.; Burke, T.R.; Grunberger, D.; Aggarwal, B.B. Caffeic Acid Phenethyl Ester is a Potent and Specific Inhibitor of Activation of Nuclear Transcription Factor NF-κ B. Proc. Natl. Acad. Sci. USA 1996, 93, 9090–9095. [Google Scholar] [CrossRef]
- Tambuwala, M.M.; Kesharwani, P.; Shukla, R.; Thompson, P.D.; McCarron, P.A. Caffeic acid phenethyl ester (CAPE) reverses fibrosis caused by chronic colon inflammation in murine model of colitis. Pathol. Res. Pract. 2018, 214, 1909–1911. [Google Scholar] [CrossRef] [PubMed]
- Yordanov, Y. Caffeic acid phenethyl ester (CAPE): Cornerstone pharmacological studies and drug delivery systems. Pharmacia 2019, 66, 223–231. [Google Scholar] [CrossRef]
- Wei, X.; Dai, J.; Du, Y.; Liu, L.; Li, R.; Wang, Z.; Wang, L.; Huang, Y.; Chen, P.; Zhou, Z.; et al. Caffeic acid phenethyl ester loaded in a targeted delivery system based on a solid-in-oil-in-water multilayer emulsion: Characterization, stability, and fate of the emulsion during in vivo digestion. Food Res. Int. 2022, 161, 111756. [Google Scholar] [CrossRef]
- Cai, X.; Han, Y.; Gu, M.; Song, M.; Wu, X.; Li, Z.; Li, F.; Goulette, T.; Xiao, H. Dietary cranberry suppressed colonic inflammation and alleviated gut microbiota dysbiosis in dextran sodium sulfate-treated mice. Food Funct. 2019, 10, 6331–6341. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, B.; Ross, R.P.; Jin, Y.; Stanton, C.; Zhao, J.; Zhang, H.; Chen, W. Orally Administered CLA Ameliorates DSS-Induced Colitis in Mice via Intestinal Barrier Improvement, Oxidative Stress Reduction, and Inflammatory Cytokine and Gut Microbiota Modulation. J. Agric. Food Chem. 2019, 67, 13282–13298. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Lin, Q.; Yang, T.; Zeng, L.; Shi, L.; Chen, Y.; Luo, F. Oat beta-glucan ameliorates dextran sulfate sodium (DSS)-induced ulcerative colitis in mice. Food Funct. 2015, 6, 3454–3463. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Yang, X.; Yuan, Y.; Jia, Y.; Liu, G.; Lin, N.; Xiao, H.; Zhang, L.; Chen, J. Toxicity, gut microbiota and metabolome effects after copper exposure during early life in SD rats. Toxicology 2020, 433–434, 152395. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Chen, S.; Zhu, G.; Huang, R.; Yin, Y.; Ren, W. Betaine Inhibits Interleukin-1beta Production and Release: Potential Mechanisms. Front. Immunol. 2018, 9, 2670. [Google Scholar] [CrossRef] [PubMed]
- Atreya, R.; Neurath, M.F. Involvement of IL-6 in the pathogenesis of inflammatory bowel disease and colon cancer. Clin. Rev. Allergy Immunol. 2005, 28, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.; Fantini, M.C.; Schramm, C.; Lehr, H.A.; Wirtz, S.; Nikolaev, A.; Burg, J.; Strand, S.; Kiesslich, R.; Huber, S.; et al. TGF-beta suppresses tumor progression in colon cancer by inhibition of IL-6 trans-signaling. Immunity 2004, 21, 491–501. [Google Scholar] [CrossRef]
- Walia, B.; Wang, L.; Merlin, D.; Sitaraman, S.V. TGF-beta down-regulates IL-6 signaling in intestinal epithelial cells: Critical role of SMAD-2. FASEB J. 2003, 17, 2130–2132. [Google Scholar] [CrossRef]
- Ouyang, W.; Rutz, S.; Crellin, N.K.; Valdez, P.A.; Hymowitz, S.G. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 2011, 29, 71–109. [Google Scholar] [CrossRef]
- Mitchell, J.P.; Carmody, R.J. NF-kappaB and the Transcriptional Control of Inflammation. Int. Rev. Cell Mol. Biol. 2018, 335, 41–84. [Google Scholar] [CrossRef]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef]
- Blaak, E.E.; Canfora, E.E.; Theis, S.; Frost, G.; Groen, A.K.; Mithieux, G.; Nauta, A.; Scott, K.; Stahl, B.; van Harsselaar, J.; et al. Short chain fatty acids in human gut and metabolic health. Benef. Microbes 2020, 11, 411–455. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Arias-Tellez, M.J.; Solis-Urra, P.; Saez-Lara, M.J.; Gil, A. Use of Probiotics in Inflammatory Bowel Disease. In Microbiome and Metabolome in Diagnosis, Therapy, and Other Strategic Applications; Elsevier: Amsterdam, The Netherlands, 2019; pp. 149–154. [Google Scholar] [CrossRef]
- McNabney, S.M.; Henagan, T.M. Short Chain Fatty Acids in the Colon and Peripheral Tissues: A Focus on Butyrate, Colon Cancer, Obesity and Insulin Resistance. Nutrients 2017, 9, 1348. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef] [PubMed]
- Rios-Covian, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; de Los Reyes-Gavilan, C.G.; Salazar, N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef]
- Rodriguez-Nogales, A.; Algieri, F.; Garrido-Mesa, J.; Vezza, T.; Utrilla, M.P.; Chueca, N.; Garcia, F.; Rodriguez-Cabezas, M.E.; Galvez, J. Intestinal anti-inflammatory effect of the probiotic Saccharomyces boulardii in DSS-induced colitis in mice: Impact on microRNAs expression and gut microbiota composition. J. Nutr. Biochem. 2018, 61, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Mentella, M.C.; Scaldaferri, F.; Pizzoferrato, M.; Gasbarrini, A.; Miggiano, G.A.D. Nutrition, IBD and Gut Microbiota: A Review. Nutrients 2020, 12, 944. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Bibiloni, R.; Mangold, M.; Madsen, K.L.; Fedorak, R.N.; Tannock, G.W. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn’s disease and ulcerative colitis patients. J. Med. Microbiol. 2006, 55, 1141–1149. [Google Scholar] [CrossRef]
- Mukhopadhya, I.; Hansen, R.; El-Omar, E.M.; Hold, G.L. IBD-what role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 219–230. [Google Scholar] [CrossRef]
- Vinella, D.; Fischer, F.; Vorontsov, E.; Gallaud, J.; Malosse, C.; Michel, V.; Cavazza, C.; Robbe-Saule, M.; Richaud, P.; Chamot-Rooke, J.; et al. Evolution of Helicobacter: Acquisition by Gastric Species of Two Histidine-Rich Proteins Essential for Colonization. PLoS Pathog. 2015, 11, e1005312. [Google Scholar] [CrossRef] [PubMed]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Xu, J.; Xu, H.; Guo, X.; Wang, J.; Zhu, M.; Zhao, H.; Li, J.; Huang, H.; Zhou, Y. IDDF2023-ABS-0100 Odoribacter. splanchnicus alleviate colitis by regulating neutrophil extracellular traps formation. Gut 2023, 72, A79–A81. [Google Scholar] [CrossRef]
- Berry, D.; Schwab, C.; Milinovich, G.; Reichert, J.; Ben Mahfoudh, K.; Decker, T.; Engel, M.; Hai, B.; Hainzl, E.; Heider, S.; et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 2012, 6, 2091–2106. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.B.; Carroll-Portillo, A.; Lin, H.C. Desulfovibrio in the Gut: The Enemy within? Microorganisms 2023, 11, 1772. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Sheng, L.; Zhong, J.; Tao, X.; Zhu, W.; Ma, J.; Yan, J.; Zhao, A.; Zheng, X.; Wu, G.; et al. Desulfovibrio vulgaris, a potent acetic acid-producing bacterium, attenuates nonalcoholic fatty liver disease in mice. Gut Microbes 2021, 13, e1930874. [Google Scholar] [CrossRef] [PubMed]
- Axelson, M.; Ellis, E.; Mörk, B.; Garmark, K.; Abrahamsson, A.; Björkhem, I.; Ericzon, B.-G.; Einarsson, C. Bile acid synthesis in cultured human hepatocytes: Support for an alternative biosynthetic pathway to cholic acid. Hepatology 2000, 31, 1305–1312. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Xie, G.; Jia, W. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2017, 15, 111–128. [Google Scholar] [CrossRef]
- Dong, S.; Zhu, M.; Wang, K.; Zhao, X.; Hu, L.; Jing, W.; Lu, H.; Wang, S. Dihydromyricetin improves DSS-induced colitis in mice via modulation of fecal-bacteria-related bile acid metabolism. Pharmacol. Res. 2021, 171, 105767. [Google Scholar] [CrossRef]
- Sitkin, S.; Demyanova, E.; Vakhitov, T.; Pokrotnieks, J. Altered Sphingolipid Metabolism and its Interaction with the Intestinal Microbiome Is Another Key to the Pathogenesis of Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2019, 25, e157–e158. [Google Scholar] [CrossRef]
- Kamal-Eldin, A.; Frank, J.; Razdan, A.; Tengblad, S.; Basu, S.; Vessby, B. Effects of Dietary Phenolic Compounds on Tocopherol Cholesterol, and Fatty Acids in Rats. Lipids 2000, 35, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Brito, B.S.; Silva, I.N.N.; Nóbrega, V.G.; da Silva, M.; Gomes, H.D.N.; Fortes, F.M.; Pimentel, A.M.; Mota, J.; Almeida, N.; et al. Frequency of Hepatobiliary Manifestations and Concomitant Liver Disease in Inflammatory Bowel Disease Patients. BioMed Res. Int. 2019, 2019, 7604939. [Google Scholar] [CrossRef] [PubMed]


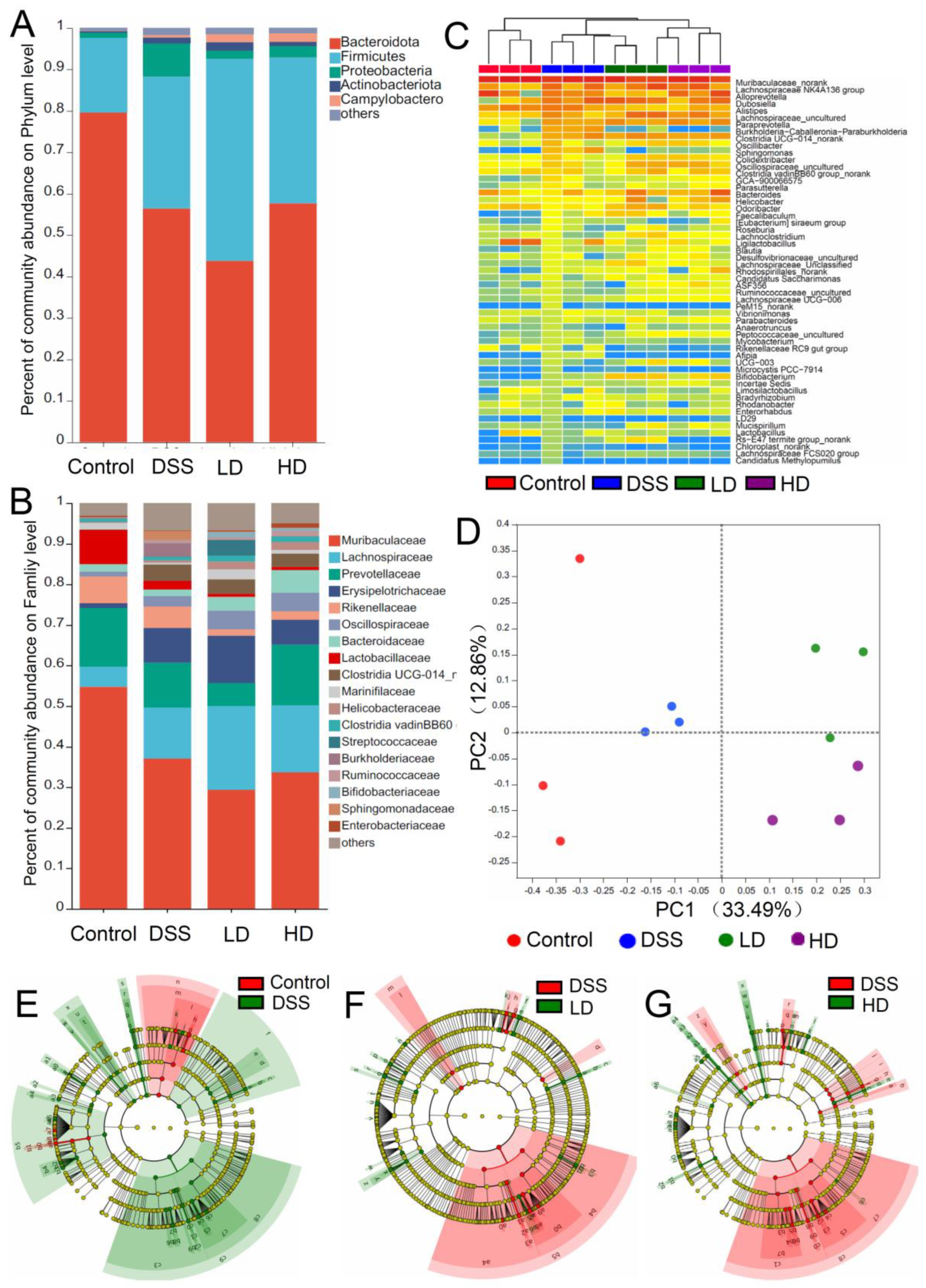
| Sobs | Ace | Shannon | Simpson | Chao 1 | |
|---|---|---|---|---|---|
| Control | 828.667 ± 29.036 a | 994.972 ± 37.129 a | 4.344 ± 0.108 a | 0.053 ± 0.011 a | 1005.394 ± 36.64 a |
| DSS | 546 ± 6.807 d | 660.252 ± 43.466 b | 4.235 ± 0.112 a | 0.05 ± 0.005 a | 669.807 ± 53.923 c |
| LD | 653 ± 21.362 c | 882.753 ± 85.217 a | 4.617 ± 0.222 a | 0.031 ± 0.01 a | 779.725 ± 49.722 bc |
| HD | 748 ± 17.898 b | 834.798 ± 45.16 ab | 4.452 ± 0.171 a | 0.036 ± 0.008 a | 847.809 ± 39.88 b |
| m/z | Excat Mass (Da) | Elemental Composition | Selected Ion | Postulated Identity | KEGG ID | VIP Values | DSS vs. Control | LD vs. DSS | HD vs. DSS |
|---|---|---|---|---|---|---|---|---|---|
| 104.1062 | 131.0946 | C6H13NO2 | [M-CO+H]+ | L-Isoleucine | C00407 | 1.9553 | — | ↓ | ↓ |
| 114.0904 | 130.1106 | C6H14N2O | [M-NH3+H]+ | N-Acetylputrescine | C02714 | 3.1699 | ↑ | ↓ | ↓ |
| 199.1473 | 270.162 | C18H22O2 | [M-C3H4O2+H]+ | Estrone | C00468 | 1.3757 | ↓ | — | ↑ |
| 201.1631 | 272.1776 | C18H24O2 | [M-C3H4O2+H]+ | Estradiol | C00951 | 1.5846 | ↓ | — | — |
| 241.1955 | 286.1933 | C19H26O2 | [M-HCOOH+H]+ | Androstenedione | C00280 | 1.3566 | — | — | ↓ |
| 313.2163 | 290.2246 | C19H30O2 | [M+Na]+ | Androstenediol | C04295 | 1.1001 | ↓ | ↓ | ↓ |
| 339.2877 | 320.2715 | C21H36O2 | [M-H2O+H]+ | Allopregnane-3α,20α-diol | C18042 | 2.2882 | ↓ | — | ↓ |
| 421.3475 | 398.3549 | C28H46O | [M+Na]+ | Episterol | C15777 | 1.1393 | ↓ | ↓ | ↑ |
| 423.3622 | 400.3705 | C28H48O | [M+Na]+ | Campesterol | C01789 | 1.5486 | ↓ | ↓ | ↑ |
| 439.3199 | 416.329 | C27H44O3 | [M+Na]+ | Calcitriol | C01673 | 1.2781 | ↓ | — | ↑ |
| 282.2787 | 299.2824 | C18H37NO2 | [M-H2O+H]+ | Sphingoid | C00319 | 1.4562 | ↓ | — | ↑ |
| 324.2879 | 301.2981 | C18H39NO2 | [M+Na]+ | Sphinganine | C00836 | 1.497 | ↓ | — | ↑ |
| 281.247 | 280.2402 | C18H32O2 | [M+H]+ | Linoleic acid | C01595 | 1.7064 | ↑ | — | — |
| 255.065 | 254.0573 | C7H14N2O6S | [M+H]+ | 5-L-Glutamyl-taurine | C05844 | 1.2138 | ↓ | ↑ | ↑ |
| 185.0800 | 184.0736 | C9H12O4 | [M+H]+ | HMPG | C05594 | 2.1462 | ↓ | ↑ | ↑ |
| 193.0491 | 170.0579 | C8H10O4 | [M+Na]+ | DHPG | C05576 | 1.6714 | ↑ | ↓ | ↓ |
| 149.1067 | 176.095 | C10H12N2O | [M-CO+H]+ | 5-Hydroxytryptamine | C00780 | 1.1039 | ↓ | — | ↓ |
| 116.0486 | 161.0477 | C9H7NO2 | [M-HCOOH+H]+ | 4,6-Dihydroxyquinoline | C05639 | 1.5739 | — | ↓ | ↓ |
| 121.0644 | 120.0575 | C8H8O | [M+H]+ | Phenylacetaldehyde | C00601 | 1.2324 | ↑ | ↓ | ↓ |
| 277.2176 | 276.2089 | C18H28O2 | [M+H]+ | Stearidonic acid | C16300 | 1.1778 | — | ↑ | ↑ |
| 311.2593 | 338.2457 | C20H34O4 | [M-CO+H]+ | 5,6-DHET | C14772 | 1.2515 | ↓ | ↑ | — |
| 391.2843 | 408.2876 | C24H40O5 | [M-H2O+H]+ | Cholic acid | C00695 | 1.8347 | ↓ | — | ↑ |
| 457.3308 | 434.3396 | C6H13NO2 | [M+Na]+ | 3,7,12-trihydroxycholestan-26-al | C01301 | 1.7466 | ↓ | — | ↑ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Dai, J.; Liu, R.; Wan, G.; Gu, S.; Du, Y.; Yang, X.; Wang, L.; Huang, Y.; Chen, P.; et al. S/O/W Emulsion with CAPE Ameliorates DSS-Induced Colitis by Regulating NF-κB Pathway, Gut Microbiota and Fecal Metabolome in C57BL/6 Mice. Nutrients 2024, 16, 1145. https://doi.org/10.3390/nu16081145
Wei X, Dai J, Liu R, Wan G, Gu S, Du Y, Yang X, Wang L, Huang Y, Chen P, et al. S/O/W Emulsion with CAPE Ameliorates DSS-Induced Colitis by Regulating NF-κB Pathway, Gut Microbiota and Fecal Metabolome in C57BL/6 Mice. Nutrients. 2024; 16(8):1145. https://doi.org/10.3390/nu16081145
Chicago/Turabian StyleWei, Xuelin, Juan Dai, Ruijia Liu, Guochao Wan, Shiyu Gu, Yuwei Du, Xinyue Yang, Lijun Wang, Yukun Huang, Pengfei Chen, and et al. 2024. "S/O/W Emulsion with CAPE Ameliorates DSS-Induced Colitis by Regulating NF-κB Pathway, Gut Microbiota and Fecal Metabolome in C57BL/6 Mice" Nutrients 16, no. 8: 1145. https://doi.org/10.3390/nu16081145
APA StyleWei, X., Dai, J., Liu, R., Wan, G., Gu, S., Du, Y., Yang, X., Wang, L., Huang, Y., Chen, P., Chen, X., Yang, X., & Wang, Q. (2024). S/O/W Emulsion with CAPE Ameliorates DSS-Induced Colitis by Regulating NF-κB Pathway, Gut Microbiota and Fecal Metabolome in C57BL/6 Mice. Nutrients, 16(8), 1145. https://doi.org/10.3390/nu16081145







