Gut Microbial Dysbiosis Differs in Two Distinct Cachectic Tumor-Bearing Models Consuming the Same Diet
Abstract
1. Introduction
2. Materials and Methods
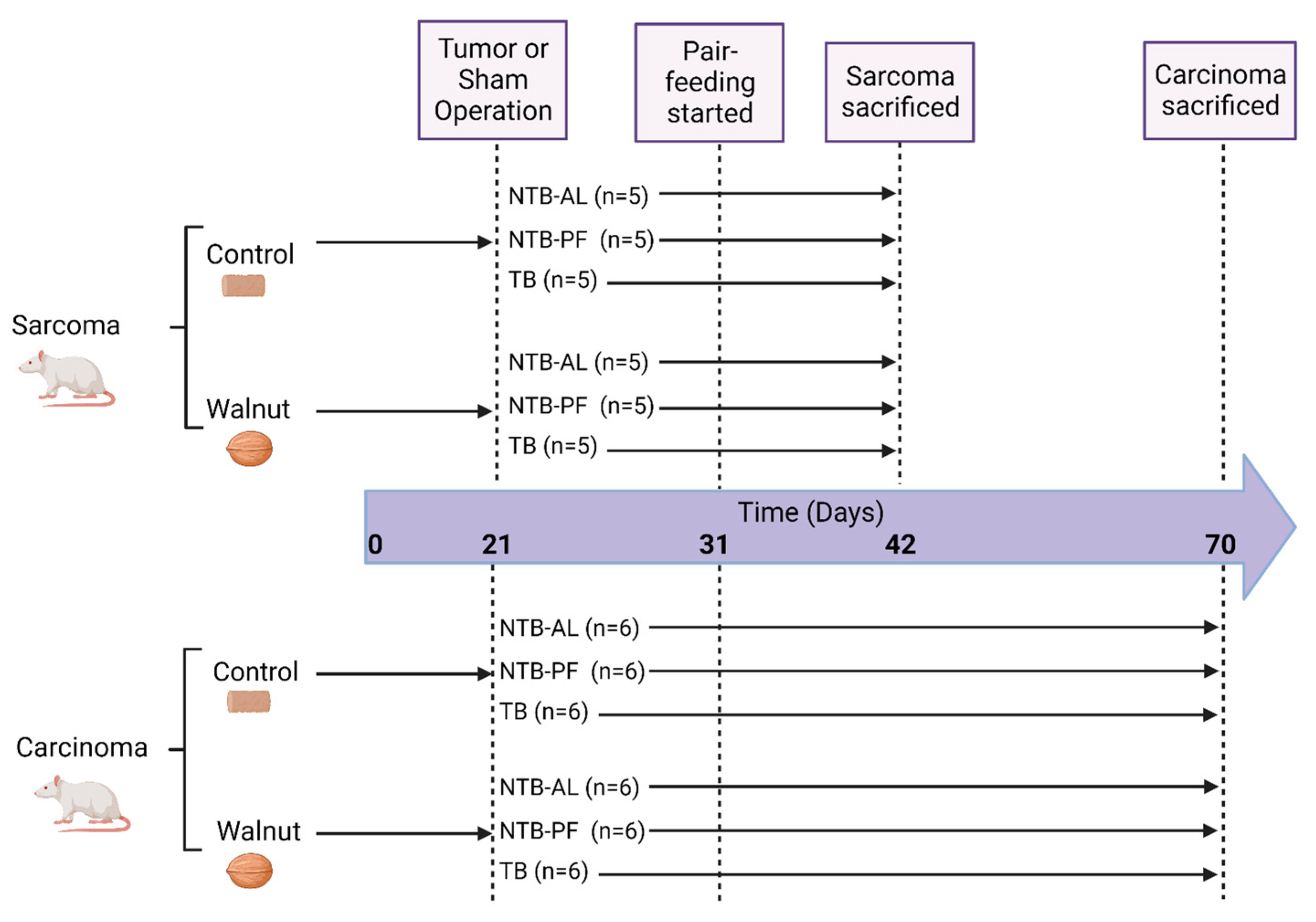
2.1. Study Design
2.2. Diets
2.3. DNA Isolation and PCR Amplification
2.4. Sequencing
2.5. Quality Filtering/Picking
2.6. Microbial Community Analysis
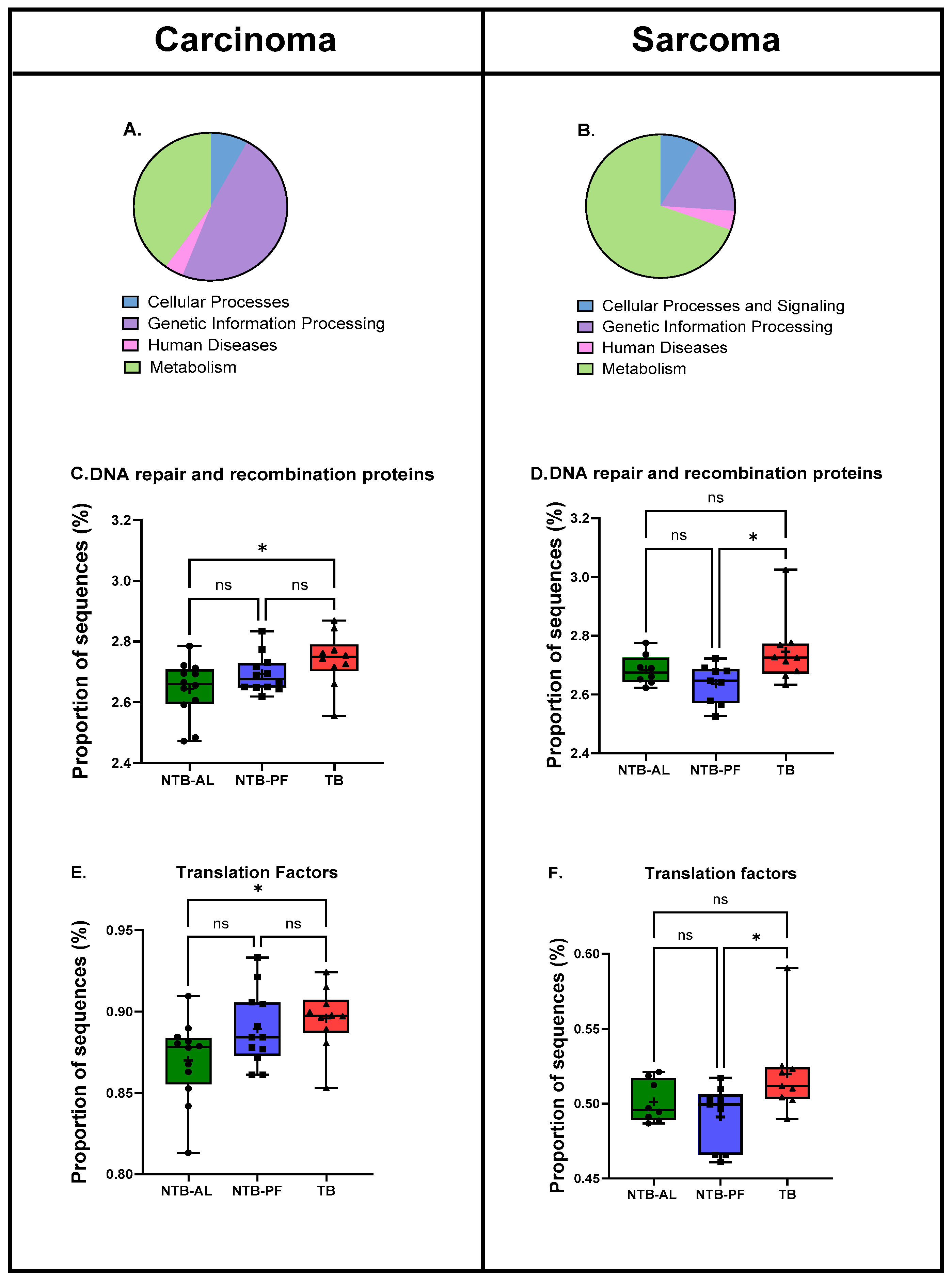
2.7. Predicted Functional Pathways
2.8. Statistical Analysis
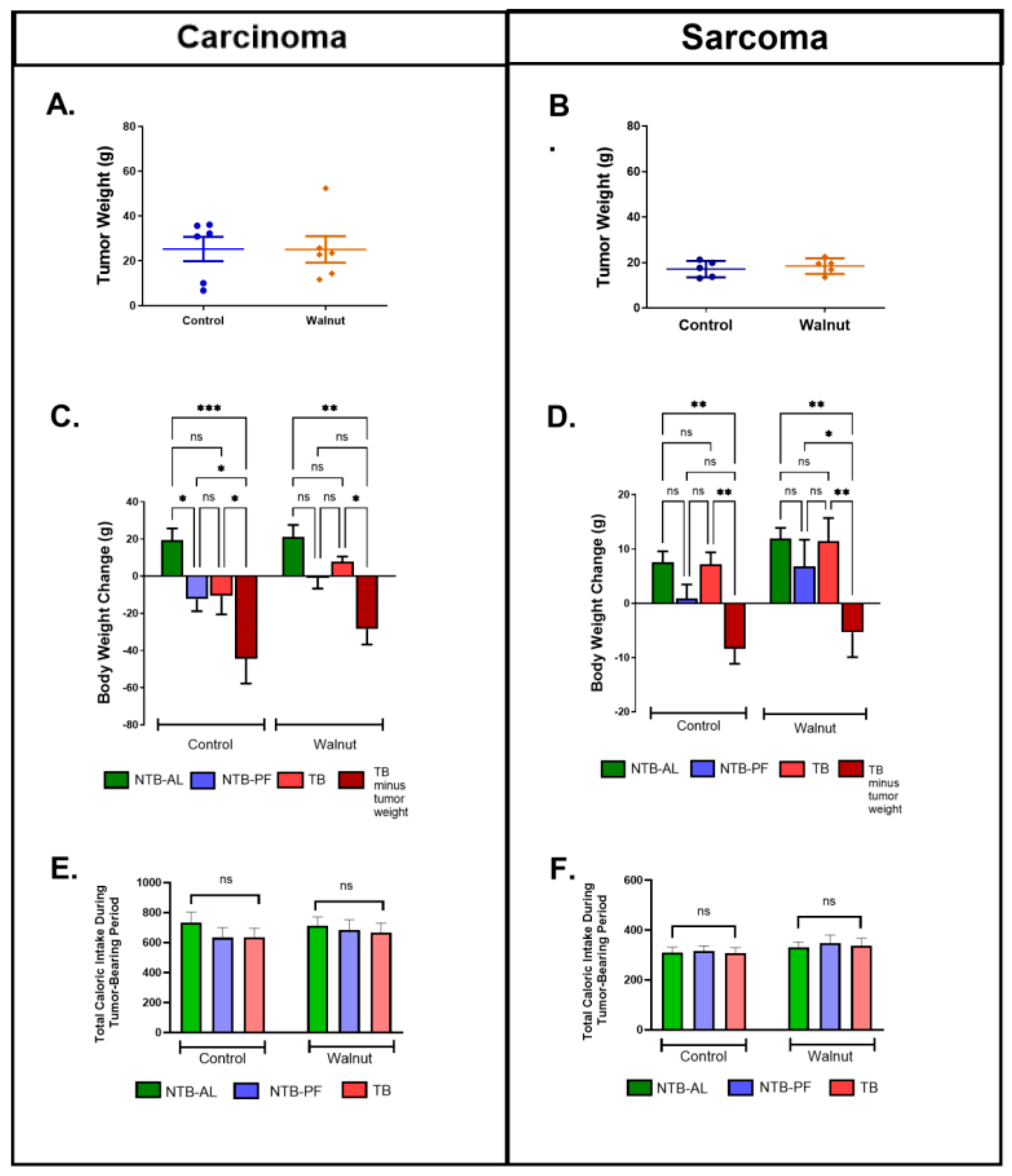
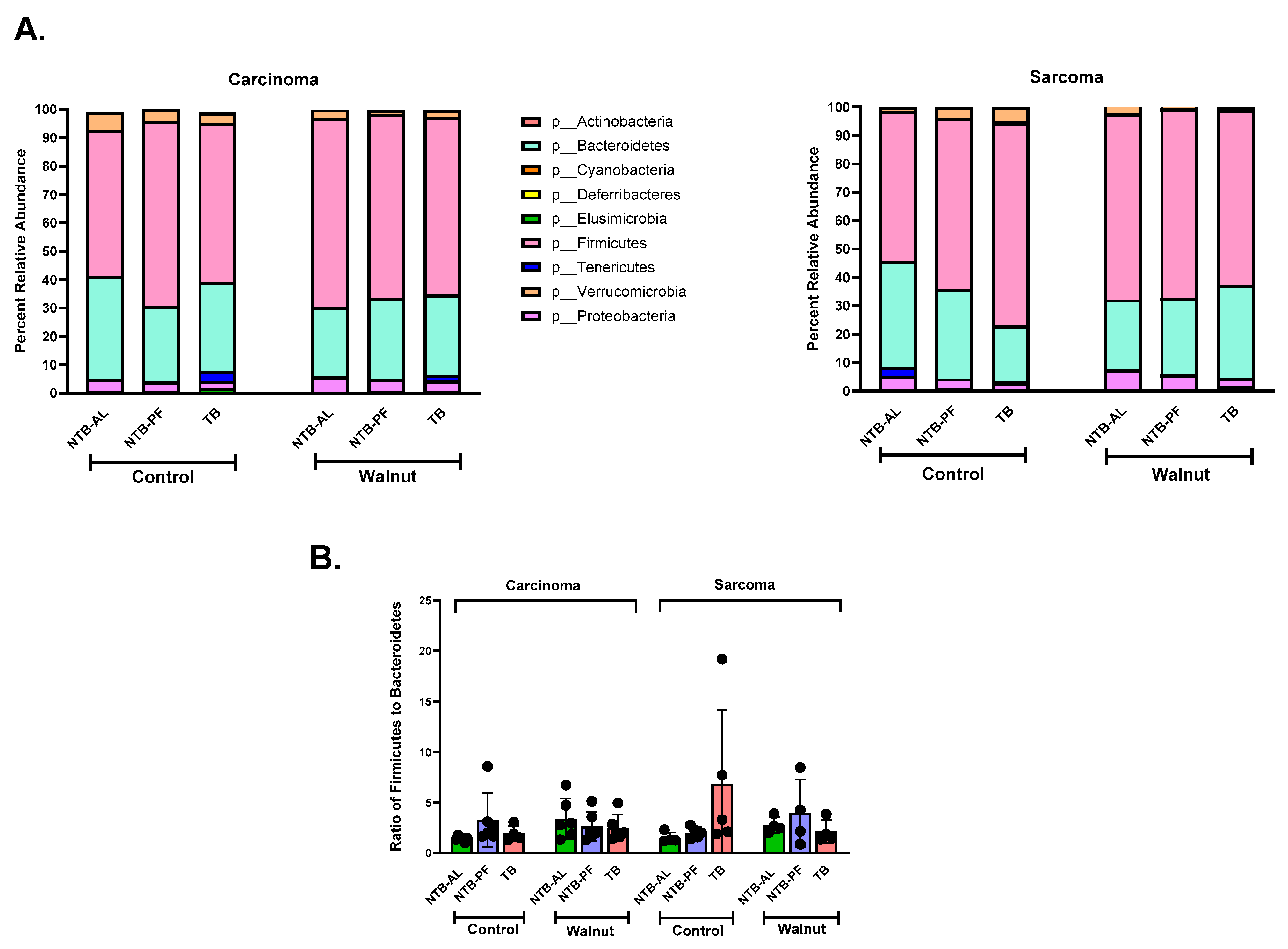
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Tumor Type | Dependent Variable | Pr > ChiSq from Kruskal–Wallis Test | Rank | m = Number of Dependent Variables | CV for BH False Detect Rate 1 |
|---|---|---|---|---|---|
| Higher in the Control group | |||||
| Sarcoma | p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__Bacteroidaceae|g__Bacteroides|Other | 0.0024 | 7 | 20 | 0.018 |
| Sarcoma | p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__Bacteroidaceae|g__Bacteroides|s__ovatus | 0.0001 | 1 | 20 | 0.003 |
| Sarcoma | p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__[Barnesiellaceae] | 0.0179 | 11 | 20 | 0.028 |
| Sarcoma | p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__Prevotellaceae|g__Prevotella|s__ | 0.0008 | 3 | 20 | 0.008 |
| Sarcoma | p__Firmicutes|c__Bacilli|o__Lactobacillales|f__Lactobacillaceae|g__Lactobacillus|s__reuteri | 0.0135 | 10 | 20 | 0.025 |
| Sarcoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales | 0.0023 | 5 | 20 | 0.013 |
| Sarcoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae | 0.0023 | 5 | 20 | 0.013 |
| Sarcoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae|g__Anaeroplasma | 0.0023 | 5 | 20 | 0.013 |
| Sarcoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae|g__Anaeroplasma|s__ | 0.0023 | 5 | 20 | 0.013 |
| Carcinoma | p__Actinobacteria|c__Actinobacteria|o__Actinomycetales | 0.0025 | 16 | 45 | 0.0178 |
| Carcinoma | p__Actinobacteria|c__Actinobacteria|o__Actinomycetales|f__MicrocoControlaceae | 0.0031 | 17 | 45 | 0.0189 |
| Carcinoma | p__Actinobacteria|c__Actinobacteria|o__Actinomycetales|f__MicrocoControlaceae|Other | 0.0031 | 17 | 45 | 0.0189 |
| Carcinoma | p__Actinobacteria|c__Actinobacteria|o__Actinomycetales|f__MicrocoControlaceae|Other|Other | 0.0031 | 17 | 45 | 0.0189 |
| Carcinoma | p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__Bacteroidaceae | 0.0032 | 20 | 45 | 0.0222 |
| Carcinoma | p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__Bacteroidaceae|g__Bacteroides | 0.0032 | 20 | 45 | 0.0222 |
| Carcinoma | p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__Bacteroidaceae|g__Bacteroides|s__ | 0.0152 | 43 | 45 | 0.0478 |
| Carcinoma | p__Firmicutes|c__Bacilli|o__Lactobacillales|f__Carnobacteriaceae | 0.0066 | 34 | 45 | 0.0378 |
| Carcinoma | p__Firmicutes|c__Bacilli|o__Lactobacillales|f__Carnobacteriaceae|g__Granulicatella | 0.0066 | 34 | 45 | 0.0378 |
| Carcinoma | p__Firmicutes|c__Bacilli|o__Lactobacillales|f__Carnobacteriaceae|g__Granulicatella|s__ | 0.0066 | 34 | 45 | 0.0378 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__Ruminococcus|s__bromii | 0.0167 | 44 | 45 | 0.0489 |
| Carcinoma | p__Proteobacteria|c__Alphaproteobacteria|o__RF32 | 0.0062 | 30 | 45 | 0.0333 |
| Carcinoma | p__Proteobacteria|c__Alphaproteobacteria|o__RF32|f__ | 0.0062 | 30 | 45 | 0.0333 |
| Carcinoma | p__Proteobacteria|c__Alphaproteobacteria|o__RF32|f__|g__ | 0.0062 | 30 | 45 | 0.0333 |
| Carcinoma | p__Proteobacteria|c__Alphaproteobacteria|o__RF32|f__|g__|s__ | 0.0062 | 30 | 45 | 0.0333 |
| Carcinoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales | 0.0033 | 22 | 45 | 0.0244 |
| Carcinoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae | 0.0033 | 22 | 45 | 0.0244 |
| Carcinoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae|g__Anaeroplasma | 0.0033 | 22 | 45 | 0.0244 |
| Carcinoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae|g__Anaeroplasma|s__ | 0.0033 | 22 | 45 | 0.0244 |
| Higher in the Walnut group | |||||
| Sarcoma | p__Firmicutes|c__Bacilli | 0.0020 | 4 | 20 | 0.010 |
| Sarcoma | p__Firmicutes|c__Bacilli|o__Lactobacillales | 0.0270 | 17 | 20 | 0.043 |
| Sarcoma | p__Firmicutes|c__Bacilli|o__Lactobacillales|f__Lactobacillaceae | 0.0308 | 18 | 20 | 0.045 |
| Sarcoma | p__Firmicutes|c__Bacilli|o__Lactobacillales|f__Lactobacillaceae|g__Lactobacillus | 0.0308 | 18 | 20 | 0.045 |
| Sarcoma | p__Firmicutes|c__Bacilli|o__Lactobacillales|f__Lactobacillaceae|g__Lactobacillus|s__ | 0.0396 | 20 | 20 | 0.050 |
| Sarcoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__Ruminococcus|Other | 0.0006 | 2 | 20 | 0.005 |
| Carcinoma | p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__ | 0.0009 | 11 | 45 | 0.0122 |
| Carcinoma | p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__|g__ | 0.0009 | 11 | 45 | 0.0122 |
| Carcinoma | p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__|g__|s__ | 0.0009 | 11 | 45 | 0.0122 |
| Carcinoma | p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__Prevotellaceae|g__Prevotella|s__ | 0.0045 | 28 | 45 | 0.0311 |
| Carcinoma | p__Firmicutes|c__Bacilli | 0.0183 | 45 | 45 | 0.0500 |
| Carcinoma | p__Firmicutes|c__Bacilli|o__Turicibacterales | 0.0113 | 39 | 45 | 0.0433 |
| Carcinoma | p__Firmicutes|c__Bacilli|o__Turicibacterales|f__Turicibacteraceae | 0.0113 | 39 | 45 | 0.0433 |
| Carcinoma | p__Firmicutes|c__Bacilli|o__Turicibacterales|f__Turicibacteraceae|g__Turicibacter | 0.0113 | 39 | 45 | 0.0433 |
| Carcinoma | p__Firmicutes|c__Bacilli|o__Turicibacterales|f__Turicibacteraceae|g__Turicibacter|s__ | 0.0113 | 39 | 45 | 0.0433 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__CoprocoControlus|Other | 0.0005 | 10 | 45 | 0.0111 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Moryella | 0.0018 | 14 | 45 | 0.0156 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Moryella|s__ | 0.0018 | 14 | 45 | 0.0156 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Roseburia | 0.0002 | 2 | 45 | 0.0022 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Roseburia|Other | 0.0002 | 2 | 45 | 0.0022 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|Other | 0.0004 | 7 | 45 | 0.0078 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|Other|Other | 0.0004 | 7 | 45 | 0.0078 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|Other|Other|Other | 0.0004 | 7 | 45 | 0.0078 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Peptococcaceae|Other | 0.0093 | 37 | 45 | 0.0411 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Peptococcaceae|Other|Other | 0.0093 | 37 | 45 | 0.0411 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__Oscillospira | 0.0002 | 2 | 45 | 0.0022 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__Oscillospira|Other | 0.0003 | 6 | 45 | 0.0067 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__Oscillospira|s__ | 0.0002 | 2 | 45 | 0.0022 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__Ruminococcus|Other | 0.0001 | 1 | 45 | 0.0011 |
| Carcinoma | p__Proteobacteria|c__Alphaproteobacteria | 0.005 | 29 | 45 | 0.0322 |
| Carcinoma | p__Proteobacteria|c__Deltaproteobacteria|o__Desulfovibrionales|f__Desulfovibrionaceae|Other | 0.004 | 26 | 45 | 0.0289 |
| Carcinoma | p__Proteobacteria|c__Deltaproteobacteria|o__Desulfovibrionales|f__Desulfovibrionaceae|Other|Other | 0.004 | 26 | 45 | 0.0289 |
| Tumor Type | Dependent Variable | Pr > ChiSq from Kruskal–Wallis Test | Rank | m = Number of Dependent Variables | CV for BH False Detect Rate 1 | Group with the Greatest Amount | p-Value |
|---|---|---|---|---|---|---|---|
| NTB-AL vs. NTB-PF | |||||||
| p__Firmicutes | |||||||
| Sarcoma | c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__ | 0.0004 | 1 | 16 | 0.003125 | NTB-AL | 0.0146 |
| Sarcoma | c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__|s__ | 0.0004 | 1 | 16 | 0.003125 | NTB-AL | 0.0146 |
| Sarcoma | c__Clostridia|o__Clostridiales|f__Veillonellaceae|g__Veillonella | 0.0005 | 3 | 16 | 0.009375 | NTB-AL | 0.0057 |
| Sarcoma | c__Clostridia|o__Clostridiales|f__Veillonellaceae|g__Veillonella|s__dispar | 0.0005 | 3 | 16 | 0.009375 | NTB-AL | 0.0057 |
| p__Actinobacteria | |||||||
| Carcinoma | c__Coriobacteriia | 0.0013 | 7 | 41 | 0.0085 | NTB-AL | 0.0154 |
| Carcinoma | c__Coriobacteriia|o__Coriobacteriales | 0.0013 | 7 | 41 | 0.0085 | NTB-AL | 0.0154 |
| Carcinoma | c__Coriobacteriia|o__Coriobacteriales|f__Coriobacteriaceae | 0.0013 | 7 | 41 | 0.0085 | NTB-AL | 0.0154 |
| p__Bacteroidetes | |||||||
| Carcinoma | c__Bacteroidia|o__Bacteroidales|f__Porphyromonadaceae|g__Parabacteroides | 0.017 | 24 | 41 | 0.0293 | NTB-AL | 0.0216 |
| Carcinoma | c__Bacteroidia|o__Bacteroidales|f__Porphyromonadaceae|g__Parabacteroides|s__ | 0.017 | 24 | 41 | 0.0293 | NTB-AL | 0.0216 |
| Carcinoma | c__Bacteroidia|o__Bacteroidales|f__Rikenellaceae | 0.0284 | 38 | 41 | 0.0463 | NTB-AL | 0.0406 |
| p__Firmicutes | |||||||
| Carcinoma | c__Bacilli|o__Lactobacillales | 0.0174 | 26 | 41 | 0.0317 | NTB-AL | 0.0406 |
| p__Proteobacteria | |||||||
| Carcinoma | c__Gammaproteobacteria|o__Pasteurellales|f__Pasteurellaceae|g__Haemophilus | 0.0102 | 22 | 41 | 0.0268 | NTB-AL | 0.0097 |
| Carcinoma | c__Gammaproteobacteria|o__Pasteurellales|f__Pasteurellaceae|g__Haemophilus|s__parainfluenzae | 0.0102 | 22 | 41 | 0.0268 | NTB-AL | 0.0097 |
| NTB-AL vs. TB | |||||||
| p__Firmicutes | |||||||
| Sarcoma | c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Blautia | 0.0019 | 6 | 16 | 0.01875 | TB | 0.0331 |
| Sarcoma | c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Blautia|Other | 0.0009 | 5 | 16 | 0.015625 | TB | 0.0193 |
| Sarcoma | c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__ | 0.0004 | 1 | 16 | 0.003125 | TB | 0.0427 |
| Sarcoma | c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__|s__ | 0.0004 | 1 | 16 | 0.003125 | TB | 0.0427 |
| p__Elusimicrobia | |||||||
| Carcinoma | p__Elusimicrobia | 0.0199 | 27 | 41 | 0.0329 | TB | 0.039 |
| Carcinoma | c__Elusimicrobia | 0.0199 | 27 | 41 | 0.0329 | TB | 0.039 |
| Carcinoma | c__Elusimicrobia|o__Elusimicrobiales | 0.0199 | 27 | 41 | 0.0329 | TB | 0.039 |
| Carcinoma | c__Elusimicrobia|o__Elusimicrobiales|f__Elusimicrobiaceae | 0.0199 | 27 | 41 | 0.0329 | TB | 0.039 |
| Carcinoma | c__Elusimicrobia|o__Elusimicrobiales|f__Elusimicrobiaceae|g__Elusimicrobium | 0.0199 | 27 | 41 | 0.0329 | TB | 0.039 |
| Carcinoma | c__Elusimicrobia|o__Elusimicrobiales|f__Elusimicrobiaceae|g__Elusimicrobium|s__ | 0.0199 | 27 | 41 | 0.0329 | TB | 0.039 |
| p__Firmicutes | |||||||
| Carcinoma | c__Bacilli|o__Lactobacillales | 0.0174 | 26 | 41 | 0.0317 | NTB-AL | 0.0463 |
| Carcinoma | c__Bacilli|o__Lactobacillales|f__Lactobacillaceae|g__Lactobacillus|s__ | 0.0354 | 39 | 41 | 0.0476 | NTB-AL | 0.0463 |
| p__Proteobacteria | |||||||
| Carcinoma | c__Betaproteobacteria | 0.0015 | 11 | 41 | 0.0134 | NTB-AL | 0.0227 |
| Carcinoma | c__Betaproteobacteria|o__Burkholderiales | 0.0015 | 11 | 41 | 0.0134 | NTB-AL | 0.0227 |
| Carcinoma | c__Betaproteobacteria|o__Burkholderiales|f__Alcaligenaceae | 0.0015 | 11 | 41 | 0.0134 | NTB-AL | 0.0227 |
| Carcinoma | c__Betaproteobacteria|o__Burkholderiales|f__Alcaligenaceae|g__Sutterella | 0.0015 | 11 | 41 | 0.0134 | NTB-AL | 0.0227 |
| Carcinoma | c__Betaproteobacteria|o__Burkholderiales|f__Alcaligenaceae|g__Sutterella|s__ | 0.0015 | 11 | 41 | 0.0134 | NTB-AL | 0.0227 |
| p__Tenericutes | |||||||
| Carcinoma | p__Tenericutes | 0.0014 | 10 | 41 | 0.0122 | TB | 0.0055 |
| Carcinoma | c__Mollicutes | 0.0273 | 33 | 41 | 0.0402 | TB | 0.0055 |
| Carcinoma | c__Mollicutes|o__Anaeroplasmatales | 0.0047 | 18 | 41 | 0.0220 | TB | 0.0273 |
| Carcinoma | c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae | 0.0047 | 18 | 41 | 0.0220 | TB | 0.0273 |
| Carcinoma | c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae|g__Anaeroplasma | 0.0047 | 18 | 41 | 0.0220 | TB | 0.0273 |
| Carcinoma | c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae|g__Anaeroplasma|s__ | 0.0047 | 18 | 41 | 0.0220 | TB | 0.0273 |
| NTB-PF vs. TB | |||||||
| p__Firmicutes | |||||||
| Sarcoma | c__Bacilli|o__Lactobacillales | 0.0211 | 12 | 16 | 0.0375 | TB | 0.0194 |
| Sarcoma | c__Bacilli|o__Lactobacillales|f__Lactobacillaceae | 0.0268 | 13 | 16 | 0.040625 | TB | 0.0249 |
| Sarcoma | c__Bacilli|o__Lactobacillales|f__Lactobacillaceae|g__Lactobacillus | 0.0268 | 13 | 16 | 0.040625 | TB | 0.0249 |
| Sarcoma | c__Bacilli|o__Lactobacillales|f__Lactobacillaceae|g__Lactobacillus|s__ | 0.0268 | 13 | 16 | 0.040625 | TB | 0.0249 |
| Sarcoma | c__Bacilli|o__Lactobacillales|f__Lactobacillaceae|g__Lactobacillus|s__reuteri | 0.0297 | 16 | 16 | 0.05 | TB | 0.0249 |
| Sarcoma | c__Bacilli|o__Lactobacillales|f__StreptocoControlaceae|g__StreptocoControlus | 0.0063 | 8 | 16 | 0.025 | TB | 0.0087 |
| Sarcoma | c__Bacilli|o__Lactobacillales|f__StreptocoControlaceae|g__StreptocoControlus|s__ | 0.0061 | 7 | 16 | 0.021875 | TB | 0.0087 |
| Sarcoma | c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Blautia | 0.0019 | 6 | 16 | 0.01875 | TB | 0.0065 |
| Sarcoma | c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Blautia|Other | 0.0009 | 5 | 16 | 0.015625 | TB | 0.0036 |
| Sarcoma | c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__ | 0.0004 | 1 | 16 | 0.003125 | TB | 0.0027 |
| Sarcoma | c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__|s__ | 0.0004 | 1 | 16 | 0.003125 | TB | 0.0027 |
| Sarcoma | c__Clostridia|o__Clostridiales|f__Veillonellaceae|g__Veillonella | 0.0005 | 3 | 16 | 0.009375 | TB | 0.0019 |
| Sarcoma | c__Clostridia|o__Clostridiales|f__Veillonellaceae|g__Veillonella|s__dispar | 0.0005 | 3 | 16 | 0.009375 | TB | 0.0019 |
| Sarcoma | c__Erysipelotrichi | 0.0085 | 9 | 16 | 0.028125 | TB | 0.0402 |
| Sarcoma | c__Erysipelotrichi|o__Erysipelotrichales | 0.0085 | 9 | 16 | 0.028125 | TB | 0.0402 |
| Sarcoma | c__Erysipelotrichi|o__Erysipelotrichales|f__Erysipelotrichaceae | 0.0085 | 9 | 16 | 0.028125 | TB | 0.0402 |
| p__Actinobacteria | |||||||
| Carcinoma | c__Coriobacteriia | 0.0013 | 7 | 41 | 0.0085 | TB | 0.0035 |
| Carcinoma | c__Coriobacteriia|o__Coriobacteriales | 0.0013 | 7 | 41 | 0.0085 | TB | 0.0035 |
| Carcinoma | c__Coriobacteriia|o__Coriobacteriales|f__Coriobacteriaceae | 0.0013 | 7 | 41 | 0.0085 | TB | 0.0035 |
| Carcinoma | c__Bacteroidia|o__Bacteroidales|f__Rikenellaceae|g__AF12 | 0.0033 | 16 | 41 | 0.0195 | TB | 0.0028 |
| Carcinoma | c__Bacteroidia|o__Bacteroidales|f__Rikenellaceae|g__AF12|s__ | 0.0033 | 16 | 41 | 0.0195 | TB | 0.0028 |
| p__Proteobacteria | |||||||
| Carcinoma | c__Betaproteobacteria | 0.0015 | 11 | 41 | 0.0134 | NTB-PF | 0.0044 |
| Carcinoma | c__Betaproteobacteria|o__Burkholderiales | 0.0015 | 11 | 41 | 0.0134 | NTB-PF | 0.0044 |
| Carcinoma | c__Betaproteobacteria|o__Burkholderiales|f__Alcaligenaceae | 0.0015 | 11 | 41 | 0.0134 | NTB-PF | 0.0044 |
| Carcinoma | c__Betaproteobacteria|o__Burkholderiales|f__Alcaligenaceae|g__Sutterella | 0.0015 | 11 | 41 | 0.0134 | NTB-PF | 0.0044 |
| Carcinoma | c__Betaproteobacteria|o__Burkholderiales|f__Alcaligenaceae|g__Sutterella|s__ | 0.0015 | 11 | 41 | 0.0134 | NTB-PF | 0.0044 |
| p__Tenericutes | |||||||
| Carcinoma | p__Tenericutes | 0.0014 | 10 | 41 | 0.0122 | TB | 0.0035 |
| Carcinoma | c__Mollicutes | 0.0273 | 33 | 41 | 0.0402 | TB | 0.0028 |
| Carcinoma | c__Mollicutes|o__RF39 | 0.0273 | 33 | 41 | 0.0402 | TB | 0.039 |
| Carcinoma | c__Mollicutes|o__RF39|f__ | 0.0273 | 33 | 41 | 0.0402 | TB | 0.039 |
| Carcinoma | c__Mollicutes|o__RF39|f__|g__ | 0.0273 | 33 | 41 | 0.0402 | TB | 0.039 |
| Carcinoma | c__Mollicutes|o__RF39|f__|g__|s__ | 0.0273 | 33 | 41 | 0.0402 | TB | 0.039 |
| Carcinoma | c__Mollicutes|o__Anaeroplasmatales | 0.0047 | 18 | 41 | 0.0220 | TB | 0.0099 |
| Carcinoma | c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae | 0.0047 | 18 | 41 | 0.0220 | TB | 0.0099 |
| Carcinoma | c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae|g__Anaeroplasma | 0.0047 | 18 | 41 | 0.0220 | TB | 0.0099 |
| Carcinoma | c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae|g__Anaeroplasma|s__ | 0.0047 | 18 | 41 | 0.0220 | TB | 0.0099 |
References
- Aziz, T.; Hussain, N.; Hameed, Z.; Lin, L. Elucidating the role of diet in maintaining gut health to reduce the risk of obesity, cardiovascular and other age-related inflammatory diseases: Recent challenges and future recommendations. Gut Microbes 2024, 16, 2297864. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Chen, W.D.; Wang, Y.D. Gut Microbiota: An Integral Moderator in Health and Disease. Front. Microbiol. 2018, 9, 151. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chen, F.; Wu, G. Potential effects of gut microbiota on host cancers: Focus on immunity, DNA damage, cellular pathways, and anticancer therapy. ISME J. 2023, 17, 1535–1551. [Google Scholar] [CrossRef] [PubMed]
- Petruzzelli, M.; Wagner, E.F. Mechanisms of metabolic dysfunction in cancer-associated cachexia. Genes Dev. 2016, 30, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Suijkerbuijk, S.J.E.; van Rheenen, J. From good to bad: Intravital imaging of the hijack of physiological processes by cancer cells. Dev. Biol. 2017, 428, 328–337. [Google Scholar] [CrossRef] [PubMed]
- DeWys, W.D. Anorexia in cancer patients. Cancer Res. 1977, 37, 2354–2358. [Google Scholar]
- DeWys, D.; Begg, C.; Lavin, P.T.; Band, P.R.; Bennett, J.M.; Bertino, J.R.; Cohen, M.H.; Douglass, H.O.; Engstrom, P.F.; Ezdinli, E.Z.; et al. Prognostic effect of weight loss prior to chemotherapy in cancer patients. Am. J. Med. 1980, 69, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Veuthey, B.; Gonzalez, D.; Cardenas, J.P. Role of microbial secreted proteins in gut microbiota-host interactions. Front. Cell Infect. Microbiol. 2022, 12, 964710. [Google Scholar] [CrossRef] [PubMed]
- Chanmugam, P.; Feng, L.; Liou, S.; Jang, B.C.; Boudreau, M.; Yu, G.; Lee, J.H.; Kwon, H.J.; Beppu, T.; Yoshida, M. Radicicol, a protein tyrosine kinase inhibitor, suppresses the expression of mitogen-inducible cyclooxygenase in macrophages stimulated with lipopolysaccharide and in experimental glomerulonephritis. J. Biol. Chem. 1995, 270, 5418–5426. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Black, P.R.; Brooks, D.C.; Bessey, P.Q.; Wolfe, R.R.; Wilmore, D.W. Mechanisms of insulin resistance following injury. Ann. Surg. 1982, 196, 420–435. [Google Scholar] [CrossRef]
- Zhao, L.Y.; Mei, J.X.; Yu, G.; Lei, L.; Zhang, W.H.; Liu, K.; Chen, X.L.; Kolat, D.; Yang, K.; Hu, J.K. Role of the gut microbiota in anticancer therapy: From molecular mechanisms to clinical applications. Signal Transduct. Target. Ther. 2023, 8, 201. [Google Scholar] [CrossRef]
- Herremans, K.M.; Riner, A.N.; Cameron, M.E.; Trevino, J.G. The Microbiota and Cancer Cachexia. Int. J. Mol. Sci. 2019, 20, 6267. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Zogg, H.; Wei, L.; Bartlett, A.; Ghoshal, U.C.; Rajender, S.; Ro, S. Gut Microbial Dysbiosis in the Pathogenesis of Gastrointestinal Dysmotility and Metabolic Disorders. J. Neurogastroenterol. Motil. 2021, 27, 19–34. [Google Scholar] [CrossRef]
- Potgens, S.A.; Brossel, H.; Sboarina, M.; Catry, E.; Cani, P.D.; Neyrinck, A.M.; Delzenne, N.M.; Bindels, L.B. Klebsiella oxytoca expands in cancer cachexia and acts as a gut pathobiont contributing to intestinal dysfunction. Sci. Rep. 2018, 8, 12321. [Google Scholar] [CrossRef]
- Feng, L.; Zhang, W.; Shen, Q.; Miao, C.; Chen, L.; Li, Y.; Gu, X.; Fan, M.; Ma, Y.; Wang, H.; et al. Bile acid metabolism dysregulation associates with cancer cachexia: Roles of liver and gut microbiome. J. Cachexia Sarcopenia Muscle 2021, 12, 1553–1569. [Google Scholar] [CrossRef]
- Bindels, L.B.; Beck, R.; Schakman, O.; Martin, J.C.; De Backer, F.; Sohet, F.M.; Dewulf, E.M.; Pachikian, B.D.; Neyrinck, A.M.; Thissen, J.P.; et al. Restoring specific lactobacilli levels decreases inflammation and muscle atrophy markers in an acute leukemia mouse model. PLoS ONE 2012, 7, e37971. [Google Scholar] [CrossRef] [PubMed]
- Bindels, L.B.; Neyrinck, A.M.; Claus, S.P.; Le Roy, C.I.; Grangette, C.; Pot, B.; Martinez, I.; Walter, J.; Cani, P.D.; Delzenne, N.M. Synbiotic approach restores intestinal homeostasis and prolongs survival in leukaemic mice with cachexia. ISME J. 2016, 10, 1456–1470. [Google Scholar] [CrossRef]
- de Maria, Y.; Aciole Barbosa, D.; Menegidio, F.B.; Santos, K.; Humberto, A.C.; Alencar, V.C.; Silva, J.F.S.; Costa de Oliveira, R.; Batista, M.L., Jr.; Nunes, L.R.; et al. Analysis of mouse faecal dysbiosis, during the development of cachexia, induced by transplantation with Lewis lung carcinoma cells. Microbiology 2021, 167. [Google Scholar] [CrossRef]
- Ziemons, J.; Smidt, M.L.; Damink, S.O.; Rensen, S.S. Gut microbiota and metabolic aspects of cancer cachexia. Best Pract. Res. Clin. Endocrinol. Metab. 2021, 35, 101508. [Google Scholar] [CrossRef]
- Bindels, L.B.; Neyrinck, A.M.; Loumaye, A.; Catry, E.; Walgrave, H.; Cherbuy, C.; Leclercq, S.; Van Hul, M.; Plovier, H.; Pachikian, B.; et al. Increased gut permeability in cancer cachexia: Mechanisms and clinical relevance. Oncotarget 2018, 9, 18224–18238. [Google Scholar] [CrossRef] [PubMed]
- Ubachs, J.; Ziemons, J.; Soons, Z.; Aarnoutse, R.; van Dijk, D.P.J.; Penders, J.; van Helvoort, A.; Smidt, M.L.; Kruitwagen, R.; Baade-Corpelijn, L.; et al. Gut microbiota and short-chain fatty acid alterations in cachectic cancer patients. J. Cachexia Sarcopenia Muscle 2021, 12, 2007–2021. [Google Scholar] [CrossRef] [PubMed]
- Pedroza Matute, S.; Iyavoo, S. Exploring the gut microbiota: Lifestyle choices, disease associations, and personal genomics. Front. Nutr. 2023, 10, 1225120. [Google Scholar] [CrossRef] [PubMed]
- Byerley, L.O.; Samuelson, D.; Blanchard, E.T.; Luo, M.; Lorenzen, B.N.; Banks, S.; Ponder, M.A.; Welsh, D.A.; Taylor, C.M. Changes in the gut microbial communities following addition of walnuts to the diet. J. Nutr. Biochem. 2017, 48, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Hardman, W.E.; Ion, G.; Akinsete, J.A.; Witte, T.R. Dietary walnut suppressed mammary gland tumorigenesis in the C(3)1 TAg mouse. Nutr. Cancer 2011, 63, 960–970. [Google Scholar] [CrossRef]
- Reiter, R.J.; Tan, D.X.; Manchester, L.C.; Korkmaz, A.; Fuentes-Broto, L.; Hardman, W.E.; Rosales-Corral, S.A.; Qi, W. A walnut-enriched diet reduces the growth of LNCaP human prostate cancer xenografts in nude mice. Cancer Investig. 2013, 31, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Vazquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2013, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Baeza, Y.; Gonzalez, A.; Smarr, L.; McDonald, D.; Morton, J.T.; Navas-Molina, J.A.; Knight, R. Bringing the Dynamic Microbiome to Life with Animations. Cell Host Microbe 2017, 21, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Argiles, J.M.; Lopez-Soriano, F.J.; Stemmler, B.; Busquets, S. Cancer-associated cachexia—Understanding the tumour macroenvironment and microenvironment to improve management. Nat. Rev. Clin. Oncol. 2023, 20, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Argiles, J.M.; Busquets, S.; Stemmler, B.; Lopez-Soriano, F.J. Cancer cachexia: Understanding the molecular basis. Nat. Rev. Cancer 2014, 14, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Varian, B.J.; Gourishetti, S.; Poutahidis, T.; Lakritz, J.R.; Levkovich, T.; Kwok, C.; Teliousis, K.; Ibrahim, Y.M.; Mirabal, S.; Erdman, S.E. Beneficial bacteria inhibit cachexia. Oncotarget 2016, 7, 11803–11816. [Google Scholar] [CrossRef] [PubMed]
- Byerley, L.O.; Chang, H.M.; Lorenzen, B.; Guidry, J.; Hardman, W.E. Impact of dietary walnuts, a nutraceutical option, on circulating markers of metabolic dysregulation in a rodent cachectic tumor model. Biomed. Pharmacother. 2022, 155, 113728. [Google Scholar] [CrossRef]
- Jeong, S.M.; Jin, E.J.; Wei, S.; Bae, J.H.; Ji, Y.; Jo, Y.; Jeong, J.H.; Im, S.J.; Ryu, D. The impact of cancer cachexia on gut microbiota composition and short-chain fatty acid metabolism in a murine model. BMB Rep. 2023, 56, 404–409. [Google Scholar] [CrossRef]
- Ni, Y.; Lohinai, Z.; Heshiki, Y.; Dome, B.; Moldvay, J.; Dulka, E.; Galffy, G.; Berta, J.; Weiss, G.J.; Sommer, M.O.A.; et al. Distinct composition and metabolic functions of human gut microbiota are associated with cachexia in lung cancer patients. ISME J. 2021, 15, 3207–3220. [Google Scholar] [CrossRef] [PubMed]
- Koh, Y.C.; Kuo, L.H.; Tung, Y.C.; Weerawatanakorn, M.; Pan, M.H. Identification of Indicative Gut Microbial Guilds in a Natural Aging Mouse Model. ACS Omega 2023, 8, 36569–36580. [Google Scholar] [CrossRef] [PubMed]
- Delbrassinne, L.; Mahillon, J. Bacillus: Occurrence. In Encyclopedia of Food and Health; Caballero, B., Finglas, P.M., Toldrá, F., Eds.; Academic Press: Waltham, MA, USA, 2016. [Google Scholar]
- La Reau, A.J.; Suen, G. The Ruminococci: Key symbionts of the gut ecosystem. J. Microbiol. 2018, 56, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Bamberger, C.; Rossmeier, A.; Lechner, K.; Wu, L.; Waldmann, E.; Fischer, S.; Stark, R.G.; Altenhofer, J.; Henze, K.; Parhofer, K.G. A Walnut-Enriched Diet Affects Gut Microbiome in Healthy Caucasian Subjects: A Randomized, Controlled Trial. Nutrients 2018, 10, 244. [Google Scholar] [CrossRef]
- Holscher, H.D.; Guetterman, H.M.; Swanson, K.S.; An, R.; Matthan, N.R.; Lichtenstein, A.H.; Novotny, J.A.; Baer, D.J. Walnut Consumption Alters the Gastrointestinal Microbiota, Microbially Derived Secondary Bile Acids, and Health Markers in Healthy Adults: A Randomized Controlled Trial. J. Nutr. 2018, 148, 861–867. [Google Scholar] [CrossRef]

| Tumor Type | Dependent Variable | Pr > ChiSq | Rank | m =Number of Dependent Variables | CV for BH False Detect Rate 1 |
|---|---|---|---|---|---|
| Higher in the control diet group | |||||
| Sarcoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales | 0.0023 | 5 | 20 | 0.013 |
| Sarcoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae|g__Anaeroplasma | 0.0023 | 5 | 20 | 0.013 |
| Sarcoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae|g__Anaeroplasma|s__ | 0.0023 | 5 | 20 | 0.013 |
| Carcinoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales | 0.0033 | 22 | 45 | 0.0244 |
| Carcinoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae|g__Anaeroplasma | 0.0033 | 22 | 45 | 0.0244 |
| Carcinoma | p__Tenericutes|c__Mollicutes|o__Anaeroplasmatales|f__Anaeroplasmataceae|g__Anaeroplasma|s__ | 0.0033 | 22 | 45 | 0.0244 |
| Higher in the walnut diet group | |||||
| Sarcoma | p__Firmicutes|c__Bacilli | 0.0020 | 4 | 20 | 0.010 |
| Sarcoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__Ruminococcus|Other | 0.0006 | 2 | 20 | 0.005 |
| Carcinoma | p__Firmicutes|c__Bacilli | 0.0183 | 45 | 45 | 0.0500 |
| Carcinoma | p__Firmicutes|c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__Ruminococcus|Other | 0.0001 | 1 | 45 | 0.0011 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Byerley, L.O.; Lorenzen, B.; Chang, H.-M.; Hartman, W.G.; Keenan, M.J.; Page, R.; Luo, M.; Dowd, S.E.; Taylor, C.M. Gut Microbial Dysbiosis Differs in Two Distinct Cachectic Tumor-Bearing Models Consuming the Same Diet. Nutrients 2024, 16, 1076. https://doi.org/10.3390/nu16071076
Byerley LO, Lorenzen B, Chang H-M, Hartman WG, Keenan MJ, Page R, Luo M, Dowd SE, Taylor CM. Gut Microbial Dysbiosis Differs in Two Distinct Cachectic Tumor-Bearing Models Consuming the Same Diet. Nutrients. 2024; 16(7):1076. https://doi.org/10.3390/nu16071076
Chicago/Turabian StyleByerley, Lauri O., Brittany Lorenzen, Hsiao-Man Chang, William G. Hartman, Michael J. Keenan, Ryan Page, Meng Luo, Scot E. Dowd, and Christopher M. Taylor. 2024. "Gut Microbial Dysbiosis Differs in Two Distinct Cachectic Tumor-Bearing Models Consuming the Same Diet" Nutrients 16, no. 7: 1076. https://doi.org/10.3390/nu16071076
APA StyleByerley, L. O., Lorenzen, B., Chang, H.-M., Hartman, W. G., Keenan, M. J., Page, R., Luo, M., Dowd, S. E., & Taylor, C. M. (2024). Gut Microbial Dysbiosis Differs in Two Distinct Cachectic Tumor-Bearing Models Consuming the Same Diet. Nutrients, 16(7), 1076. https://doi.org/10.3390/nu16071076






