Identification and Characterization of Neuroprotective Properties of Thaumatin-like Protein 1a from Annurca Apple Flesh Polyphenol Extract
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Annurca Apple Flesh Polyphenol Extraction and Polyphenol Content Evaluation
2.3. Purification and Molecular Characterization of P33 Protein from AFPE
2.3.1. Desalting of P33 Protein by RP-HPLC
2.3.2. High-Resolution Nano-LC–Tandem Mass Spectrometry
2.3.3. Separation of AFPE Components by Gel-Filtration Chromatography
2.3.4. Qualitative Analysis of Extracted Polyphenols by RP-HPLC Analysis
2.3.5. SDS-PAGE Protein Analysis
3. Enzymatic Assays
3.1. Cholinesterase Assay
3.2. Monoamine Oxidase Assay
4. Aβ Self-Aggregation Assay
4.1. Inhibition of Aβ 1–40 Self-Aggregation
4.2. Reversion of Pre-Aggregated Aβ 1–40 Peptides
5. Cell Cultures and Treatments
Cell Viability Assay and Morphological Analysis
6. Statistical Analysis
7. Results
7.1. AFPE Protein Analysis, Biochemical Fractionation, and Identification of ~33 kDa Thaumatin-like Protein 1a
7.2. Identification of Some Polyphenols in Fractionated AFPE
7.3. Effects of Peaks 1–4 on Cholinesterase Activity
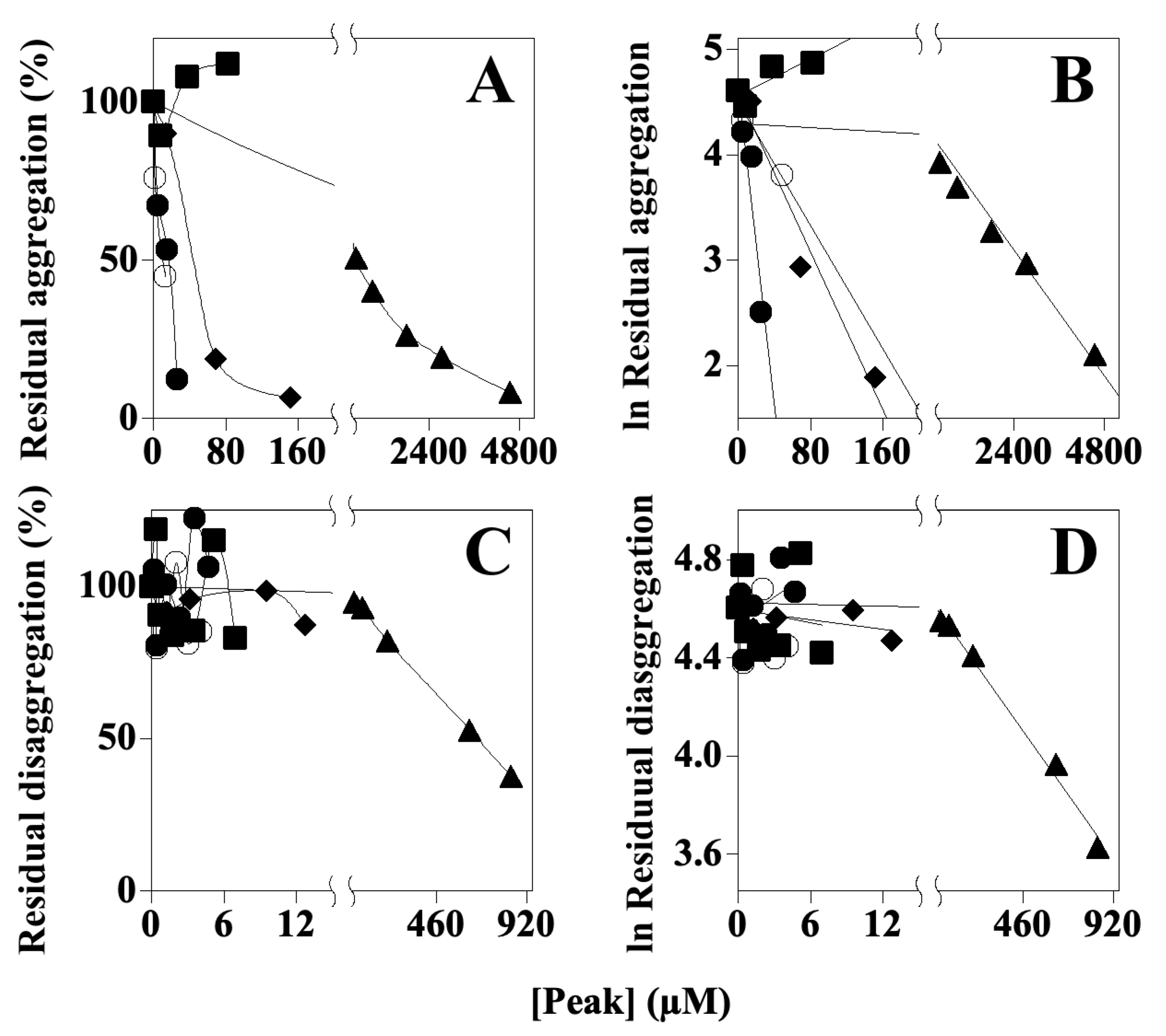
7.4. Effects of AFPE and Peaks 1–4 on Aβ 1–40 Self-Aggregation and Disaggregation Processes
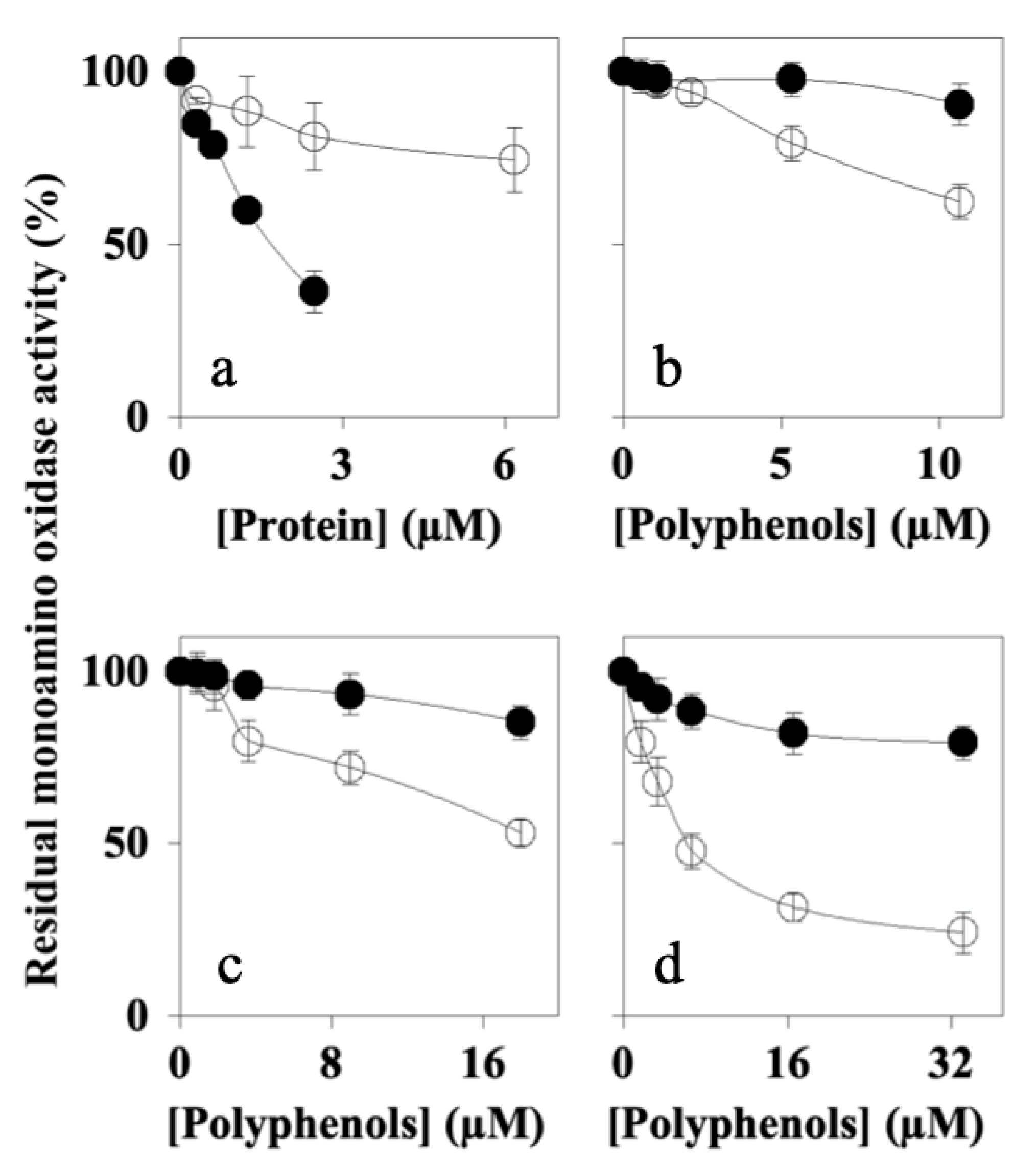
7.5. Effects of Peaks 1–4 on MAO-A and MAO-B Activity
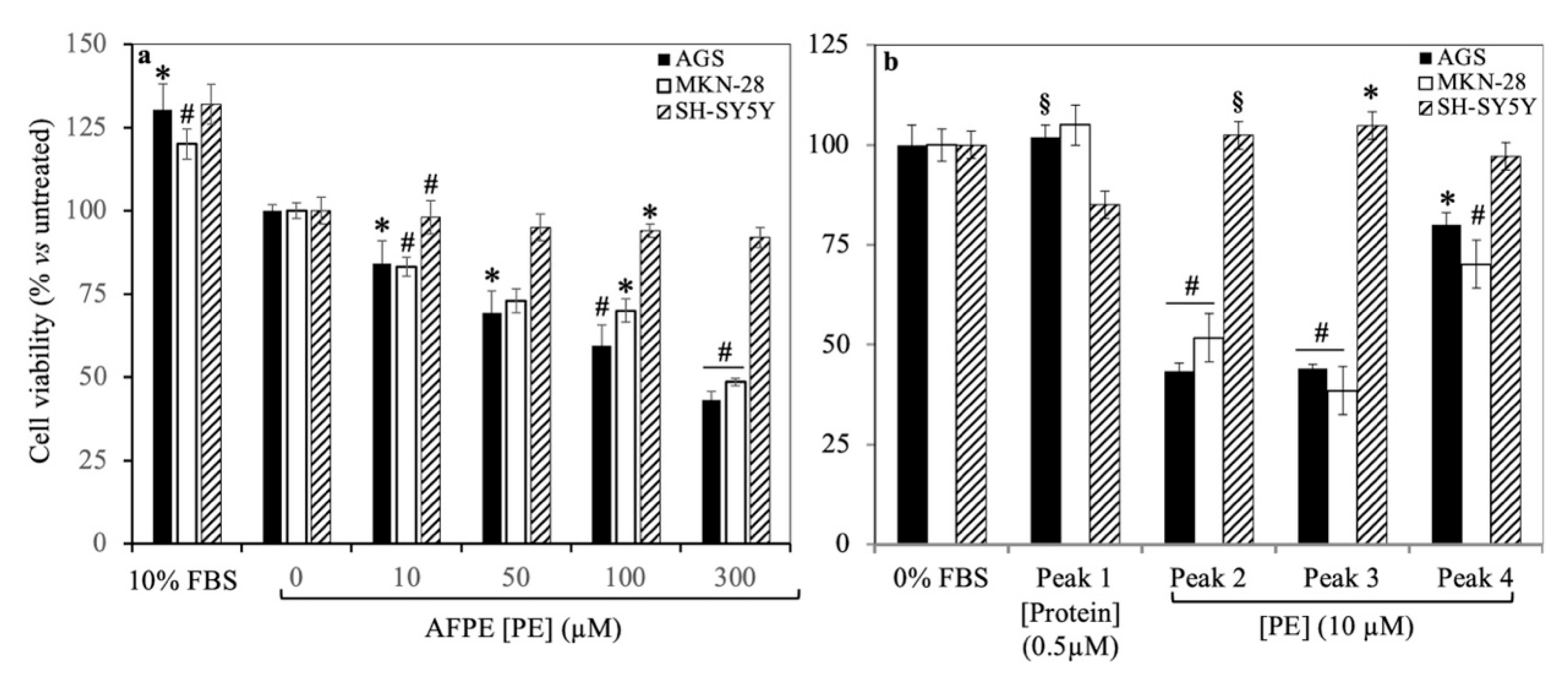
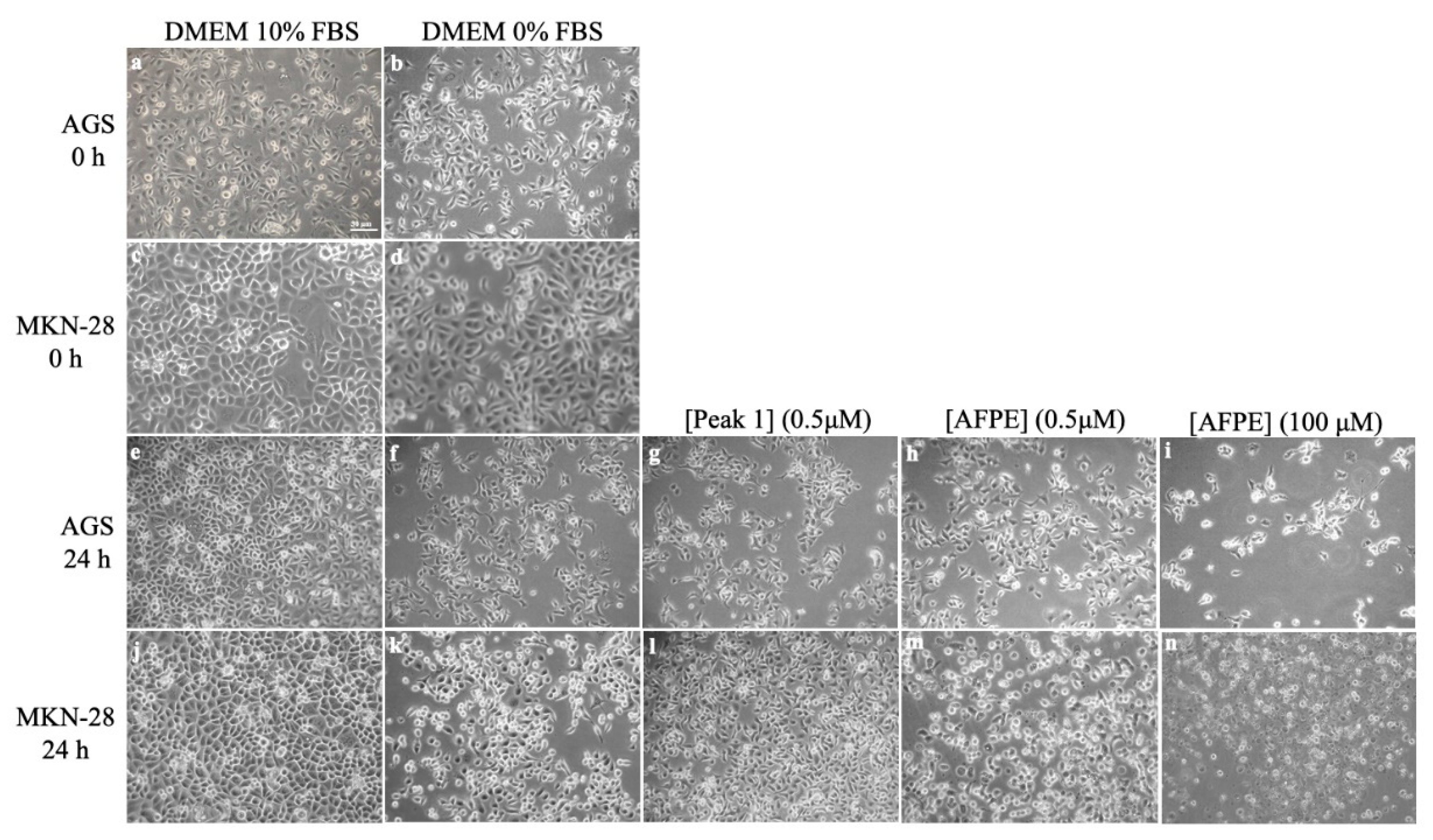
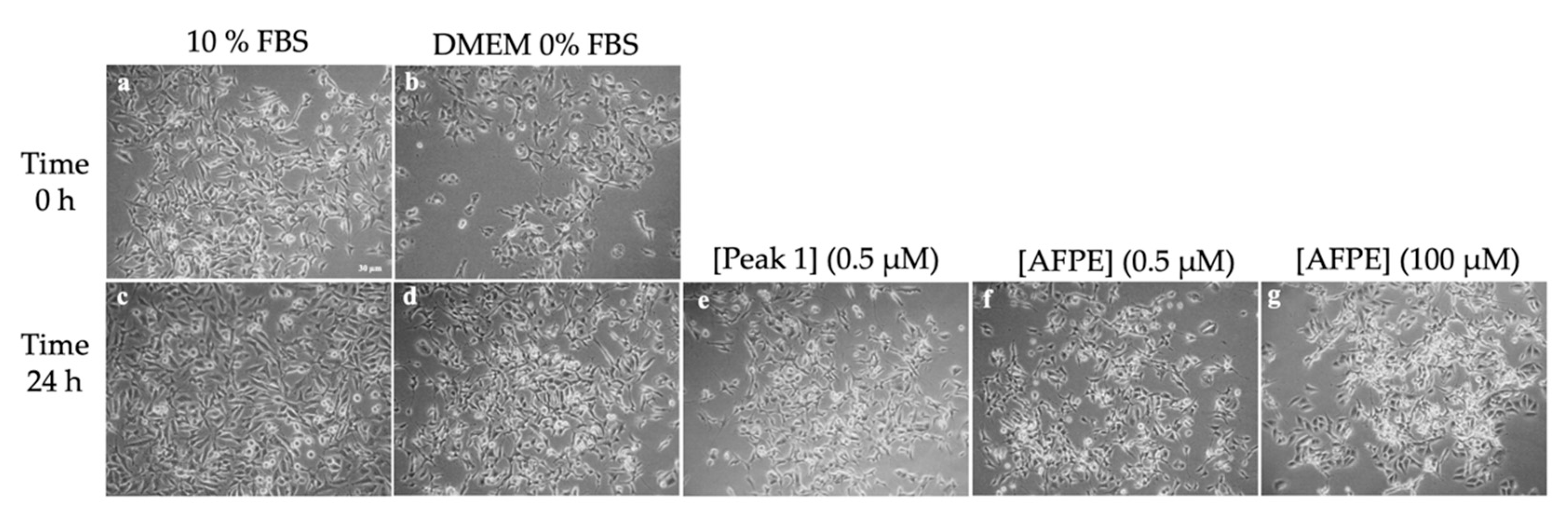
8. Effects of AFPE and Peaks 1–4 on the Cell Viability and Morphology of AGS and MKN-28 Human Gastric Adenocarcinoma Cells and SH-SY5Y Human Neuroblastoma Cells
9. Discussion
10. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mayne, K.; White, J.A.; McMurran, C.E.; Rivera, F.J.; de la Fuente, A.G. Aging and Neurodegenerative Disease: Is the Adaptive Immune System a Friend or Foe? Front. Aging Neurosci. 2020, 12, 572090. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, S.; Safia; Haque, E.; Mir, S.S. Neurodegenerative Diseases: Multifactorial Conformational Diseases and Their Therapeutic Interventions. J. Neurodegener. Dis. 2013, 2013, 563481. [Google Scholar] [CrossRef] [PubMed]
- Szeto, J.Y.; Lewis, S.J. Current Treatment Options for Alzheimer’s Disease and Parkinson’s Disease Dementia. Curr. Neuropharmacol. 2016, 14, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Alcaro, S.; Arcone, R.; Costa, G.; De Vita, D.; Iannone, M.; Ortuso, F.; Procopio, A.; Pasceri, R.; Rotiroti, D.; Scipione, L. Simple choline esters as potential anti-Alzheimer agents. Curr. Pharm. Des. 2010, 16, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, S.; Ansari, M.A.; Fatease, A.A.; Safhi, A.Y.; Hani, U.; Jahan, R.; Alomary, M.N.; Ansari, M.N.; Ahmed, N.; Wahab, S.; et al. Plant-Derived Bioactive Compounds in the Management of Neurodegenerative Disorders: Challenges, Future Directions and Molecular Mechanisms Involved in Neuroprotection. Pharmaceutics 2023, 23, 749. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Guo, M.S.; Zhang, Y.; Yu, L.; Wu, J.M.; Tang, Y.; Ai, W.; Zhu, F.D.; Law, B.Y.K.; Chen, Q.; et al. Dietary Plant Polyphenols as the Potential Drugs in Neurodegenerative Diseases: Current Evidence, Advances, and Opportunities. Oxidative Med. Cell. Longev. 2022, 2022, 5288698. [Google Scholar] [CrossRef]
- Durães, F.; Pinto, M.; Sousa, E. Old Drugs as New Treatments for Neurodegenerative Diseases. Pharmaceuticals 2018, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Arcone, R.; D’Errico, A.; Nasso, R.; Rullo, R.; Poli, A.; Di Donato, P.; Masullo, M. Inhibition of Enzymes Involved in Neurodegenerative Disorders and Abeta(1-40) Aggregation by Citrus limon Peel Polyphenol Extract. Molecules 2023, 28, 6332. [Google Scholar] [CrossRef]
- Armeli, F.; Bonucci, A.; Maggi, E.; Pinto, A.; Businaro, R. Mediterranean Diet and Neurodegenerative Diseases: The Neglected Role of Nutrition in the Modulation of the Endocannabinoid System. Biomolecules 2021, 11, 790. [Google Scholar] [CrossRef]
- Franco, G.A.; Interdonato, L.; Cordaro, M.; Cuzzocrea, S.; Di Paola, R. Bioactive Compounds of the Mediterranean Diet as Nutritional Support to Fight Neurodegenerative Disease. Int. J. Mol. Sci. 2023, 24, 7318. [Google Scholar] [CrossRef]
- Gardener, H.; Caunca, M.R. Mediterranean Diet in Preventing Neurodegenerative Diseases. Curr. Nutr. Rep. 2018, 7, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Pagliara, V.; De Rosa, M.; Di Donato, P.; Nasso, R.; D’Errico, A.; Cammarota, F.; Poli, A.; Masullo, M.; Arcone, R. Inhibition of Interleukin-6-Induced Matrix Metalloproteinase-2 Expression and Invasive Ability of Lemon Peel Polyphenol Extract in Human Primary Colon Cancer Cells. Molecules 2021, 26, 7076. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.; Liu, R.H. Apple phytochemicals and their health benefits. Nutr. J. 2004, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Fratianni, F.; De Giulio, A.; Sada, A.; Nazzaro, F. Biochemical characteristics and biological properties of Annurca apple cider. J. Med. Food 2012, 15, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Guarino, C.; Arena, S.; De Simone, L.; D’Ambrosio, C.; Santoro, S.; Rocco, M.; Scaloni, A.; Marra, M. Proteomic analysis of the major soluble components in Annurca apple flesh. Mol. Nutr. Food Res. 2007, 51, 255–262. [Google Scholar] [CrossRef] [PubMed]
- de Jesús-Pires, C.; Ferreira-Neto, J.R.C.; Pacifico Bezerra-Neto, J.; Kido, E.A.; de Oliveira Silva, R.L.; Pandolfi, V.; Wanderley-Nogueira, A.C.; Binneck, E.; da Costa, A.F.; Pio-Ribeiro, G.; et al. Plant Thaumatin-like Proteins: Function, Evolution and Biotechnological Applications. Curr. Protein Pept. Sci. 2020, 21, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Krebitz, M.; Wagner, B.; Ferreira, F.; Peterbauer, C.; Campillo, N.; Witty, M.; Kolarich, D.; Steinkellner, H.; Scheiner, O.; Breiteneder, H. Plant-based heterologous expression of Mal d 2, a thaumatin-like protein and allergen of apple (Malus domestica), and its characterization as an antifungal protein. J. Mol. Biol. 2003, 329, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Oberhuber, C.; Ma, Y.; Marsh, J.; Rigby, N.; Smole, U.; Radauer, C.; Alessandri, S.; Briza, P.; Zuidmeer, L.; Maderegger, B.; et al. Purification and characterisation of relevant natural and recombinant apple allergens. Mol. Nutr. Food Res. 2008, 52 (Suppl. S2), S208–S219. [Google Scholar] [CrossRef]
- Pasquariello, M.S.; Palazzo, P.; Tuppo, L.; Liso, M.; Petriccione, M.; Rega, P.; Tartaglia, A.; Tamburrini, M.; Alessandri, C.; Ciardiello, M.A. Analysis of the potential allergenicity of traditional apple cultivars by Multiplex Biochip-Based Immunoassay. Food Chem. 2012, 135, 219–227. [Google Scholar] [CrossRef]
- Liu, J.J.; Sturrock, R.; Ekramoddoullah, A.K. The superfamily of thaumatin-like proteins: Its origin, evolution, and expression towards biological function. Plant Cell. Rep. 2010, 29, 419–436. [Google Scholar] [CrossRef]
- van der Wel, H.; Loeve, K. Isolation and characterization of thaumatin I and II, the sweet-tasting proteins from Thaumatococcus daniellii Benth. Eur. J. Biochem. 1972, 31, 221–225. [Google Scholar] [CrossRef]
- Fadahunsi, O.; Adegbola, P.; Olorunnisola, S.; Akinloye, O. Phytochemistry, nutritional composition, and pharmacological activities of Thaumatococcus daniellii (Benth): A review. BioTechnologia 2021, 102, 101–117. [Google Scholar] [CrossRef]
- Firsov, A.; Shaloiko, L.; Kozlov, O.; Vinokurov, L.; Vainstein, A.; Dolgov, S. Purification and characterization of recombinant supersweet protein thaumatin II from tomato fruit. Protein Expr. Purif. 2016, 123, 1–5. [Google Scholar] [CrossRef]
- Smole, U.; Bublin, M.; Radauer, C.; Ebner, C.; Breiteneder, H. Mal d 2, the thaumatin-like allergen from apple, is highly resistant to gastrointestinal digestion and thermal processing. Int. Arch. Allergy Immunol. 2008, 147, 289–298. [Google Scholar] [CrossRef]
- Narasimhan, M.L.; Coca, M.A.; Jin, J.; Yamauchi, T.; Ito, Y.; Kadowaki, T.; Kim, K.K.; Pardo, J.M.; Damsz, B.; Hasegawa, P.M.; et al. Osmotin is a homolog of mammalian adiponectin and controls apoptosis in yeast through a homolog of mammalian adiponectin receptor. Mol. Cell 2005, 17, 171–180. [Google Scholar] [CrossRef]
- Viktorova, J.; Krasny, L.; Kamlar, M.; Novakova, M.; Mackova, M.; Macek, T. Osmotin, a pathogenesis-related protein. Curr. Protein Pept. Sci. 2012, 13, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.; Yoon, G.H.; Shah, S.A.; Lee, H.Y.; Kim, M.O. Osmotin attenuates amyloid beta-induced memory impairment, tau phosphorylation and neurodegeneration in the mouse hippocampus. Sci. Rep. 2015, 5, 11708. [Google Scholar] [CrossRef] [PubMed]
- Amin, F.U.; Hoshiar, A.K.; Do, T.D.; Noh, Y.; Shah, S.A.; Khan, M.S.; Yoon, J.; Kim, M.O. Osmotin-loaded magnetic nanoparticles with electromagnetic guidance for the treatment of Alzheimer’s disease. Nanoscale 2017, 9, 10619–10632. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Choe, K.; Lee, H.J.; Park, T.J.; Kim, M.O. Neuroprotective effects of osmotin in Parkinson’s disease-associated pathology via the AdipoR1/MAPK/AMPK/mTOR signaling pathways. J. Biomed. Sci. 2023, 30, 66. [Google Scholar] [CrossRef]
- Shah, S.A.; Lee, H.Y.; Bressan, R.A.; Yun, D.J.; Kim, M.O. Novel osmotin attenuates glutamate-induced synaptic dysfunction and neurodegeneration via the JNK/PI3K/Akt pathway in postnatal rat brain. Cell Death Dis. 2014, 5, e1026. [Google Scholar] [CrossRef]
- Arcone, R.; Nasso, R.; Pagliara, V.; D’Errico, A.; Motti, M.L.; D’Angelo, S.; Carbonara, G.; Masullo, M. Inhibition of Interleukin-6 Dependent Metalloproteinases-9/2 Expression in Cancer Cells by Diet Polyphenols. Antiinflamm. Antiallergy Agents Med. Chem. 2023, 22, 30–41. [Google Scholar] [CrossRef]
- Nasso, R.; Pagliara, V.; D’Angelo, S.; Rullo, R.; Masullo, M.; Arcone, R. Annurca Apple Polyphenol Extract Affects Acetyl-Cholinesterase and Mono-Amine Oxidase In Vitro Enzyme Activity. Pharmaceuticals 2021, 14, 62. [Google Scholar] [CrossRef]
- Borah, A.; Paul, R.; Choudhury, S.; Choudhury, A.; Bhuyan, B.; Das Talukdar, A.; Dutta Choudhury, M.; Mohanakumar, K.P. Neuroprotective potential of silymarin against CNS disorders: Insight into the pathways and molecular mechanisms of action. CNS Neurosci. Ther. 2013, 19, 847–853. [Google Scholar] [CrossRef]
- Singleton, V.L.; Joseph, A.; Rossi, J. Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar] [CrossRef]
- Shirazi, O.U.; Khattak, M.M.A.K.; Shukri, N.A.M. Determination of total phenolic, flavonoid content and free radical scavenging activities of common herbs and spices. J. Pharmacogn. Phytochem. 2014, 3, 104–108. [Google Scholar]
- Iglesias, R.; Russo, R.; Landi, N.; Valletta, M.; Chambery, A.; Di Maro, A.; Bolognesi, A.; Ferreras, J.M.; Citores, L. Structure and Biological Properties of Ribosome-Inactivating Proteins and Lectins from Elder (Sambucus nigra L.) Leaves. Toxins 2022, 14, 611. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Ellman, G.L.; Courtney, K.D.; Andres, V., Jr.; Feather-Stone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Saidemberg, D.M.; Ferreira, M.A.; Takahashi, T.N.; Gomes, P.C.; Cesar-Tognoli, L.M.; da Silva-Filho, L.C.; Tormena, C.F.; da Silva, G.V.; Palma, M.S. Monoamine oxidase inhibitory activities of indolylalkaloid toxins from the venom of the colonial spider Parawixia bistriata: Functional characterization of PwTX-I. Toxicon 2009, 54, 717–724. [Google Scholar] [CrossRef]
- Zheng, Q.; Song, J.; Doncaster, K.; Rowland, E.; Byers, D.M. Qualitative and quantitative evaluation of protein extraction protocols for apple and strawberry fruit suitable for two-dimensional electrophoresis and mass spectrometry analysis. J. Agric. Food Chem. 2007, 55, 1663–1673. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Arcone, R.; Chinali, A.; Pozzi, N.; Parafati, M.; Maset, F.; Pietropaolo, C.; De Filippis, V. Conformational and biochemical characterization of a biologically active rat recombinant Protease Nexin-1 expressed in E. coli. Biochim. Biophys. Acta 2009, 1794, 602–614. [Google Scholar] [CrossRef]
- Pagliara, V.; Nasso, R.; Di Donato, P.; Finore, I.; Poli, A.; Masullo, M.; Arcone, R. Lemon Peel Polyphenol Extract Reduces Interleukin-6-Induced Cell Migration, Invasiveness, and Matrix Metalloproteinase-9/2 Expression in Human Gastric Adenocarcinoma MKN-28 and AGS Cell Lines. Biomolecules 2019, 9, 833. [Google Scholar] [CrossRef]
- Oh, D.H.; Song, K.J.; Shin, Y.U.; Chung, W.I. Isolation of a cDNA encoding a 31-kDa, pathogenesis-related 5/thaumatin-like (PR5/TL) protein abundantly expressed in apple fruit (Nalus domestica cv. Fuji). Biosci. Biotechnol. Biochem. 2000, 64, 355–362. [Google Scholar] [CrossRef][Green Version]
- Alborghetti, M.; Nicoletti, F. Different Generations of Type-B Monoamine Oxidase Inhibitors in Parkinson’s Disease: From Bench to Bedside. Curr. Neuropharmacol. 2019, 17, 861–873. [Google Scholar] [CrossRef]
- Kumar, B.; Dwivedi, A.R.; Sarkar, B.; Gupta, S.K.; Krishnamurthy, S.; Mantha, A.K.; Parkash, J.; Kumar, V. 4,6-Diphenylpyrimidine Derivatives as Dual Inhibitors of Monoamine Oxidase and Acetylcholinesterase for the Treatment of Alzheimer’s Disease. ACS Chem. Neurosci. 2019, 10, 252–265. [Google Scholar] [CrossRef]
- Nurulita, N.A.; Kusuma, A.M.; Darsini, D.T.P.; Delvia, W.; Yulianti, V.T. The Cytoprotective and Cell Recovery Properties of Apple Extracts on H2O2 induced-NIH3T3 Cells: An Anti Aging Candidate. Indones. J. Cancer Chemoprevention 2018, 9, 78–85. [Google Scholar] [CrossRef]
- Alghamdi, S.S.; Suliman, R.S.; Aljammaz, N.A.; Kahtani, K.M.; Aljatli, D.A.; Albadrani, G.M. Natural Products as Novel Neuroprotective Agents; Computational Predictions of the Molecular Targets, ADME Properties, and Safety Profile. Plants 2022, 11, 549. [Google Scholar] [CrossRef]
- Katayama, S.; Corpuz, H.M.; Nakamura, S. Potential of plant-derived peptides for the improvement of memory and cognitive function. Peptides 2021, 142, 170571. [Google Scholar] [CrossRef]
- Wani, S.S.; Dar, P.A.; Zargar, S.M.; Dar, T.A. Therapeutic Potential of Medicinal Plant Proteins: Present Status and Future Perspectives. Curr. Protein Pept. Sci. 2020, 21, 443–487. [Google Scholar] [CrossRef]
- Cefarelli, G.; D’Abrosca, B.; Fiorentino, A.; Izzo, A.; Mastellone, C.; Pacifico, S.; Piscopo, V. Free-radical-scavenging and antioxidant activities of secondary metabolites from reddened cv. Annurca apple fruits. J. Agric. Food Chem. 2006, 54, 803–809. [Google Scholar] [CrossRef]
- Koseoğlu, A.; Al-Taie, A. The potential chemo-preventive roles of Malus domestica against the risk of colorectal cancer: A suggestive insight into clinical application. Clin. Nutr. ESPEN 2022, 52, 360–364. [Google Scholar] [CrossRef]
- Tenore, G.C.; Campiglia, P.; Stiuso, P.; Ritieni, A.; Novellino, E. Nutraceutical potential of polyphenolic fractions from Annurca apple (M. pumila Miller cv Annurca). Food Chem. 2013, 140, 614–622. [Google Scholar] [CrossRef]
- Chowdhury, S.; Kumar, S. Alpha-Terpinyl Acetate: A Natural Monoterpenoid from Elettaria cardamomum as Multi-Target Directed Ligand in Alzheimer’s Disease. J. Funct. Foods 2020, 68, 103892. [Google Scholar] [CrossRef]
- Miele, M.; Costantini, S.; Colonna, G. Structural and functional similarities between osmotin from Nicotiana tabacum seeds and human adiponectin. PLoS ONE 2011, 6, e16690. [Google Scholar] [CrossRef]
- Ishikawa, M.; Kitayama, J.; Yamauchi, T.; Kadowaki, T.; Maki, T.; Miyato, H.; Yamashita, H.; Nagawa, H. Adiponectin inhibits the growth and peritoneal metastasis of gastric cancer through its specific membrane receptors AdipoR1 and AdipoR2. Cancer Sci. 2007, 98, 1120–1127. [Google Scholar] [CrossRef]
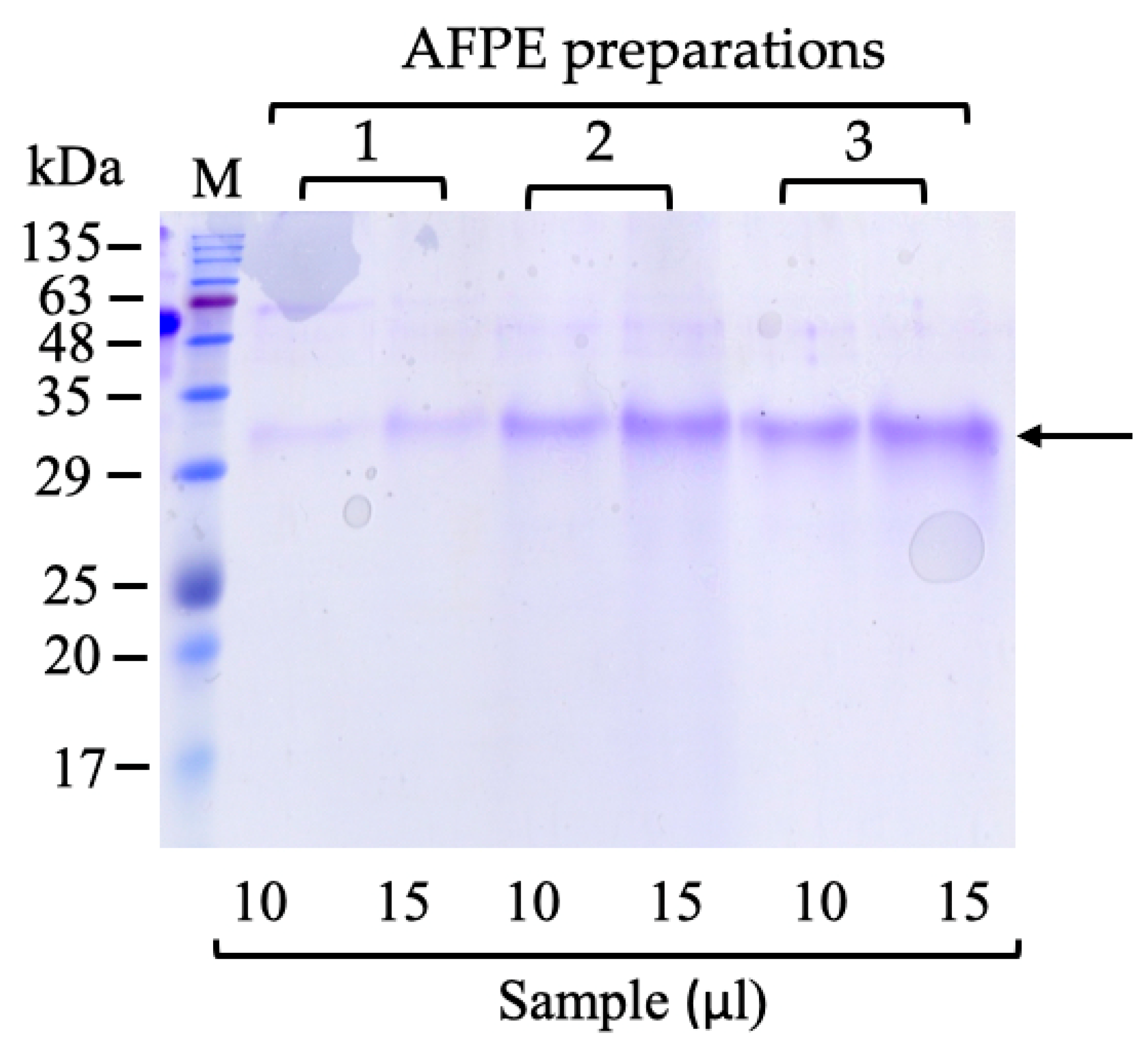
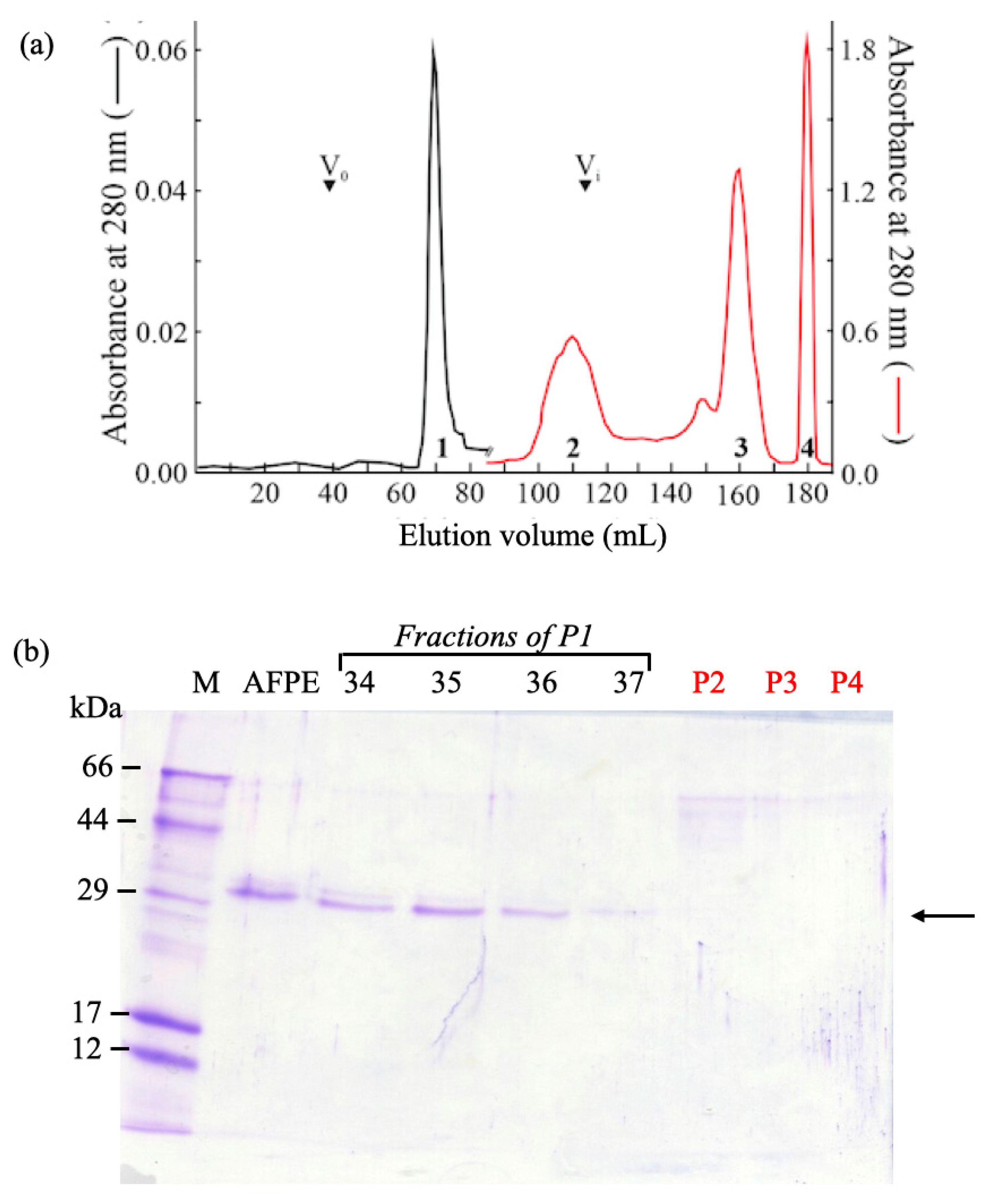
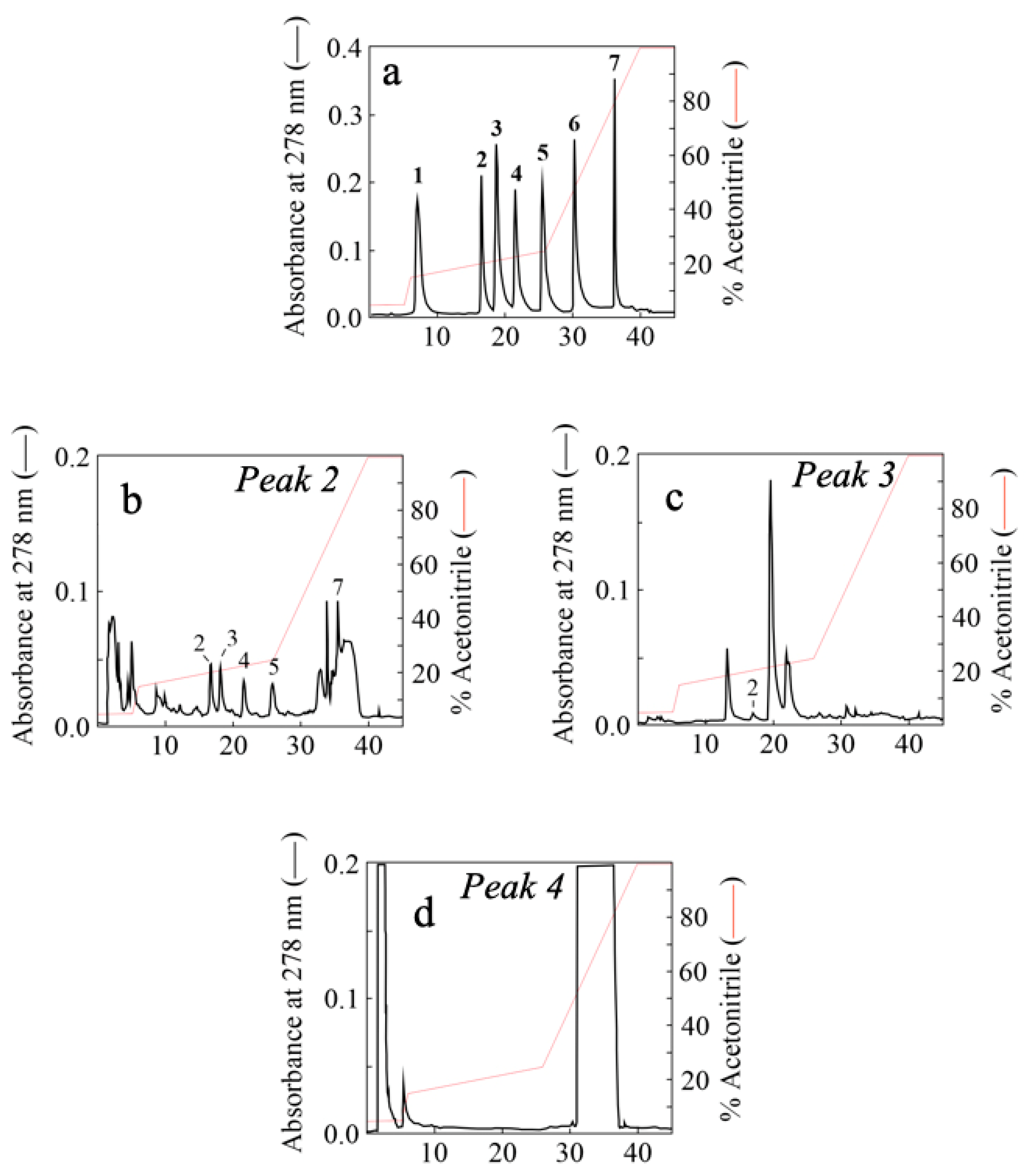
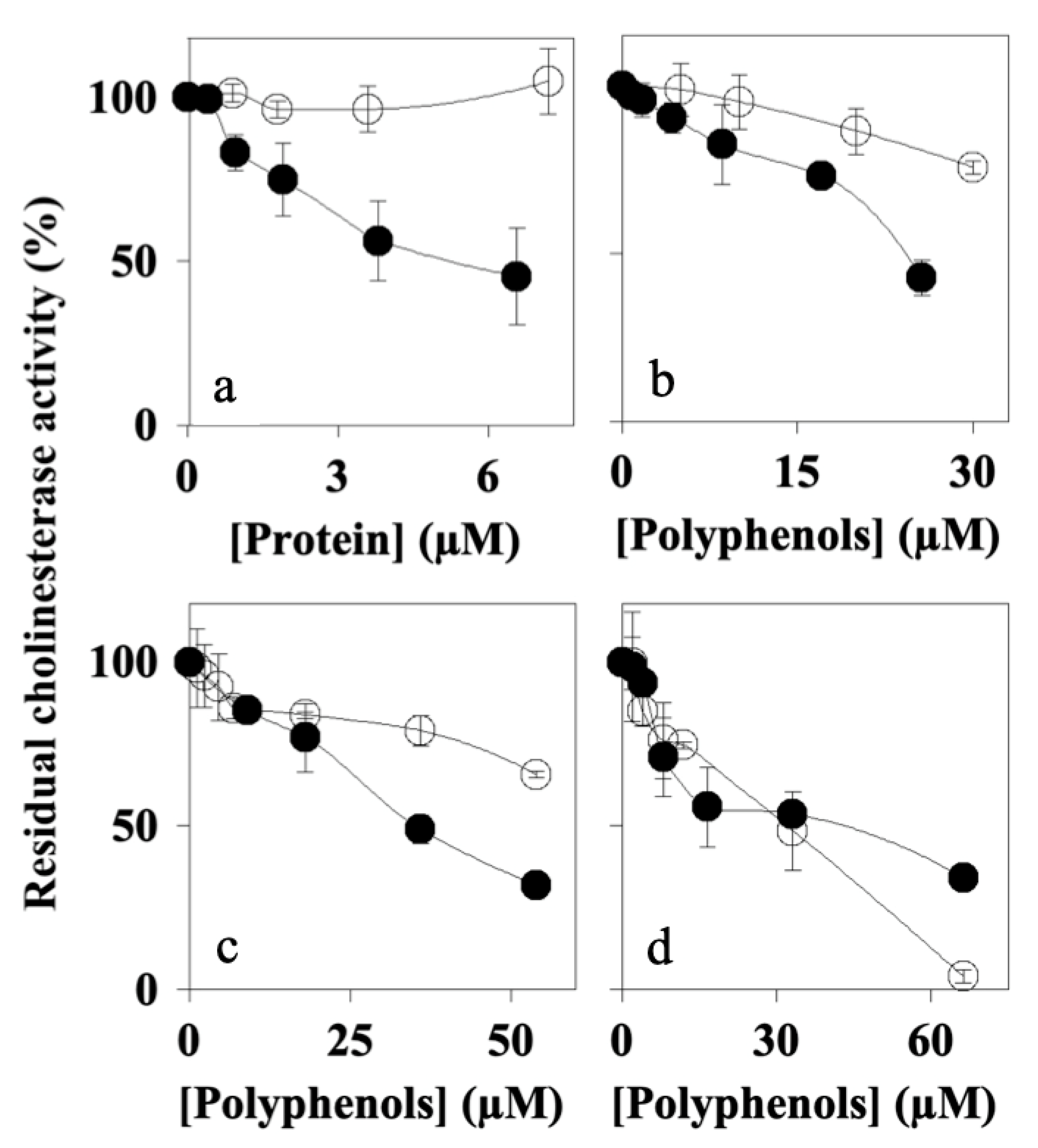
| AFPE Preparations | Proteins (mM) | Polyphenols (mM) | Proteins/Polyphenols (%) |
|---|---|---|---|
| 1 | 0.017 | 10.00 | 0.17 |
| 2 | 0.029 | 8.60 | 0.34 |
| 3 | 0.033 | 15.00 | 0.22 |
| Mean ± S.D. | 0.026 ± 0.008 | 11.20 ± 3.40 | 0.24 ± 0.09 |
| AFPE | Peak 1 | Peak 2 | Peak 3 | Peak 4 | |
|---|---|---|---|---|---|
| Enzyme | PE (µM) | Protein (µM) | PE (µM) | PE (µM) | PE (µM) |
| AChE | 859 ± 18 # | 5.53 ± 0.09 | 23.43 ± 0.99 | 32.30 ± 0.17 | 43.67 ± 2.33 |
| BuChE | 966 ± 72 # | N.D. | 73.14 ± 1.29 | 101.35 ± 3.60 | 12.70 ± 0.60 |
| MAO-A | 145 ± 30 # | 1.71 ± 0.05 | 84.61 ± 5.64 | 79.60 ± 0.50 | 62.92 ± 2.48 |
| MAO-B | 199 ± 70 # | 16.43 ± 1.06 | 15.77 ± 0.12 | 19.63 ± 0.34 | 6.33 ± 0.02 |
| Fibrillation process | |||||
| Inhibition of Aβ 1–40 fibril formation | 1396.00 ± 3.05 | 9.16 ± 0.52 | 44.47 ± 1.54 | N.D. | 37.11 ± 0.41 |
| Reversion of Aβ 1–40 fibrils | 616.70 ± 2.65 | N.D. | N.D. | N.D. | 104.87 ± 43.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Errico, A.; Nasso, R.; Di Maro, A.; Landi, N.; Chambery, A.; Russo, R.; D’Angelo, S.; Masullo, M.; Arcone, R. Identification and Characterization of Neuroprotective Properties of Thaumatin-like Protein 1a from Annurca Apple Flesh Polyphenol Extract. Nutrients 2024, 16, 307. https://doi.org/10.3390/nu16020307
D’Errico A, Nasso R, Di Maro A, Landi N, Chambery A, Russo R, D’Angelo S, Masullo M, Arcone R. Identification and Characterization of Neuroprotective Properties of Thaumatin-like Protein 1a from Annurca Apple Flesh Polyphenol Extract. Nutrients. 2024; 16(2):307. https://doi.org/10.3390/nu16020307
Chicago/Turabian StyleD’Errico, Antonio, Rosarita Nasso, Antimo Di Maro, Nicola Landi, Angela Chambery, Rosita Russo, Stefania D’Angelo, Mariorosario Masullo, and Rosaria Arcone. 2024. "Identification and Characterization of Neuroprotective Properties of Thaumatin-like Protein 1a from Annurca Apple Flesh Polyphenol Extract" Nutrients 16, no. 2: 307. https://doi.org/10.3390/nu16020307
APA StyleD’Errico, A., Nasso, R., Di Maro, A., Landi, N., Chambery, A., Russo, R., D’Angelo, S., Masullo, M., & Arcone, R. (2024). Identification and Characterization of Neuroprotective Properties of Thaumatin-like Protein 1a from Annurca Apple Flesh Polyphenol Extract. Nutrients, 16(2), 307. https://doi.org/10.3390/nu16020307










