Identification of Sweetness Preference-Related Single-Nucleotide Polymorphisms for Polygenic Risk Scores Associated with Obesity
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Populations
2.2. Assessment of Obesity and Covariates
2.3. Dietary Assessment and Phenotype Definitions
2.4. Genotyping and Calculating PRS
2.5. Functional Annotation and Gene Mapping
2.6. Statistical Analysis
3. Results
3.1. Sweetness Preference GWAS Analysis
3.2. Characteristics According to PRS between Males and Females
3.3. Genotype of SNP rs4861982 and Obesity
3.4. Correlation Effects of PRS and Environmental Factors on Obesity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boutari, C.; Mantzoros, C.S. A 2022 Update on the Epidemiology of Obesity and a Call to Action: As Its Twin COVID-19 Pandemic Appears to Be Receding, the Obesity and Dysmetabolism Pandemic Continues to Rage On. Metabolism 2022, 133, 155217. [Google Scholar] [CrossRef] [PubMed]
- Herrera, B.M.; Keildson, S.; Lindgren, C.M. Genetics and Epigenetics of Obesity. Maturitas 2011, 69, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Gerspach, A.C.; Steinert, R.E.; Schönenberger, L.; Graber-Maier, A.; Beglinger, C. The Role of the Gut Sweet Taste Receptor in Regulating GLP-1, PYY, and CCK Release in Humans. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E317–E325. [Google Scholar] [CrossRef] [PubMed]
- Prandi, S.; Voigt, A.; Meyerhof, W.; Behrens, M. Expression Profiling of Tas2r Genes Reveals a Complex Pattern along the Mouse GI Tract and the Presence of Tas2r131 in a Subset of Intestinal Paneth Cells. Cell. Mol. Life Sci. 2018, 75, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Hwang, L.-D.; Lin, C.; Gharahkhani, P.; Cuellar-Partida, G.; Ong, J.-S.; An, J.; Gordon, S.D.; Zhu, G.; MacGregor, S.; Lawlor, D.A.; et al. New Insight into Human Sweet Taste: A Genome-Wide Association Study of the Perception and Intake of Sweet Substances. Am. J. Clin. Nutr. 2019, 109, 1724–1737. [Google Scholar] [CrossRef]
- Hwang, L.-D.; Gharahkhani, P.; Breslin, P.A.S.; Gordon, S.D.; Zhu, G.; Martin, N.G.; Reed, D.R.; Wright, M.J. Bivariate Genome-Wide Association Analysis Strengthens the Role of Bitter Receptor Clusters on Chromosomes 7 and 12 in Human Bitter Taste. BMC Genom. 2018, 19, 678. [Google Scholar] [CrossRef]
- Coltell, O.; Sorlí, J.V.; Asensio, E.M.; Fernández-Carrión, R.; Barragán, R.; Ortega-Azorín, C.; Estruch, R.; González, J.I.; Salas-Salvadó, J.; Lamon-Fava, S.; et al. Association between Taste Perception and Adiposity in Overweight or Obese Older Subjects with Metabolic Syndrome and Identification of Novel Taste-Related Genes. Am. J. Clin. Nutr. 2019, 109, 1709–1723. [Google Scholar] [CrossRef]
- Wohlgemuth, K.J.; Arieta, L.R.; Brewer, G.J.; Hoselton, A.L.; Gould, L.M.; Smith-Ryan, A.E. Sex Differences and Considerations for Female Specific Nutritional Strategies: A Narrative Review. J. Int. Soc. Sports Nutr. 2021, 18, 27. [Google Scholar] [CrossRef]
- Faruque, S.; Tong, J.; Lacmanovic, V.; Agbonghae, C.; Minaya, D.M.; Czaja, K. The Dose Makes the Poison: Sugar and Obesity in the United States—A Review. Pol. J. Food Nutr. Sci. 2019, 69, 219–233. [Google Scholar] [CrossRef]
- Sen, T.; Cawthon, C.R.; Ihde, B.T.; Hajnal, A.; DiLorenzo, P.M.; de La Serre, C.B.; Czaja, K. Diet-Driven Microbiota Dysbiosis Is Associated with Vagal Remodeling and Obesity. Physiol. Behav. 2017, 173, 305–317. [Google Scholar] [CrossRef]
- Hunter, D.J.; Kraft, P.; Jacobs, K.B.; Cox, D.G.; Yeager, M.; Hankinson, S.E.; Wacholder, S.; Wang, Z.; Welch, R.; Hutchinson, A.; et al. A Genome-Wide Association Study Identifies Alleles in FGFR2 Associated with Risk of Sporadic Postmenopausal Breast Cancer. Nat. Genet. 2007, 39, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, M.C.; Monda, K.L.; Yu, K.; Paynter, N.; Azzato, E.M.; Bennett, S.N.; Berndt, S.I.; Boerwinkle, E.; Chanock, S.; Chatterjee, N.; et al. Genome-Wide Meta-Analysis Identifies Regions on 7p21 (AHR) and 15q24 (CYP1A2) as Determinants of Habitual Caffeine Consumption. PLoS Genet. 2011, 7, e1002033. [Google Scholar] [CrossRef]
- Jensen, M.K.; Pers, T.H.; Dworzynski, P.; Girman, C.J.; Brunak, S.; Rimm, E.B. Protein Interaction-Based Genome-Wide Analysis of Incident Coronary Heart Disease. Circ. Cardiovasc. Genet. 2011, 4, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, B.E.; Haskell, W.L.; Whitt, M.C.; Irwin, M.L.; Swartz, A.M.; Strath, S.J.; O’Brien, W.L.; Bassett, D.R.J.; Schmitz, K.H.; Emplaincourt, P.O.; et al. Compendium of Physical Activities: An Update of Activity Codes and MET Intensities. Med. Sci. Sports Exerc. 2000, 32, S498–S504. [Google Scholar] [CrossRef] [PubMed]
- Willett, W.C.; Sampson, L.; Stampfer, M.J.; Rosner, B.; Bain, C.; Witschi, J.; Hennekens, C.H.; Speizer, F.E. Reproducibility and validity of a Semiquantitative food frequency questionnaire. Am. J. Epidemiol. 1985, 122, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Zhong, V.W.; Kuang, A.; Danning, R.D.; Kraft, P.; van Dam, R.M.; Chasman, D.I.; Cornelis, M.C. A Genome-Wide Association Study of Bitter and Sweet Beverage Consumption. Hum. Mol. Genet. 2019, 28, 2449–2457. [Google Scholar] [CrossRef]
- Cornelis, M.C.; Tordoff, M.G.; El-Sohemy, A.; van Dam, R.M. Recalled Taste Intensity, Liking and Habitual Intake of Commonly Consumed Foods. Appetite 2017, 109, 182–189. [Google Scholar] [CrossRef]
- Lindström, S.; Loomis, S.; Turman, C.; Huang, H.; Huang, J.; Aschard, H.; Chan, A.T.; Choi, H.; Cornelis, M.; Curhan, G.; et al. A Comprehensive Survey of Genetic Variation in 20,691 Subjects from Four Large Cohorts. PLoS ONE 2017, 12, e0173997. [Google Scholar] [CrossRef]
- Jiang, L.; Penney, K.L.; Giovannucci, E.; Kraft, P.; Wilson, K.M. A Genome-Wide Association Study of Energy Intake and Expenditure. PLoS ONE 2018, 13, e0201555. [Google Scholar] [CrossRef]
- Colditz, G.A.; Hankinson, S.E. The Nurses’ Health Study: Lifestyle and Health among Women. Nat. Rev. Cancer 2005, 5, 388–396. [Google Scholar] [CrossRef]
- Wang, T.; Heianza, Y.; Sun, D.; Huang, T.; Ma, W.; Rimm, E.B.; Manson, J.E.; Hu, F.B.; Willett, W.C.; Qi, L. Improving Adherence to Healthy Dietary Patterns, Genetic Risk, and Long Term Weight Gain: Gene-Diet Interaction Analysis in Two Prospective Cohort Studies. BMJ 2018, 360, j5644. [Google Scholar] [CrossRef] [PubMed]
- Skol, A.D.; Scott, L.J.; Abecasis, G.R.; Boehnke, M. Joint Analysis Is More Efficient than Replication-Based Analysis for Two-Stage Genome-Wide Association Studies. Nat. Genet. 2006, 38, 209–213. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A General Framework for Estimating the Relative Pathogenicity of Human Genetic Variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of Functional Variation in Personal Genomes Using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic Mining of Putative Causal Variants, Cell Types, Regulators and Target Genes for Human Complex Traits and Disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and Efficient Meta-Analysis of Genomewide Association Scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef]
- Khalili, H.; Ananthakrishnan, A.N.; Konijeti, G.G.; Higuchi, L.M.; Fuchs, C.S.; Richter, J.M.; Chan, A.T. Measures of Obesity and Risk of Crohn’s Disease and Ulcerative Colitis. Inflamm. Bowel Dis. 2015, 21, 361–368. [Google Scholar] [CrossRef]
- Bertrand, K.A.; Giovannucci, E.; Rosner, B.A.; Zhang, S.M.; Laden, F.; Birmann, B.M. Dietary Fat Intake and Risk of Non-Hodgkin Lymphoma in 2 Large Prospective Cohorts. Am. J. Clin. Nutr. 2017, 106, 650–656. [Google Scholar] [CrossRef][Green Version]
- Cornelis, M.C.; Byrne, E.M.; Esko, T.; Nalls, M.A.; Ganna, A.; Paynter, N.; Monda, K.L.; Amin, N.; Fischer, K.; Renstrom, F.; et al. Genome-Wide Meta-Analysis Identifies Six Novel Loci Associated with Habitual Coffee Consumption. Mol. Psychiatry 2015, 20, 647–656. [Google Scholar] [CrossRef]
- Seidell, J.C.; Halberstadt, J. Obesity: The Obesity Epidemic in the USA—No End in Sight? Nat. Rev. Endocrinol. 2016, 12, 499–500. [Google Scholar] [CrossRef] [PubMed]
- Deckelbaum, R.J.; Williams, C.L. Childhood Obesity: The Health Issue. Obes. Res. 2001, 9 (Suppl. 4), 239S–243S. [Google Scholar] [CrossRef]
- Turner, T.N.; Sharma, K.; Oh, E.C.; Liu, Y.P.; Collins, R.L.; Sosa, M.X.; Auer, D.R.; Brand, H.; Sanders, S.J.; Moreno-De-Luca, D.; et al. Loss of δ-Catenin Function in Severe Autism. Nature 2015, 520, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Steinke, A.; Meier-Stiegen, S.; Drenckhahn, D.; Asan, E. Molecular Composition of Tight and Adherens Junctions in the Rat Olfactory Epithelium and Fila. Histochem. Cell Biol. 2008, 130, 339–361. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.; Marinescu, R.C.; Overhauser, J.; Kosik, K.S. Hemizygosity of Delta-Catenin (CTNND2) Is Associated with Severe Mental Retardation in Cri-Du-Chat Syndrome. Genomics 2000, 63, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhao, D.; Wu, Y.; Xu, C.; Zhang, F. Cyclic Stretch Induced Gene Expression of Extracellular Matrix and Adhesion Molecules in Human Periodontal Ligament Cells. Arch. Oral. Biol. 2015, 60, 447–455. [Google Scholar] [CrossRef]
- Ho, C.; Zhou, J.; Medina, M.; Goto, T.; Jacobson, M.; Bhide, P.G.; Kosik, K.S. Delta-Catenin Is a Nervous System-Specific Adherens Junction Protein Which Undergoes Dynamic Relocalization during Development. J. Comp. Neurol. 2000, 420, 261–276. [Google Scholar] [CrossRef]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of Protein-Coding Genetic Variation in 60,706 Humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef]
- Sayols-Baixeras, S.; Subirana, I.; Fernández-Sanlés, A.; Sentí, M.; Lluís-Ganella, C.; Marrugat, J.; Elosua, R. DNA Methylation and Obesity Traits: An Epigenome-Wide Association Study. The REGICOR Study. Epigenetics 2017, 12, 909–916. [Google Scholar] [CrossRef]
- Giordano, M.V.; Lucas, H.D.S.; Fiorelli, R.K.A.; Giordano, L.A.; Giordano, M.G.; Baracat, E.C.; Júnior, J.M.S. Expression Levels of BCL2 and MKI67 in Endometrial Polyps in Postmenopausal Women and Their Correlation with Obesity. Mol. Clin. Oncol. 2020, 13, 69. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Ambrosi, J.; Catalán, V.; Diez-Caballero, A.; Martinez-Cruz, L.A.; Gil, M.J.; García-Foncillas, J.; Cienfuegos, J.A.; Salvador, J.; Mato, J.M.; Frühbeck, G. Gene Expression Profile of Omental Adipose Tissue in Human Obesity. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2004, 18, 215–217. [Google Scholar] [CrossRef]
- Shintani, T.; Higashi, S.; Takeuchi, Y.; Gaudio, E.; Trapasso, F.; Fusco, A.; Noda, M. The R3 Receptor-like Protein Tyrosine Phosphatase Subfamily Inhibits Insulin Signalling by Dephosphorylating the Insulin Receptor at Specific Sites. J. Biochem. 2015, 158, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Suzuki, R.; Takeuchi, Y.; Shirasawa, T.; Noda, M. Deletion or Inhibition of PTPRO Prevents Ectopic Fat Accumulation and Induces Healthy Obesity with Markedly Reduced Systemic Inflammation. Life Sci. 2023, 313, 121292. [Google Scholar] [CrossRef] [PubMed]
- Riedmann, C.; Ma, Y.; Melikishvili, M.; Godfrey, S.G.; Zhang, Z.; Chen, K.C.; Rouchka, E.C.; Fondufe-Mittendorf, Y.N. Inorganic Arsenic-Induced Cellular Transformation Is Coupled with Genome Wide Changes in Chromatin Structure, Transcriptome and Splicing Patterns. BMC Genom. 2015, 16, 212. [Google Scholar] [CrossRef]
- Eick, S.M.; Steinmaus, C. Arsenic and Obesity: A Review of Causation and Interaction. Curr. Environ. Heal. Rep. 2020, 7, 343–351. [Google Scholar] [CrossRef]
- Gold, E.B. The Timing of the Age at Which Natural Menopause Occurs. Obstet. Gynecol. Clin. N. Am. 2011, 38, 425–440. [Google Scholar] [CrossRef]
- Pines, A. Male Menopause: Is It a Real Clinical Syndrome? Climacteric 2011, 14, 15–17. [Google Scholar] [CrossRef]
- Chiolero, A.; Faeh, D.; Paccaud, F.; Cornuz, J. Consequences of Smoking for Body Weight, Body Fat Distribution, and Insulin Resistance. Am. J. Clin. Nutr. 2008, 87, 801–809. [Google Scholar] [CrossRef]
- Romero-Corral, A.; Somers, V.K.; Sierra-Johnson, J.; Thomas, R.J.; Collazo-Clavell, M.L.; Korinek, J.; Allison, T.G.; Batsis, J.A.; Sert-Kuniyoshi, F.H.; Lopez-Jimenez, F. Accuracy of Body Mass Index in Diagnosing Obesity in the Adult General Population. Int. J. Obes. 2008, 32, 959–966. [Google Scholar] [CrossRef]

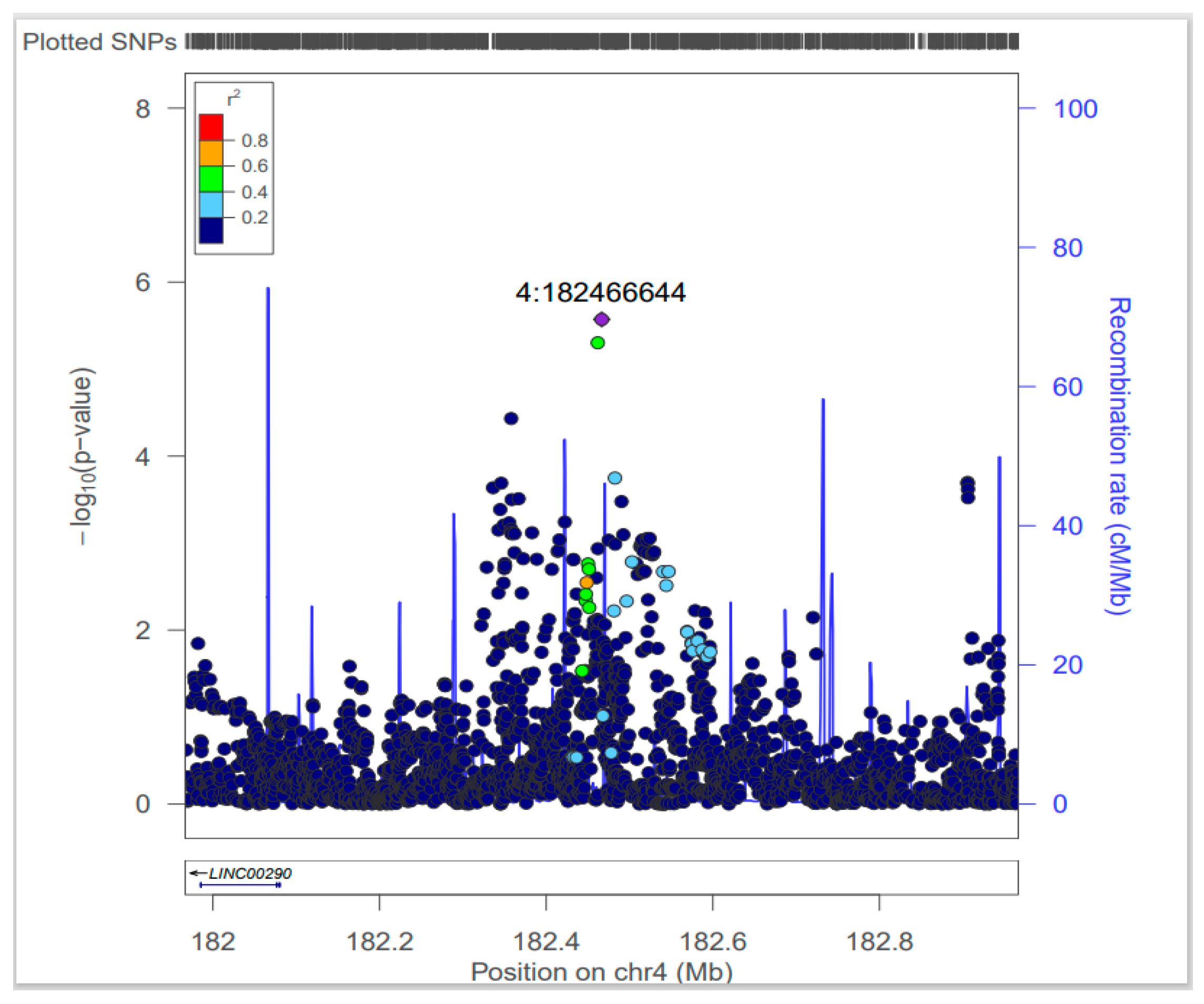

| Lead SNPs | Adjacent Gene | CHR | BP | Minor Allele | Major Allele | MAF | OR (95% CI) | P-Value | Q | I2 | CADD | RDB | eQTL (Tissue) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4861982 | LINC00290 | 4 | 182466644 | T | G | 0.15 | 1.18 (1.10–1.26) | 2.68 × 10−6 | 0.45 | 0 | 0.35 | 6 | LINC00290 (Adrenal gland, Brain) |
| rs3891675 | CTNND2 | 5 | 11537389 | G | A | 0.33 | 1.13 (1.07–1.19) | 8.95 × 10−6 | 0.10 | 38.22 | 1.55 | NA | |
| rs17512228 | PDZD2 | 5 | 32071267 | T | C | 0.07 | 1.25 (1.13–1.37) | 9.23 × 10−6 | 0.41 | 3.01 | 1.12 | 5 | |
| rs2032890 | ERAP1 | 5 | 96121152 | C | A | 0.30 | 0.88 (0.84–0.93) | 9.43 × 10−6 | 0.79 | 0 | 7.63 | 4 | CAST (Adipose_Visceral_Omentum, Brain) ERAP1 (Adipose_Subcutaneous) |
| rs11771792 | AUTS2 | 7 | 68458785 | C | T | 0.17 | 1.17 (1.09–1.25) | 5.72 × 10−6 | 0.65 | 0 | 1.81 | 5 | |
| rs2778038 | WBP1L | 10 | 104515069 | C | A | 0.22 | 1.15 (1.08–1.22) | 7.57 × 10−6 | 0.75 | 0 | 6.52 | 6 | AS3MT (Adipose_Subcutaneous, Brain), WBP1L (Muscle_Skeletal, Brain) SFXN2 (Adipose_Visceral) |
| rs11596125 | MKI67 | 10 | 130587803 | A | G | 0.24 | 1.15 (1.09–1.22) | 1.45 × 10−6 | 0.25 | 20.74 | 8.36 | 5 | |
| rs11606257 | APIP | 11 | 34933982 | C | T | 0.07 | 1.25 (1.14–1.38) | 3.34 × 10−6 | 0.40 | 4.52 | 13.03 | 7 | PDHX (Muscle_Skeletal) |
| rs220850 | CADM1 | 11 | 115248355 | C | T | 0.46 | 0.89 (0.85–0.94) | 7.33 × 10−6 | 0.48 | 0 | 4.66 | NA | CADM1 (Lung) |
| rs1457538 | PTPRO | 12 | 15584196 | G | C | 0.26 | 0.86 (0.81–0.91) | 9.68 × 10−8 | 0.36 | 8.49 | 1.96 | 7 | PTPRO (Adipose tissue) |
| rs915378 | MIR656 | 14 | 101713910 | T | C | 0.37 | 0.89 (0.84–0.94) | 7.92 × 10−6 | 0.18 | 28.3 | 3.14 | 5 |
| PRS of NHS1 | PRS of HPFS | |||||
|---|---|---|---|---|---|---|
| Low (n = 3990) | Intermediate (n = 4022) | High (n = 4086) | Low (n = 2559) | Intermediate (n = 2463) | High (n = 2533) | |
| Age (years) | 57.2 (6.8) | 56.9 (6.8) | 57.2 (6.9) | 57.6 (8.5) | 57.3 (8.7) | 57.8 (8.9) |
| Caucasian (%) | 99.7 | 99.8 | 99.7 | 94.6 | 94.9 | 95.6 |
| BMI (kg/m2) | 23.6 (2.7) | 23.7 (2.7) | 23.6 (2.6) | 24.9 (2.3) | 24.8 (2.2) | 24.9 (2.2) |
| Weight (kg) | 63.7 (8.5) | 63.8 (8.4) | 63.6 (8.4) | 79.5 (8.9) | 79.3 (9.0) | 79.5 (9.0) |
| Never smokers (%) | 40.9 | 45.2 | 48.2 | 50.8 | 49.5 | 49.8 |
| Past smokers (%) | 39.5 | 37.1 | 35.4 | 42.2 | 42.4 | 42.3 |
| Current smokers (%) | 19.3 | 17.5 | 16.2 | 7.1 | 8.1 | 7.9 |
| Alcohol intake (g/day) | 8.4 (11.6) | 7.3 (10.8) | 6.6 (9.8) | 12.2 (15.5) | 12.8 (16.1) | 11.9 (15.2) |
| Physical activity (MET-h/week) | 15.7 (21.6) | 15.2 (18.7) | 14.7 (20.8) | 21.8 (25.8) | 20.6 (23.0) | 20.4 (23.7) |
| Total energy intake (kcal/d) | 1673.2 (466.1) | 1754.4 (467.7) | 1865.5 (487.4) | 2002.4 (581.9) | 2004.0 (594.7) | 2058.0 (632.8) |
| AHEI | 47.4 (10.2) | 46.0 (9.9) | 44.9 (9.7) | 46.9 (10.9) | 47.1 (11.1) | 46.6 (10.7) |
| Glycemic load | 96.9 (17.9) | 99.0 (17.6) | 100.1 (16.2) | 124.0 (25.3) | 123.9 (25.2) | 124.5 (25.1) |
| Trans fat | 1.8 (0.6) | 1.9 (0.6) | 2.0 (0.6) | 2.8 (1.1) | 2.8 (1.1) | 2.9 (1.1) |
| Total fiber | 17.6 (5.0) | 17.4 (4.8) | 17.1 (4.5) | 21.2 (6.9) | 21.2 (7) | 20.7 (6.5) |
| Fruit | 73.2 (44.8) | 74.2 (44.3) | 77.7 (45.5) | 61.7 (147.1) | 67.4 (160.2) | 78.0 (187.1) |
| Vegetable | 87.3 (49.5) | 85.0 (45.5) | 85.4 (44.2) | 80.3 (214.2) | 101.9 (280.9) | 119.8 (315.1) |
| Coffee | 17.0 (30.6) | 17.4 (29.3) | 18.3 (30.1) | 10.3 (36.1) | 9.9 (35.6) | 12.2 (40.3) |
| Tea | 6.5 (6.2) | 6.1 (5.3) | 6.3 (5.4) | 7.3 (29.5) | 9.7 (35.7) | 11.5 (39.4) |
| Sweetened beverage | 36.5 (39.8) | 38.1 (42) | 38.4 (40.7) | 12.8 (24.9) | 13.9 (28.8) | 15.1 (31.9) |
| Chocolate | 1.2 (3.5) | 1.5 (3.8) | 1.8 (4.3) | 1.8 (5) | 2.6 (10.8) | 4.9 (18.1) |
| Ice cream | 1.4 (2.6) | 1.5 (2.6) | 1.7 (2.8) | 7.5 (29.9) | 7.2 (29) | 9.1 (33.2) |
| Cake | 1.6 (2.1) | 2.0 (3.2) | 2.5 (3.4) | 1.6 (15.3) | 4.1 (27.6) | 13.6 (53.3) |
| Sleep (h/day) | 7.0 (0.9) | 7.1 (0.9) | 7.0 (0.9) | 7.1 (0.8) | 7.1 (0.8) | 7.1 (0.8) |
| PRS | 631.3 (8.0) | 647.9 (3.8) | 664.6 (8.1) | 631.3 (7.5) | 647.7 (3.8) | 665.2 (9.0) |
| Cases/person years | 729/76,338 | 749/78,784 | 693/80,321 | 330/47,722 | 331/46,575 | 312/47,416 |
| Crude incidence/100 K PY | 955 | 951 | 863 | 692 | 711 | 658 |
| Model 1 (Age-Adjusted Model) 2 | Model 2 (Multivariate-Adjusted Model) 3 | |||||
|---|---|---|---|---|---|---|
| Independent Variable | HR | 95% CI | p-Value | HR | 95% CI | p-Value |
| rs4861982 (TG vs. GG) | ||||||
| NHS1 4 | 1.037 | 0.943–1.142 | 0.453 | 1.033 | 0.938–1.137 | 0.511 |
| HPFS 5 | 1.010 | 0.873–1.168 | 0.897 | 0.992 | 0.858–1.148 | 0.914 |
| Pooled 6 | 1.029 | 0.950–1.115 | 0.485 | 1.020 | 0.942–1.106 | 0.623 |
| rs4861982 (TT vs. GG) | ||||||
| NHS1 4 | 1.103 | 0.857–1.419 | 0.448 | 1.111 | 0.863–1.430 | 0.414 |
| HPFS 5 | 1.596 | 1.145–2.225 | 0.006 | 1.565 | 1.122–2.184 | 0.008 |
| Pooled 6 | 1.262 | 1.032–1.543 | 0.023 | 1.259 | 1.030–1.540 | 0.025 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, J.H.; Kang, H. Identification of Sweetness Preference-Related Single-Nucleotide Polymorphisms for Polygenic Risk Scores Associated with Obesity. Nutrients 2024, 16, 2972. https://doi.org/10.3390/nu16172972
Bae JH, Kang H. Identification of Sweetness Preference-Related Single-Nucleotide Polymorphisms for Polygenic Risk Scores Associated with Obesity. Nutrients. 2024; 16(17):2972. https://doi.org/10.3390/nu16172972
Chicago/Turabian StyleBae, Ji Hyun, and Hyunju Kang. 2024. "Identification of Sweetness Preference-Related Single-Nucleotide Polymorphisms for Polygenic Risk Scores Associated with Obesity" Nutrients 16, no. 17: 2972. https://doi.org/10.3390/nu16172972
APA StyleBae, J. H., & Kang, H. (2024). Identification of Sweetness Preference-Related Single-Nucleotide Polymorphisms for Polygenic Risk Scores Associated with Obesity. Nutrients, 16(17), 2972. https://doi.org/10.3390/nu16172972






