Blood Glucose Prediction from Nutrition Analytics in Type 1 Diabetes: A Review
Abstract
1. Introduction
2. Related Literature
2.1. Physiological Blood Glucose Prediction Models
2.2. Data-Driven Blood Glucose Prediction Models
2.3. Hybrid Blood Glucose Prediction Models
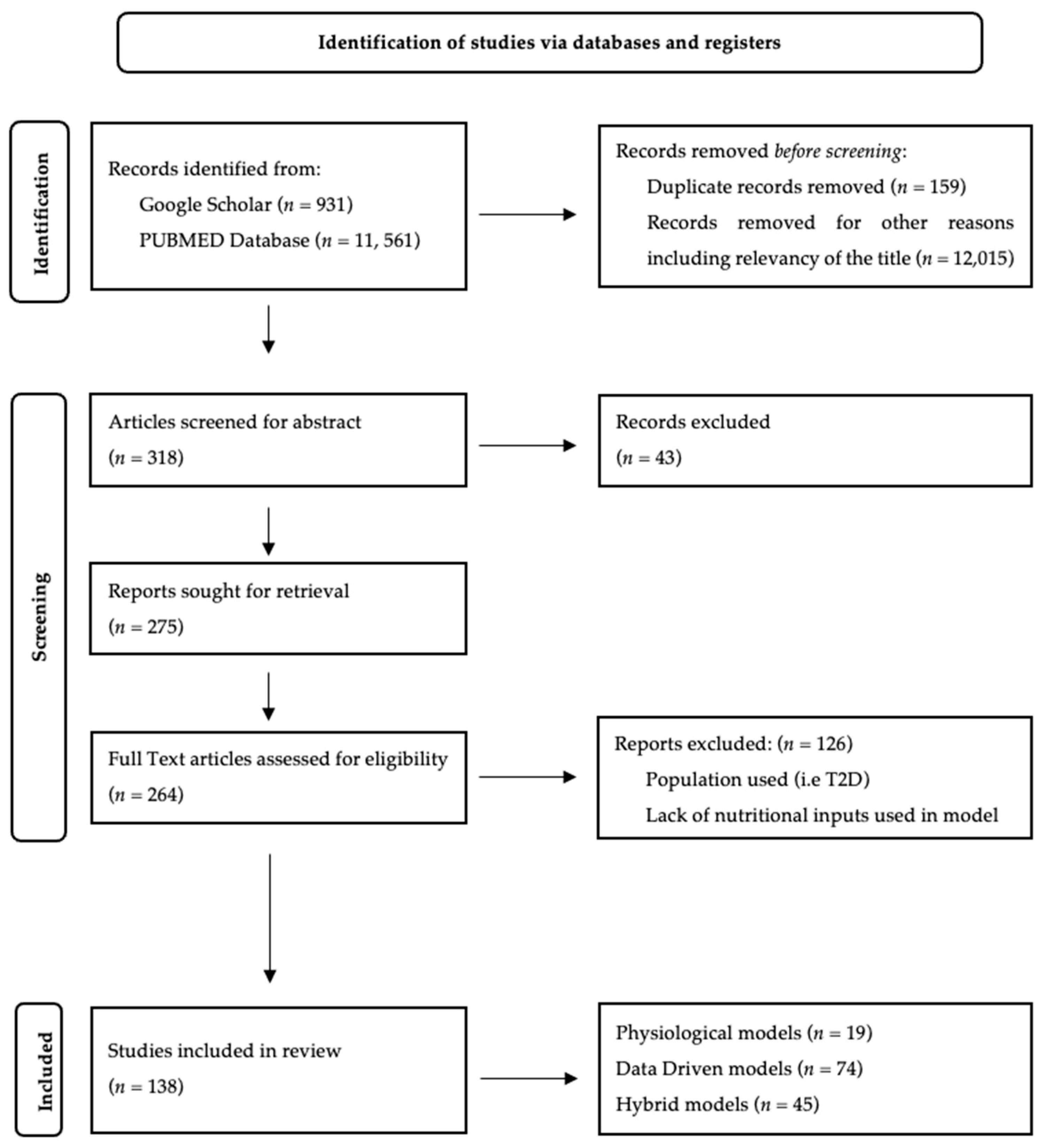
3. Methodology
3.1. Data Sources and Search Strategy
3.2. Eligibility Criteria
- Publications written in English;
- Publications peer-reviewed and published by 1 May 2024;
- Publications focused on a Type 1 Diabetes population;
- Focused on blood glucose prediction algorithms without hyper/hypo-glycemia prevention;
- Prediction algorithms, including carbohydrate content or meal inputs;
- Conference papers or academic journal articles.
3.3. Study Selection
- Author: This includes the authors listed on the publication, the reference number, and the year of publication.
- Model Type: The core model used for the prediction is outlined; this includes previously published physiological models and data-driven prediction techniques.
- Additional Aspects: Any additional techniques used to supplement the core model used.
- Sub-Systems: This feature in the physiological and hybrid model summary tables highlights any absorption profiles included in the model and details the compartment models presented.
- Prediction Horizon: Highlights the time frame used in the prediction model.
- Patients: Here, the population included is outlined. The population is classified as “real” (where the model is validated using real-world data either in an in-patient setting or T1DM outpatients and/or their data) or simulated (where simulated patient data were used for validation).
- Inputs Used: Here, the checkmark indicates the use of GCM or BG data from self-monitoring blood glucose (SMBG) methods, insulin administration, carbohydrate intake, and/or mixed meals (carbohydrate/protein/fat/fiber).
- Additional Inputs: Any other included factors are presented here, such as physical activity, insulin-to-carbohydrate ratios, and sleep.
- Accuracy: Where reported, the models’ performance is indicated, and the metrics used are included along with the measurement used.
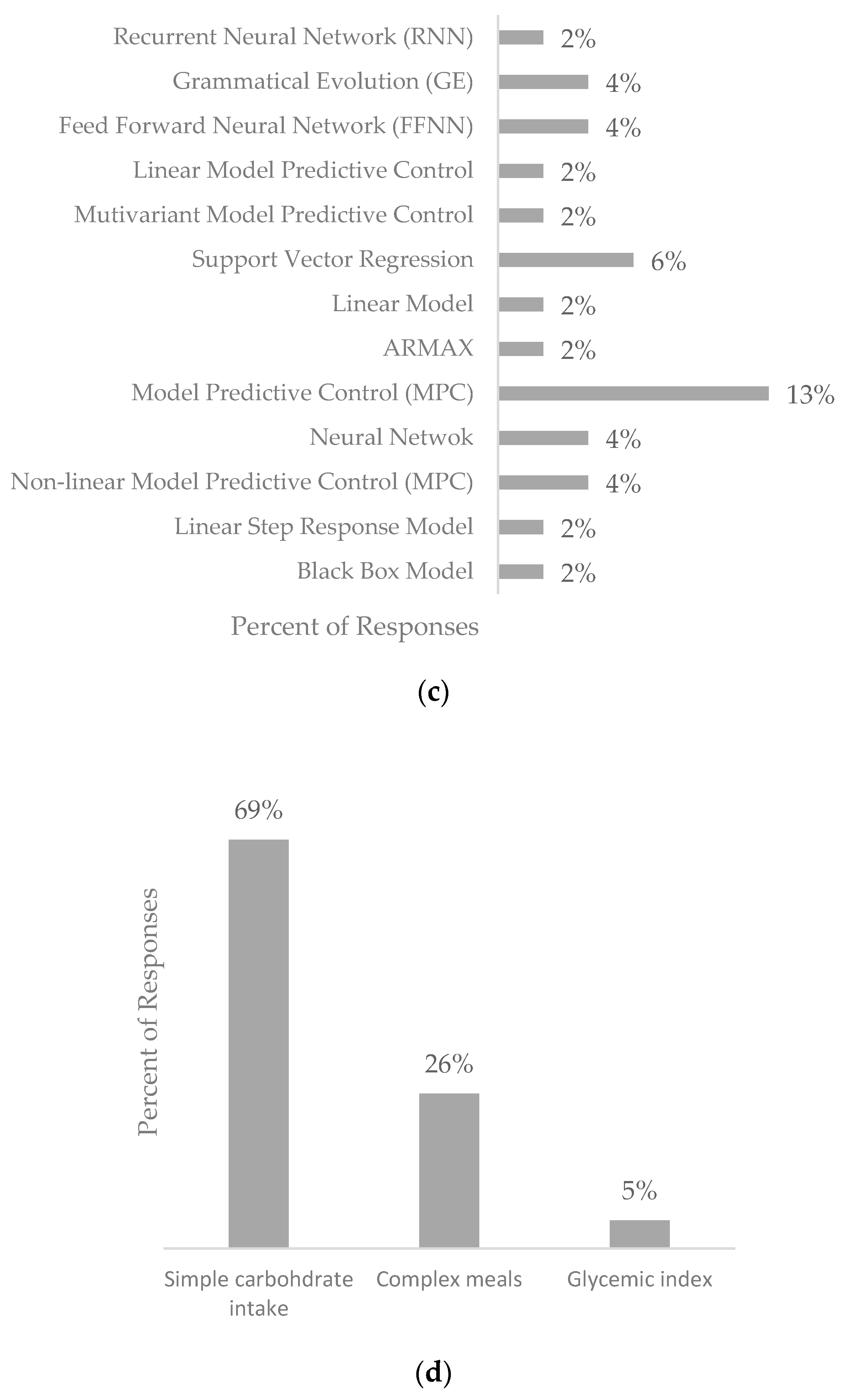
4. Results
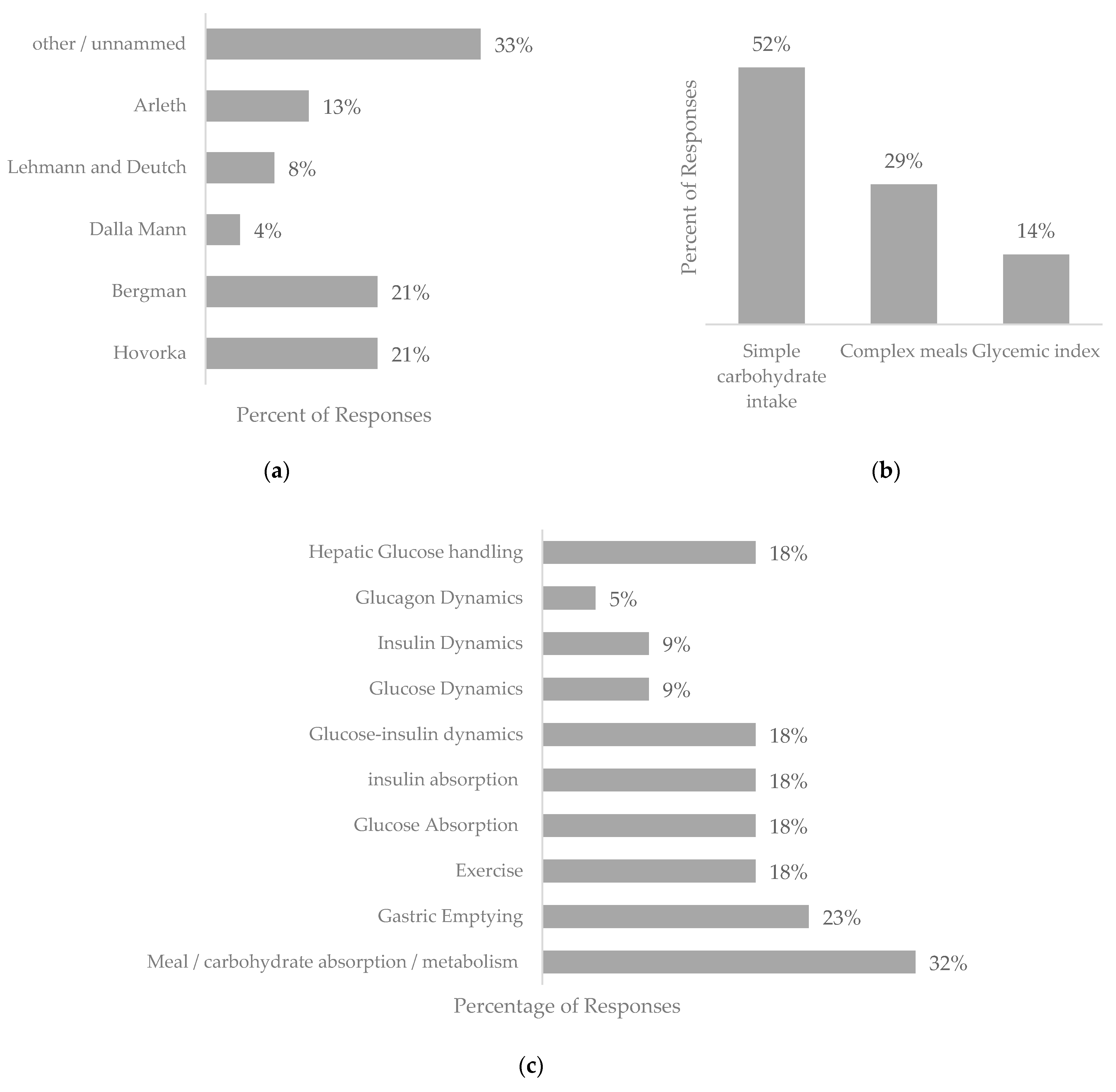
4.1. Physiological Blood Glucose Prediction Models
4.2. Data-Driven Blood Glucose Prediction Models
4.3. Hybrid Prediction Models
5. Discussion
5.1. Validity of Different Model Types
5.2. Prediction Horizons
5.3. Impact of Nutritional Intake on Prediction Models
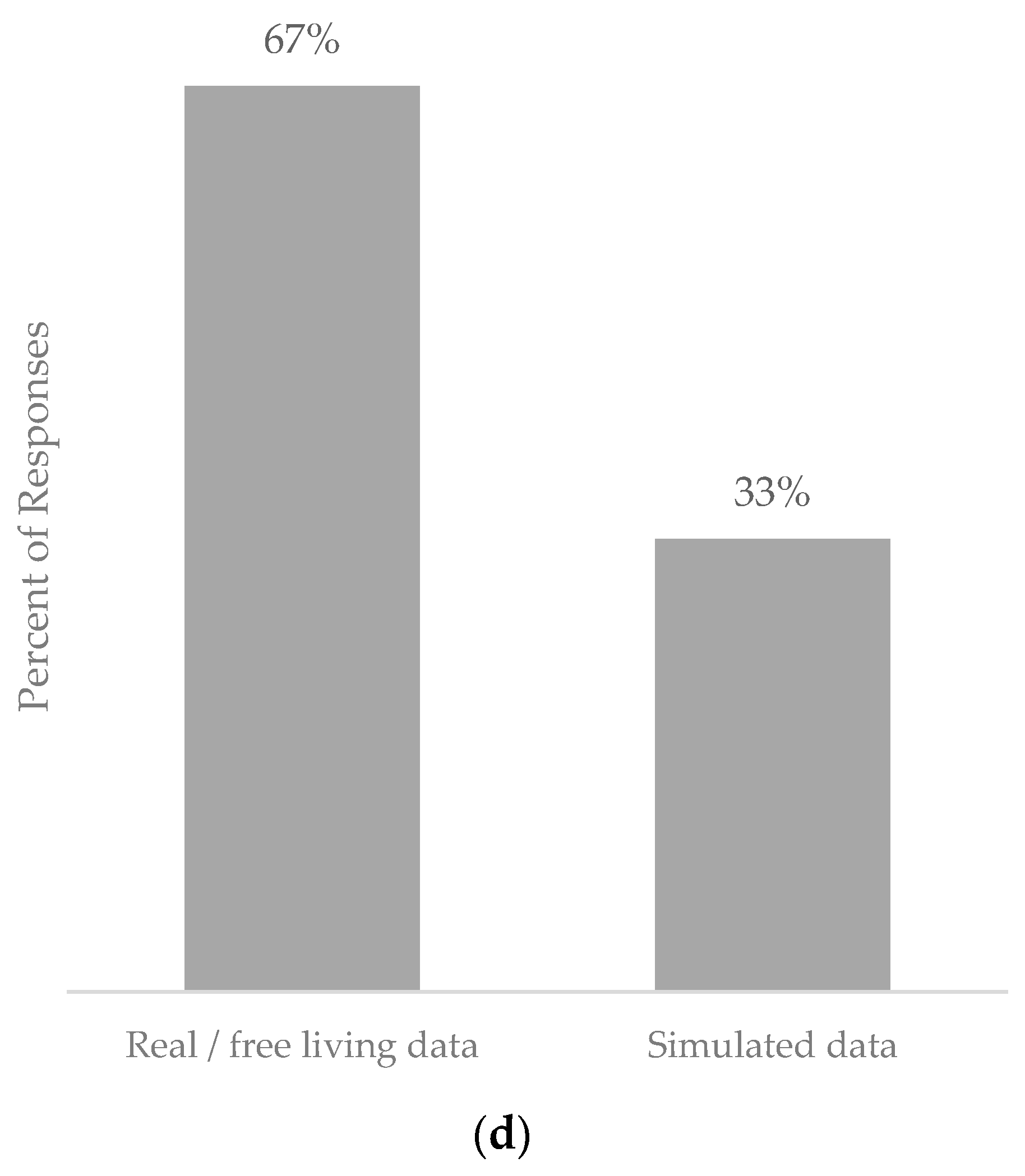
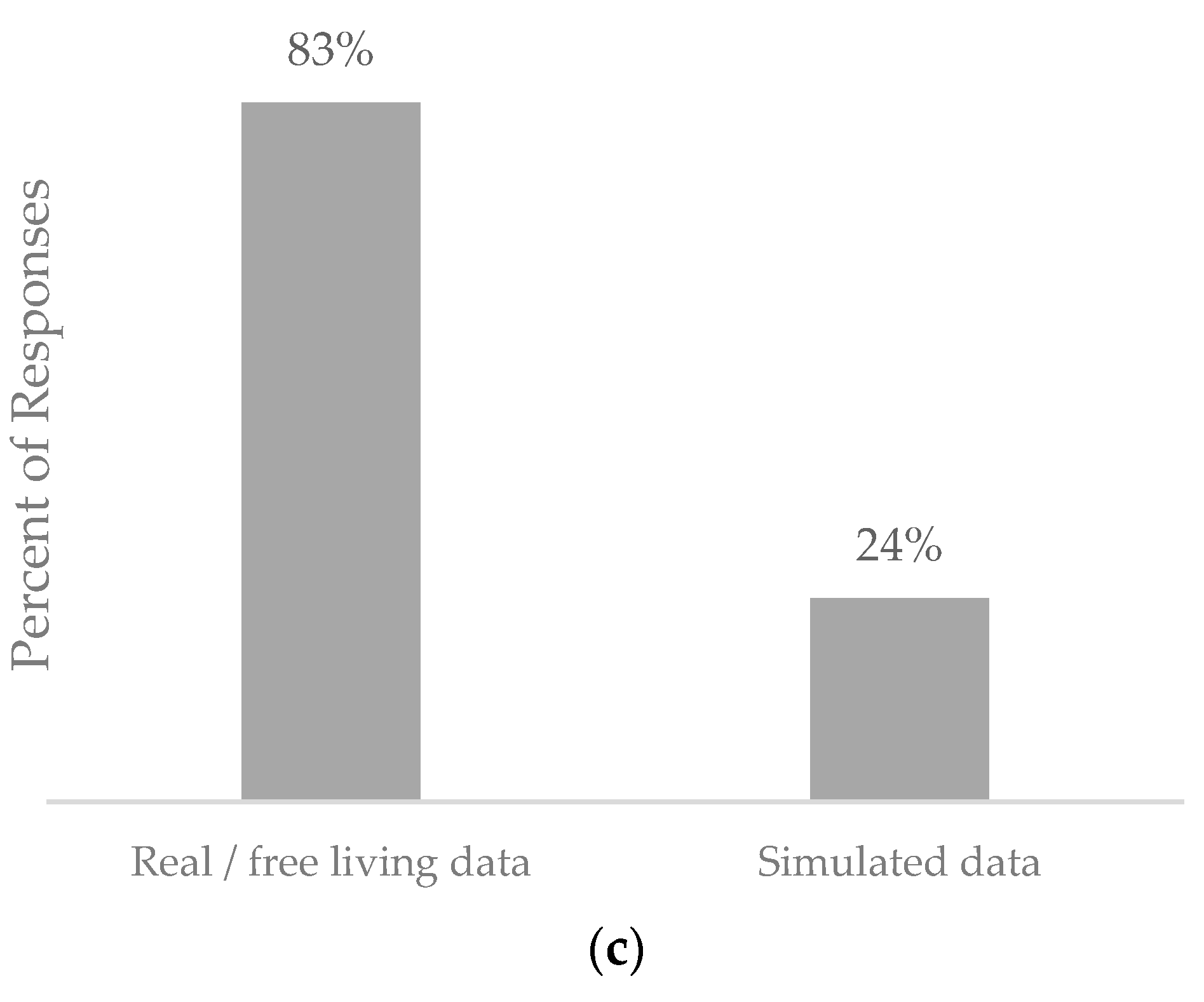
5.4. Population Used
6. Limitations and Future Work
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- JDRF: Type 1 Diabetes Facts and Figures. Available online: https://jdrf.org.uk/information-support/about-type-1-diabetes/facts-and-figures/ (accessed on 6 October 2022).
- Atkinson, M.A.; Eisenbarth, G.S.; Michels, A.W. Type 1 Diabetes. Lancet 2014, 383, 69–82. [Google Scholar] [CrossRef]
- National Institute for Health and Care Excellence. Diabetes (Type 1 and Type 2) in Children and Young People: Diagnosis and Management; National Institute for Health and Care Excellence: London, UK, 2015; p. 55. [Google Scholar]
- Jenkins, D.; Wolever, T.; Taylor, R.; Barker, H.; Fielden, H.; Baldwin, J.; Bowling, A.; Newman, H.; Jenkins, A.; Goff, D. Glycemic Index of Foods: A Physiological Basis for Carbohydrate Exchange. Am. J. Clin. Nutr. 1981, 34, 362–366. [Google Scholar] [CrossRef]
- Gabbay, M.A.L.; Rodacki, M.; Calliari, L.E.; Vianna, A.G.D.; Krakauer, M.; Pinto, M.S.; Reis, J.S.; Puñales, M.; Miranda, L.G.; Ramalho, A.C.; et al. Time in Range: A New Parameter to Evaluate Blood Glucose Control in Patients with Diabetes. Diabetol. Metab. Syndr. 2020, 12, 22. [Google Scholar] [CrossRef]
- Battelino, T.; Danne, T.; Bergenstal, R.M.; Amiel, S.A.; Beck, R.; Biester, T.; Bosi, E.; Buckingham, B.A.; Cefalu, W.T.; Close, K.L.; et al. Clinical Targets for Continuous Glucose Monitoring Data Interpretation: Recommendations from the International Consensus on Time in Range. Diabetes Care 2019, 42, 1593–1603. [Google Scholar] [CrossRef] [PubMed]
- Karim, R.A.H.; Vassányi, I.; Kósa, I. After-Meal Blood Glucose Level Prediction Using an Absorption Model for Neural Network Training. Comput. Biol. Med. 2020, 125, 103956. [Google Scholar] [CrossRef] [PubMed]
- Saiti, K.; Macaš, M.; Lhotská, L.; Štechová, K.; Pithová, P. Ensemble Methods in Combination with Compartment Models for Blood Glucose Level Prediction in Type 1 Diabetes Mellitus. Comput. Methods Programs Biomed. 2020, 196, 105628. [Google Scholar] [CrossRef] [PubMed]
- Oviedo, S.; Vehí, J.; Calm, R.; Armengol, J. A Review of Personalized Blood Glucose Prediction Strategies for T1DM Patients: Personalized Blood Glucose Prediction Strategies for T1D Patients. Int. J. Numer. Meth. Biomed. Engng. 2017, 33, e2833. [Google Scholar] [CrossRef] [PubMed]
- Karim, R.A.H.; Vassányi, I.; Kósa, I. Improved Methods for Mid-Term Blood Glucose Level Prediction Using Dietary and Insulin Logs. Medicina 2021, 57, 676. [Google Scholar] [CrossRef] [PubMed]
- Moreira, T.R.; Negreiros, F.D.D.S.; Aquino, M.D.J.N.D.; Silva, L.M.S.D.; Moreira, T.M.M.; Torres, R.A.M. Digital Technology and Its Effects on Knowledge Improvement for Diabetes Management: An Integrative Review. Int. J. Nurs. Pract. 2023, 29, e13029. [Google Scholar] [CrossRef] [PubMed]
- Annuzzi, G.; Apicella, A.; Arpaia, P.; Bozzetto, L.; Criscuolo, S.; Benedetto, E.D.; Pesola, M.; Prevete, R.; Vallefuoco, E. Impact of Nutritional Factors in Blood Glucose Prediction in Type 1 Diabetes through Machine Learning. IEEE Access 2023, 11, 17104–17115. [Google Scholar] [CrossRef]
- Butt, H.; Khosa, I.; Iftikhar, M.A. Feature Transformation for Efficient Blood Glucose Prediction in Type 1 Diabetes Mellitus Patients. Diagnostics 2023, 13, 340. [Google Scholar] [CrossRef] [PubMed]
- Woldaregay, A.Z.; Årsand, E.; Walderhaug, S.; Albers, D.; Mamykina, L.; Botsis, T.; Hartvigsen, G. Data-Driven Modeling and Prediction of Blood Glucose Dynamics: Machine Learning Applications in Type 1 Diabetes. Artif. Intell. Med. 2019, 98, 109–134. [Google Scholar] [CrossRef] [PubMed]
- Bremer, T.; Gough, D.A. Is Blood Glucose Predictable from Previous Values? A Solicitation for Data. Diabetes 1999, 48, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Organero, M. Deep Physiological Model for Blood Glucose Prediction in T1DM Patients. Sensors 2020, 20, 3896. [Google Scholar] [CrossRef] [PubMed]
- Borle, N.C.; Ryan, E.A.; Greiner, R. The Challenge of Predicting Blood Glucose Concentration Changes in Patients with Type I Diabetes. Health Inform. J. 2021, 27, 146045822097758. [Google Scholar] [CrossRef]
- Georga, E.I.; Protopappas, V.C.; Polyzos, D.; Fotiadis, D.I. Online prediction of glucose concentration in type 1 diabetes using extreme learning machines. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2015, 2015, 3262–3265. [Google Scholar] [CrossRef] [PubMed]
- Calm, R.; García-Jaramillo, M.; Bondia, J.; Sainz, M.A.; Vehí, J. Comparison of Interval and Monte Carlo Simulation for the Prediction of Postprandial Glucose under Uncertainty in Type 1 Diabetes Mellitus. Comput. Methods Programs Biomed. 2011, 104, 325–332. [Google Scholar] [CrossRef]
- Brazeau, A.S.; Mircescu, H.; Desjardins, K.; Leroux, C.; Strychar, I.; Ekoé, J.M.; Rabasa-Lhoret, R. Carbohydrate Counting Accuracy and Blood Glucose Variability in Adults with Type 1 Diabetes. Diabetes Res. Clin. Pract. 2013, 99, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Aliberti, A.; Pupillo, I.; Terna, S.; Macii, E.; Di Cataldo, S.; Patti, E.; Acquaviva, A. A Multi-Patient Data-Driven Approach to Blood Glucose Prediction. IEEE Access 2019, 7, 69311–69325. [Google Scholar] [CrossRef]
- Berry, S.E.; Valdes, A.M.; Drew, D.A.; Asnicar, F.; Mazidi, M.; Wolf, J.; Capdevila, J.; Hadjigeorgiou, G.; Davies, R.; Al Khatib, H.; et al. Human Postprandial Responses to Food and Potential for Precision Nutrition. Nat. Med. 2020, 26, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef]
- Georga, E.I.; Protopappas, V.C.; Ardigo, D.; Marina, M.; Zavaroni, I.; Polyzos, D.; Fotiadis, D.I. Multivariate Prediction of Subcutaneous Glucose Concentration in Type 1 Diabetes Patients Based on Support Vector Regression. IEEE J. Biomed. Health Inform. 2013, 17, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, N.P.; Samavedham, L.; Rangaiah, G.P. Personalized Mechanistic Models for Exercise, Meal and Insulin Interventions in Children and Adolescents with Type 1 Diabetes. J. Theor. Biol. 2014, 357, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rodríguez, I.; Chatzigiannakis, I.; Rodríguez, J.-V.; Maranghi, M.; Gentili, M.; Zamora-Izquierdo, M.-Á. Utility of Big Data in Predicting Short-Term Blood Glucose Levels in Type 1 Diabetes Mellitus through Machine Learning Techniques. Sensors 2019, 19, 4482. [Google Scholar] [CrossRef] [PubMed]
- Bunescu, R.; Struble, N.; Marling, C.; Shubrook, J.; Schwartz, F. Blood Glucose Level Prediction Using Physiological Models and Support Vector Regression. In Proceedings of the 2013 12th International Conference on Machine Learning and Applications, Miami, FL, USA, 4–7 December 2013; pp. 135–140. [Google Scholar]
- Contreras, I.; Oviedo, S.; Vettoretti, M.; Visentin, R.; Vehí, J. Personalized Blood Glucose Prediction: A Hybrid Approach Using Grammatical Evolution and Physiological Models. PLoS ONE 2017, 12, e0187754. [Google Scholar] [CrossRef]
- Felizardo, V.; Garcia, N.M.; Pombo, N.; Megdiche, I. Data-Based Algorithms and Models Using Diabetics Real Data for Blood Glucose and Hypoglycaemia Prediction–A Systematic Literature Review. Artif. Intell. Med. 2021, 118, 102120. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.P.A.; Brandão, A.A.F.; Da Silva Bevilacqua, J.; Correa Giannella, M.L.C. A Hybrid Model to Predict Glucose Oscillation for Patients with Type 1 Diabetes and Suggest Customized Recommendations. In Intelligent Systems and Applications; Bi, Y., Bhatia, R., Kapoor, S., Eds.; Advances in Intelligent Systems and Computing; Springer International Publishing: Cham, Switzerland, 2020; Volume 1038, pp. 790–801. ISBN 978-3-030-29512-7. [Google Scholar]
- Moher, D.; Shamseer, L.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A. Preferred Reporting Items for Systematic Review and Meta-Analysis Protocols (PRISMA-P) 2015 Statement. Syst. Rev. 2015, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Google Scholar. Available online: https://scholar.google.com/#d=gs_hdr_drw&t=1714405788098 (accessed on 1 June 2022).
- PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov (accessed on 1 July 2022).
- Mendeley Reference Management Software 2024. Available online: www.mendeley.com (accessed on 1 June 2023).
- Asad, M.; Khan, Y.; Qamar, U.; Bashir, S. Blood Glucose Level Prediction Using Optimized Neural Network for Virtual Patients. In Intelligent Systems and Applications; Bi, Y., Bhatia, R., Kapoor, S., Eds.; Advances in Intelligent Systems and Computing; Springer International Publishing: Cham, Switzerland, 2020; Volume 1038, pp. 671–683. ISBN 978-3-030-29512-7. [Google Scholar]
- Willmott, C.; Matsuura, K. Advantages of the Mean Absolute Error (MAE) over the Root Mean Square Error (RMSE) in Assessing Average Model Performance. Clim. Res. 2005, 30, 79–82. [Google Scholar] [CrossRef]
- Mondal, H.; Mondal, S. Clarke Error Grid Analysis on Graph Paper and Microsoft Excel. J. Diabetes Sci. Technol. 2020, 14, 499. [Google Scholar] [CrossRef] [PubMed]
- Pustozerov, E.A.; Tkachuk, A.S.; Vasukova, E.A.; Anopova, A.D.; Kokina, M.A.; Gorelova, I.V.; Pervunina, T.M.; Grineva, E.N.; Popova, P.V. Machine Learning Approach for Postprandial Blood Glucose Prediction in Gestational Diabetes Mellitus. IEEE Access 2020, 8, 219308–219321. [Google Scholar] [CrossRef]
- Zecchin, C.; Facchinetti, A.; Sparacino, G.; Cobelli, C. Jump Neural Network for Online Short-Time Prediction of Blood Glucose from Continuous Monitoring Sensors and Meal Information. Comput. Methods Programs Biomed. 2014, 113, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y. Blood Glucose Prediction Using Support Vector Regression with Particle Swarm Optimization. J. Healthc. Eng. 2018, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, Y.; Zhang, X. Blood Glucose Prediction Based on Support Vector Regression. J. Healthc. Eng. 2019, 1–9. [Google Scholar]
- Martínez-Delgado, L.; Munoz-Organero, M.; Queipo-Alvarez, P. Using Absorption Models for Insulin and Carbohydrates and Deep Leaning to Improve Glucose Level Predictions. Sensors 2021, 21, 5273. [Google Scholar] [CrossRef] [PubMed]
- Hejlesen, O.K.; Andreasseen, S.; Hovorka, R.; Cavan, D.A. DIAS-the Diabetes Advisory System: Technical and Physiological Aspects of the System and Evaluation Results Obtained so Far. Comput. Methods Programs Biomed. 1997, 54, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Arleth, T.; Andreassen, S.; Orsini-Federici, M.; Timi, A.; Massi-Benedetti, M. A Model of Glucose Absorption from Mixed Meals. IFAC Proc. Vol. 2000, 33, 307–312. [Google Scholar] [CrossRef]
- O’Keefe, J.H.; Gheewala, N.M.; O’Keefe, J.O. Dietary Strategies for Improving Post-Prandial Glucose, Lipids, Inflammation, and Cardiovascular Health. J. Am. Coll. Cardiol. 2008, 51, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Elleri, D.; Allen, J.M.; Harris, J.; Kumareswaran, K.; Nodale, M.; Leelarathna, L.; Acerini, C.L.; Haidar, A.; Wilinska, M.E.; Jackson, N.; et al. Absorption Patterns of Meals Containing Complex Carbohydrates in Type 1 Diabetes. Diabetologia 2013, 56, 1108–1117. [Google Scholar] [CrossRef]
- Cunningham, K.M.; Read, N.W. The Effect of Incorporating Fat into Different Components of a Meal on Gastric Emptying and Postprandial Blood Glucose and Insulin Responses. Br. J. Nutr. 1989, 61, 285–290. [Google Scholar] [CrossRef]
- Lubasinski, N.; Thabit, H.; Nutter, P.W.; Harper, S. What Is the Tech Missing? Nutrition Reporting in Type 1 Diabetes. Nutrients 2024, 16, 1690. [Google Scholar] [CrossRef]
- Campos-Náñez, E.; Layne, J.E.; Zisser, H.C. In Silico Modeling of Minimal Effective Insulin Doses Using the UVA/PADOVA Type 1 Diabetes Simulator. J. Diabetes Sci. Technol. 2018, 12, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Bergman, R.N.; Ider, Y.Z.; Bowden, C.R.; Cobelli, C. Quantitative estimation of insulin sensitivity. Am. J. Physiol.-Endocrinol. Metab. 1979, 236, E667. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, E.D.; Deutsch, T. A physiological model of glucose-insulin interaction in type 1 diabetes mellitus. J. Biomed. Eng. 1992, 14, 235–242. [Google Scholar] [CrossRef]
- Andreassena, S.; Hejlesena, O.K.; Hovorkab, R.; Cavan, D.A. The Diabetes Advisory System-an IT approach to the management of insulin dependent diabetes. In Medical Informatics Europe’96; IOS Press: Amsterdam, The Netherlands, 1996. [Google Scholar]
- Höfig, B.; Kistner, A.; Seibold, A.; Böhm, B. Extended physiological models for the simulation of the glucose metabolism. Math. Model. Syst. 1996, 2, 41–54. [Google Scholar] [CrossRef]
- Dalla Man, C.; Raimondo, D.M.; Rizza, R.A.; Cobelli, C. GIM, Simulation Software of Meal Glucose—Insulin Model. J. Diabetes Sci Technol 2007, 1, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Leech, J.; Lin, C.C.Y.; Chrostowski, L. Peak Blood Glucose Prediction Algorithm Following a Meal Intake. In 2007 Canadian Conference on Electrical and Computer Engineering; IEEE: Vancouver, BC, Canada, 2007; pp. 579–582. [Google Scholar] [CrossRef]
- Bock, A.; François, G.; Gillet, D. A therapy parameter-based model for predicting blood glucose concentrations in patients with type 1 diabetes. Comput. Methods Programs Biomed. 2015, 118, 107–123. [Google Scholar] [CrossRef]
- Magdelaine, N.; Chaillous, L.; Guilhem, I.; Poirier, J.Y.; Krempf, M.; Moog, C.; Carpentier, E.L. A Long-Term Model of the Glucose–Insulin Dynamics of Type 1 Diabetes. IEEE Trans. Biomed. Eng. 2015, 62, 1546–1552. [Google Scholar] [CrossRef]
- Hovorka, R.; Canonico, V.; Chassin, L.J.; Haueter, U.; Massi-Benedetti, M.; Federici, M.O.; Pieber, T.R.; Schaller, H.C.; Schaupp, L.; Vering, T.; et al. Nonlinear model predictive control of glucose concentration in subjects with type 1 diabetes. Physiol. Meas. 2004, 25, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Vehi, J.; Oliver, N.; Georgiou, P.; Herrero, P. Enhancing Blood Glucose Prediction with Meal Absorption and Physical Exercise Information. arXiv 2018, arXiv:1901.07467. [Google Scholar]
- Rozendaal, Y.J.; Maas, A.H.; van Pul, C.; Cottaar, E.J.; Haak, H.R.; Hilbers, P.A.; van Reil, N.A. Model-based analysis of postprandial glycemic response dynamics for different types of food. Clin. Nutr. Exp. 2018, 19, 32–45. [Google Scholar] [CrossRef]
- Gyuk, P.; Vassányi, I.; Kósa, I. Blood Glucose Level Prediction for Diabetics Based on Nutrition and Insulin Administration Logs Using Personalized Mathematical Models. J. Healthc. Eng. 2019, 2019, 8605206. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, G.; Yu, L.; Xu, B.; Jin, H. Improved Generalized Predictive Control Algorithm for Blood Glucose Control of Type 1 Diabetes. Artif. Organs 2019, 43, 386–398. [Google Scholar] [CrossRef]
- Tresp, V.; Briegel, T. A Solution for Missing Data in Recurrent Neural Networks with an Application to Blood Glucose Prediction. Available online: https://proceedings.neurips.cc/paper_files/paper/1997/file/c73dfe6c630edb4c1692db67c510f65c-Paper.pdf (accessed on 8 June 2023).
- Sandham, W.; Nikoletou, D.; Hamilton, D.J.; Paterson, K.; Japp, A.; Macgregor, C. Blood glucose prediction for diabetes therapy using a recurrent artificial neural network. In Proceedings of the 9th European Signal Processing Conference (EUSIPCO 1998), Rhodes, Greece, 8–11 September 1998. [Google Scholar]
- Ståhl, F.; Johansson, R.; Renard, E. Post-prandial plasma glucose prediction in type I diabetes based on Impulse Response Models. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 1324–1327. [Google Scholar] [CrossRef]
- Cescon, M.; Renard, E. Adaptive subspace-based prediction of T1DM glycemia. In Proceedings of the IEEE Conference on Decision and Control and European Control Conference, Orlando, FL, USA, 12–15 December 2011; pp. 5164–5169. [Google Scholar] [CrossRef]
- Pappada, S.M.; Cameron, B.D.; Rosman, P.M.; Bourey, R.E.; Papadimos, T.J.; Olorunto, W.; Borst, M.J. Neural Network-Based Real-Time Prediction of Glucose in Patients with Insulin-Dependent Diabetes. Diabetes Technol. Ther. 2011, 13, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Pappada, S.M.; Cameron, B.D.; Rosman, P.M. Development of a Neural Network for Prediction of Glucose Concentration in Type 1 Diabetes Patients. J. Diabetes Sci. Technol. 2008, 2, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Robertson, G.; Lehmann, E.D.; Sandham, W.; Hamilton, D. Blood Glucose Prediction Using Artificial Neural Networks Trained with the AIDA Diabetes Simulator: A Proof-of-Concept Pilot Study. J. Electr. Comput. Eng. 2011, 2011, 681786. [Google Scholar] [CrossRef]
- Daskalaki, E.; Prountzou, A.; Diem, P.; Mougiakakou, S.G. Real-Time Adaptive Models for the Personalized Prediction of Glycemic Profile in Type 1 Diabetes Patients. Diabetes Technol. Ther. 2012, 14, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Pappada, S.M.; Cameron, B.D. Neural Network Modeling Approaches for Patient Specific Glycemic Forecasting. In Patient-Specific Modeling in Tomorrow’s Medicine; Gefen, A., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 9, pp. 505–529. [Google Scholar]
- Van Heusden, K.; Dassau, E.; Zisser, H.C.; Seborg, D.E.; Doyle, F.J. Control-Relevant Models for Glucose Control Using a Priori Patient Characteristics. IEEE Trans. Biomed. Eng. 2012, 59, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Steil, G.M. Algorithms for a Closed-Loop Artificial Pancreas: The Case for Proportional-Integral-Derivative Control. J. Diabetes Sci. Technol. 2013, 7, 1621–1631. [Google Scholar] [CrossRef]
- Toffanin, C.; Messori, M.; Di Palma, F.; De Nicolao, G.; Cobelli, C.; Magni, L. Artificial Pancreas: Model Predictive Control Design from Clinical Experience. J. Diabetes Sci. Technol. 2013, 7, 1470–1483. [Google Scholar] [CrossRef] [PubMed]
- Cameron, F.; Niemeyer, G.; Wilson, D.M.; Bequette, B.W.; Benassi, K.S.; Clinton, P.; Buckingham, B.A. Inpatient Trial of an Artificial Pancreas Based on Multiple Model Probabilistic Predictive Control with Repeated Large Unannounced Meals. Diabetes Technol. Ther. 2014, 16, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Del Favero, S.; Bruttomesso, D.; Di Palma, F.; Lanzola, G.; Visentin, R.; Filippi, A.; Scotton, R.; Toffanin, C.; Messori, M.; Scarpellini, S.; et al. First Use of Model Predictive Control in Outpatient Wearable Artificial Pancreas. Diabetes Care 2014, 37, 1212–1215. [Google Scholar] [CrossRef] [PubMed]
- Efendic, H.; Kirchsteiger, H.; Freckmann, G.; del Re, L. Short-term prediction of blood glucose concentration using interval probabilistic models. In Proceedings of the 22nd Mediterranean Conference on Control and Automation, Palermo, Italy, 16–19 June 2014; pp. 1494–1499. [Google Scholar] [CrossRef]
- Kirchsteiger, H.; Johansson, R.; Renard, E.; Re, L.D. Continuous-time interval model identification of blood glucose dynamics for type 1 diabetes. Int. J. Control 2014, 87, 1454–1466. [Google Scholar] [CrossRef]
- Oviedo, S.; Contreras, I.; Vehí, J.; Visentin, R.; Vettoretti, M. Mid-term blood glucose prediction: A hybrid approach using grammatical evolution and physiological models. Training 2017, 19, 15–62. [Google Scholar]
- Plis, K.; Bunescu, R.; Marling, C.; Shubrook, J.; Schwartz, F. A Machine Learning Approach to Predicting Blood Glucose Levels for Diabetes Management. Available online: https://citeseerx.ist.psu.edu/document?repid=rep1&type=pdf&doi=7e0ad22e8314dbb89695021e505bf3d043ad255a (accessed on 8 June 2023).
- Ståhl, F.; Johansson, R.; Renard, E. Ensemble Glucose Prediction in Insulin-Dependent Diabetes. In Data-Driven Modeling for Diabetes; Marmarelis, V., Mitsis, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 37–71. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, X.; Mo, X. A Novel Adaptive-Weighted-Average Framework for Blood Glucose Prediction. Diabetes Technol. Ther. 2013, 15, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Georga, E.I.; Protopappas, V.C.; Polyzos, D.; Fotiadis, D.I. Evaluation of short-term predictors of glucose concentration in type 1 diabetes combining feature ranking with regression models. Med. Biol. Eng. Comput. 2015, 53, 1305–1318. [Google Scholar] [CrossRef]
- Bazaev, N.A.; Pozhar, K.V. Blood Glucose Prediction for “Artificial Pancreas” System. In Gluconeogenesis; Zhang, W., Ed.; InTech: 2017. Available online: https://www.intechopen.com/chapters/53941 (accessed on 8 June 2023). [CrossRef]
- Hidalgo, J.I.; Colmenar, J.M.; Kronberger, G.; Winkler, S.M.; Garnica, O.; Lanchares, J. Data Based Prediction of Blood Glucose Concentrations Using Evolutionary Methods. J. Med. Syst. 2017, 41, 142. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, M.V.; Mosimann, S.; Bally, L.; Stettler, C.; Mougiakakou, S. Deep prediction model: The case of online adaptive prediction of subcutaneous glucose. In Proceedings of the 2016 13th Symposium on Neural Networks and Applications (NEUREL), Belgrade, Serbia, 22–24 November 2016. [Google Scholar] [CrossRef]
- Zecchin, C.; Facchinetti, A.; Sparacino, G.; Cobelli, C. How Much Is Short-Term Glucose Prediction in Type 1 Diabetes Improved by Adding Insulin Delivery and Meal Content Information to CGM Data? A Proof-of-Concept Study. J. Diabetes Sci. Technol. 2016, 10, 1149–1160. [Google Scholar] [CrossRef] [PubMed]
- Reiter, M.; Reiterer, F.; Del Re, L. A probabilistic framework for blood glucose control in diabetes. In Proceedings of the 2017 American Control Conference (ACC), Seattle, WA, USA, 24–26 May 2017; pp. 1450–1455. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Zeng, F.; Wang, N.; Chen, X.; Zhang, B.; Zhao, D.; Yang, W.; Cobelli, C. “Learning” Can Improve the Blood Glucose Control Performance for Type 1 Diabetes Mellitus. Diabetes Technol. Ther. 2017, 19, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Acedo, L.; Botella, M.; Cortés, J.C.; Hidalgo, J.I.; Maqueda, E.; Villanueva, R.J. Swarm hybrid optimization for a piecewise model fitting applied to a glucose model. J. Syst. Inf. Technol. 2018, 20, 404–416. [Google Scholar] [CrossRef]
- Buckingham, B.A.; Christiansen, M.P.; Forlenza, G.P.; Wadwa, R.P.; Peyser, T.A.; Lee, J.B.; O’Connor, J.; Dassau, E.; Huyett, L.M.; Layne, J.E.; et al. Performance of the Omnipod Personalized Model Predictive Control Algorithm with Meal Bolus Challenges in Adults with Type 1 Diabetes. Diabetes Technol. Ther. 2018, 20, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Vahedi, M.R.; MacBride, K.B.; Wunsik, W.; Kim, Y. Predicting Glucose Levels in Patients with Type1 Diabetes Based on Physiological and Activity Data. In Proceedings of the 8th ACM MobiHoc 2018 Workshop on Pervasive Wireless Healthcare Workshop; ACM: Los Angeles, CA, USA, 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Akbari, M.; Chunara, R. Using Contextual Information to Improve Blood Glucose Prediction. Available online: http://proceedings.mlr.press/v106/akbari19a/akbari19a.pdf (accessed on 8 June 2023).
- Borle, N.C.; Ryan, E.A.; Greiner, R. The Challenge of Predicting Meal-to-meal Blood Glucose Concentrations for Patients with Type I Diabetes. arXiv 2019, arXiv:1903.12347. [Google Scholar]
- Griva, L.O.; Martinez, R.; Basualdo, M.S. Combining short and long-term models for predicting blood glucose concentrations on diabetic patients. In Proceedings of the 2019 XVIII Workshop on Information Processing and Control (RPIC), Salvador, Brazil, 18–20 September 2019; IEEE: Bahía Blanca, Argentina, 2019; pp. 123–128. [Google Scholar] [CrossRef]
- Georga, E.I.; Príncipe, J.C.; Fotiadis, D.I. Short-term prediction of glucose in type 1 diabetes using kernel adaptive filters. Med. Biol. Eng. Comput. 2019, 57, 27–46. [Google Scholar] [CrossRef] [PubMed]
- Litinskaia, E.L.; Rudenko, P.A.; Pozhar, K.V.; Bazaev, N.A. Validation of Short-Term Blood Glucose Prediction Algorithms. Int. J. Pharma Med. Biol. Sci. 2019, 8, 34–39. [Google Scholar] [CrossRef]
- Padmapritha, T. Prediction of Blood Glucose Level by using an LSTM based Recurrent Neural networks. In Proceedings of the 2019 IEEE International Conference on Clean Energy and Energy Efficient Electronics Circuit for Sustainable Development (INCCES), Krishnankoil, India, 18–20 December 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Saiti, K.; Macaš, M.; Štechová, K.; Pit’hová, P.; Lhotská, L. A Combined-Predictor Approach to Glycaemia Prediction for Type 1 Diabetes. In World Congress on Medical Physics and Biomedical Engineering 2018; IFMBE Proceedings; Lhotska, L., Sukupova, L., Lacković, I., Ibbott, G., Eds.; Springer: Singapore, 2019; Volume 68/3. [Google Scholar] [CrossRef]
- Alqudah, A.; Younes, A.; Alqudah, A. Towards Modeling Human Body Responsiveness to Glucose Intake and Insulin Injection based on Artificial Neural Networks. J. Comput. Inf. Technol. 2020, 6. [Google Scholar] [CrossRef]
- Amar, Y.; Shilo, S.; Oron, T.; Amar, E.; Phillip, M.; Segal, E. Clinically Accurate Prediction of Glucose Levels in Patients with Type 1 Diabetes. Diabetes Technol. Ther. 2020, 22, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Balasooriya, K.; Nanayakkara, N.D. Predicting Short-Term Changing Blood Glucose Level of Diabetes Patients using Noninvasive Data. In Proceedings of the 2020 IEEE Region 10 Conference (Tencon), Osaka, Japan, 16–19 November 2020; pp. 31–36. [Google Scholar] [CrossRef]
- Camerlingo, N.; Vettoretti, M.; Del Favero, S.; Facchinetti, A.; Sparacino, G. Mathematical Models of Meal Amount and Timing Variability with Implementation in the Type-1 Diabetes Patient Decision Simulator. J. Diabetes Sci. Technol. 2021, 15, 346–359. [Google Scholar] [CrossRef] [PubMed]
- Contador, S.; Colmenar, J.M.; Garnica, O.; Hidalgo, J.I. Short and Medium Term Blood Glucose Prediction Using Multi-objective Grammatical Evolution. In Applications of Evolutionary Computation; Castillo, P.A., Jiménez Laredo, J.L., Fernández De Vega, F., Eds.; Springer International Publishing: Cham, Switzerland, 2020; Volume 12104, pp. 494–509. [Google Scholar]
- Hidalgo, J.I.; Botella, M.; Velasco, J.M.; Garnica, O.; Cervigón, C.; Martínez, R.; Aramendi, A.; Maqueda, E.; Lanchares, J. Glucose forecasting combining Markov chain based enrichment of data, random grammatical evolution and Bagging. Appl. Soft Comput. 2020, 88, 105923. [Google Scholar] [CrossRef]
- Kriventsov, S.; Lindsey, A.; Hayeri, A. The Diabits App for Smartphone-Assisted Predictive Monitoring of Glycemia in Patients with Diabetes: Retrospective Observational Study. JMIR Diabetes 2020, 5, e18660. [Google Scholar] [CrossRef]
- Pavan, J.; Prendin, F.; Meneghetti, L.; Cappon, G.; Sparaconi, G.; Facchinetti, A.; Del Favero, S. Personalized Machine Learning Algorithm Based on Shallow Network and Error Imputation Module for an Improved Blood Glucose Prediction. Available online: https://ceur-ws.org/Vol-2675/paper16.pdf (accessed on 8 June 2023).
- Song, L.; Liu, C.; Yang, W.; Zhang, J.; Kong, X.; Zhang, B.; Chen, X.; Wang, N.; Shen, D.; Li, Z.; et al. Glucose outcomes of a learning-type artificial pancreas with an unannounced meal in type 1 diabetes. Comput. Methods Programs Biomed. 2020, 191, 105416. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Rashid, M.; Sevil, M.; Hobbs, N.; Brandt, R.; Askari, M.R.; Shahidehpour, A.; Cinar, A. Prediction of Blood Glucose Levels for People with Type 1 Diabetes Using Latent-Variable-Based Model. Available online: https://ceur-ws.org/Vol-2675/paper20.pdf (accessed on 8 June 2023).
- Zhu, T.; Yao, X.; Li, K.; Herrero, P.; Georgiou, P. Blood Glucose Prediction for Type 1 Diabetes Using Generative Adversarial Networks. In CEUR Workshop Proceedings; 2020. Available online: https://ceur-ws.org/Vol-2675/paper15.pdf (accessed on 8 June 2023).
- Alvarado, J.; Velasco, J.M.; Chavez, F.; Hidalgo, J.I.; De Vega, F.F. Blood Glucose Prediction Using a Two Phase TSK Fuzzy Rule Based System. In Proceedings of the 2021 IEEE Congress on Evolutionary Computation (CEC), Kraków, Poland, 28 June–1 July 2021; pp. 712–719. [Google Scholar] [CrossRef]
- Beauchamp, J.; Bunescu, R.; Marling, C.; Li, Z.; Liu, C. LSTMs and Deep Residual Networks for Carbohydrate and Bolus Recommendations in Type 1 Diabetes Management. Sensors 2021, 21, 3303. [Google Scholar] [CrossRef] [PubMed]
- Bhargav, S.; Kaushik, S.; Dutt, V.; Choudhury, A. Development of a weighted ensemble approach for prediction of blood glucose levels. Manch. J. Artif. Intell. Appl. Sci. 2022, 3, 6. [Google Scholar]
- Cui, R.; Hettiarachchi, C.; Nolan, C.J.; Daskalaki, E.; Suominen, H. Personalised Short-Term Glucose Prediction via Recurrent Self-Attention Network. In Proceedings of the 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 June 2021; pp. 154–159. [Google Scholar] [CrossRef]
- De Bois, M.; El-Yacoubi, M.A.; Ammi, M. Integration of clinical criteria into the training of deep models: Application to glucose prediction for diabetic people. Smart Health 2021, 21, 100193. [Google Scholar] [CrossRef]
- De Falco, I.; Cioppa, A.D.; Koutny, T.; Scafuri, U.; Tarantino, E.; Ubi, M. Grammatical Evolution-Based Approach for Extracting Interpretable Glucose-Dynamics Models. In Proceedings of the 2021 IEEE Symposium on Computers and Communications (ISCC), Athens, Greece, 5–8 September 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Rabby, M.F.; Tu, Y.; Hossen, M.I.; Maida, A.S.; Hei, X. Stacked LSTM based deep recurrent neural network with kalman smoothing for blood glucose prediction. BMC Med. Inf. Decis. Mak. 2021, 21, 101. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.; Hussain, S.; Khan, W.A. Predicting continuous blood glucose level using deep learning. In Proceedings of the 14th IEEE/ACM International Conference on Utility and Cloud Computing Companion, Leicester UK, 6–9 December 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Wang, Y. A Comparison of Machine Learning Algorithms in Blood Glucose Prediction for People with Type 1 Diabetes. In Proceedings of the 2nd International Symposium on Artificial Intelligence for Medicine Sciences, Beijing China, 29–31 October 2021; pp. 351–360. [Google Scholar] [CrossRef]
- Zaidi, S.M.A.; Chandola, V.; Ibrahim, M.; Romanski, B.; Mastrandrea, L.D.; Singh, T. Multi-step ahead predictive model for blood glucose concentrations of type-1 diabetic patients. Sci. Rep. 2021, 11, 24332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Flores, K.B.; Tran, H.T. Deep learning and regression approaches to forecasting blood glucose levels for type 1 diabetes. Biomed. Signal Process. Control 2021, 69, 102923. [Google Scholar] [CrossRef]
- Aashima Bhargav, S.; Kaushik, S.; Dutt, V. A Combination of Decision Trees with Machine Learning Ensembles for Blood Glucose Level Predictions. In Proceedings of International Conference on Data Science and Applications; Saraswat, M., Roy, S., Chowdhury, C., Gandomi, A.H., Eds.; Springer: Singapore, 2022; Volume 287, pp. 533–548. [Google Scholar]
- Daniels, J.; Herrero, P.; Georgiou, P. A Multitask Learning Approach to Personalized Blood Glucose Prediction. IEEE J. Biomed. Health Inform. 2022, 26, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Yu, X.; Ma, N.; Wu, R.; Li, H. An autonomous channel deep learning framework for blood glucose prediction. Appl. Soft Comput. 2022, 120, 108636. [Google Scholar] [CrossRef]
- Mordvanyuk, N.; Torrent-Fontbona, F.; Pez, B.L. Prediction of Glucose Level Conditions from Sequential Data. Available online: http://eia.udg.es/~nmordvanyuk/papers/Prediction-of-Glucose-Level-Conditions-from%20Sequential-DataCCIA2017.pdf (accessed on 8 June 2023).
- Jaloli, M.; Cescon, M. Long-Term Prediction of Blood Glucose Levels in Type 1 Diabetes Using a CNN-LSTM-Based Deep Neural Network. J. Diabetes Sci. Technol. 2023, 17, 1590–1601. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Rashid, M.; Askari, M.R.; Cinar, A. Adaptive personalized prior-knowledge-informed model predictive control for type 1 diabetes. Control Eng. Pract. 2023, 131, 105386. [Google Scholar] [CrossRef] [PubMed]
- Cui, R.; Nolan, C.J.; Daskalaki, E.; Suominen, H. Jointly Predicting Postprandial Hypoglycemia and Hyperglycemia Using Continuous Glucose Monitoring Data in Type 1 Diabetes. In Proceedings of the 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Sydney, Australia, 24–27 July 2023; pp. 1–7. [Google Scholar] [CrossRef]
- Langarica, S.; Rodriguez-Fernandez, M.; Doyle Iii, F.J.; Núñez, F. A Probabilistic Approach to Blood Glucose Prediction in Type 1 Diabetes Under Meal Uncertainties. IEEE J. Biomed. Health Inform. 2023, 27, 5054–5065. [Google Scholar] [CrossRef]
- Langarica, S.; de la Vega, D.; Cariman, N.; Miranda, M.; Andrade, D.C.; Núñez, F.; Rodriguez-Fernandez, M. Deep Learning-Based Glucose Prediction Models: A Guide for Practitioners and a Curated Dataset for Improved Diabetes Management. IEEE Open J. Eng. Med. Biol. 2024, 5, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Aiello, E.M.; Jaloli, M.; Cescon, M. Model Predictive Control (MPC) of an artificial pancreas with data-driven learning of multi-step-ahead blood glucose predictors. Control Eng. Pract. 2024, 144, 105810. [Google Scholar] [CrossRef]
- Hutten, H. A multicompartment model for open-loop control of glucose in insulin-dependent diabetics. Comput. Methods Programs Biomed. 1990, 32, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.S.; Doyle, F.J.; Peppas, N.A. A model-based algorithm for blood glucose control in Type I diabetic patients. IEEE Trans. Biomed. Eng. 1999, 46, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Mougiakakou, S.G.; Prountzou, K.; Nikita, K.S. A Real Time Simulation Model of Glucose-Insulin Metabolism for Type 1 Diabetes Patients. In Proceedings of the 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference, Shanghai, China, 17–18 January 2006; pp. 298–301. [Google Scholar] [CrossRef]
- Roy, A.; Parker, R.S. Dynamic Modeling of Free Fatty Acid, Glucose, and Insulin: An Extended ‘Minimal Model’. Diabetes Technol. Ther. 2006, 8, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Schlotthauer, G.; Gamero, L.G.; Torres, M.E.; Nicolini, G.A. Modeling, identification and nonlinear model predictive control of type I diabetic patient. Med. Eng. Phys. 2006, 28, 240–250. [Google Scholar] [CrossRef]
- Kildegaard, J.; Randløv, J.; Poulsen, J.U.; Hejlesen, O. A Study of Trained Clinicians’ Blood Glucose Predictions Based on Diaries of People with Type 1 Diabetes. Methods Inf. Med. 2007, 46, 553–557. [Google Scholar]
- Markakis, M.G.; Mitsis, G.D.; Papavassilopoulos, G.P.; Marmarelis, V.Z. Model Predictive Control of blood glucose in Type 1 diabetes: The Principal Dynamic Modes approach. In Proceedings of the 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 20–25 August 2008; pp. 5466–5469. [Google Scholar] [CrossRef]
- Stahl, F.; Johansson, R. Short-term diabetes blood glucose prediction based on blood glucose measurements. In Proceedings of the 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 20–25 August 2008; pp. 291–294. [Google Scholar] [CrossRef]
- Estrada, G.C.; Kirchsteiger, H.; Del Re, L.; Renard, E. Innovative approach for online prediction of blood glucose profile in type 1 diabetes patients. In Proceedings of the 2010 American Control Conference 2015–2020, Baltimore, MD, USA, 30 June–2 July 2010. [Google Scholar] [CrossRef]
- Georga, E.I.; Protopappas, V.C.; Fotiadis, D.I. Predictive Metabolic Modeling for Type 1 Diabetes Using Free-Living Data on Mobile Devices. In Wireless Mobile Communication and Healthcare; Lin, J.C., Nikita, K.S., Eds.; Springer: Berlin, Heidelberg, 2011; Volume 55, pp. 187–193. [Google Scholar]
- Percival, M.W.; Wang, Y.; Grosman, B.; Dassau, E.; Zisser, H.; Jovanovič, L.; Doyle, F.J. Development of a multi-parametric model predictive control algorithm for insulin delivery in type 1 diabetes mellitus using clinical parameters. J. Process Control 2011, 21, 391–404. [Google Scholar] [CrossRef]
- De Pereda, D.; Romero-Vivo, S.; Ricarte, B.; Bondia, J. On the prediction of glucose concentration under intra-patient variability in type 1 diabetes: A monotone systems approach. Comput. Methods Programs Biomed. 2012, 108, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- Gondhalekar, R.; Dassau, E.; Zisser, H.C.; Doyle, F.J. Periodic-Zone Model Predictive Control for Diurnal Closed-Loop Operation of an Artificial Pancreas. J. Diabetes Sci. Technol. 2013, 7, 1446–1460. [Google Scholar] [CrossRef] [PubMed]
- Cescon, M.; Johansson, R.; Renard, E. Subspace-based linear multi-step predictors in type 1 diabetes mellitus. Biomed. Signal Process. Control 2015, 22, 99–110. [Google Scholar] [CrossRef]
- Mirshekarian, S.; Bunescu, R.; Marling, C.; Schwartz, F. Using LSTMs to learn physiological models of blood glucose behavior. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Seogwipo, Republic of Korea, 11–15 July 2017; pp. 2887–2891. [Google Scholar] [CrossRef]
- Bertachi, A.; Biagi, L.; Contreras, I.; Luo, N.; Vehı, J. Prediction of blood glucose levels and nocturnal hypoglycemia using physiological models and artificial neural networks. KDH@ IJCAI 2018, 85–90. [Google Scholar]
- Contreras, I.; Bertachi, A.; Biagi, L.; Oviedo, S.; Vehı, J. Using Grammatical Evolution to Generate Short-Term Blood Glucose Prediction models. KDH@ IJCAI 2018, 91–96. [Google Scholar]
- Hajizadeh, I.; Rashid, M.; Turksoy, K.; Samadi, S.; Feng, J.; Sevil, M.; Hobbs, N.; Lazaro, C.; Maloney, Z.; Littlejohn, E.; et al. Incorporating Unannounced Meals and Exercise in Adaptive Learning of Personalized Models for Multivariable Artificial Pancreas Systems. J. Diabetes Sci. Technol. 2018, 12, 953–966. [Google Scholar] [CrossRef] [PubMed]
- De Falco, I.; Della Cioppa, A.; Giugliano, A.; Marcelli, A.; Koutny, T.; Krcma, M.; Scafuri, U.; Tarantino, E. A genetic programming-based regression for extrapolating a blood glucose-dynamics model from interstitial glucose measurements and their first derivatives. Appl. Soft Comput. 2019, 77, 316–328. [Google Scholar] [CrossRef]
- Hajizadeh, I.; Rashid, M.; Cinar, A. Plasma-insulin-cognizant adaptive model predictive control for artificial pancreas systems. J. Process Control 2019, 77, 97–113. [Google Scholar] [CrossRef]
- Knopp, J.L.; Signal, M.; Harris, D.L.; Marics, G.; Weston, P.; Harding, J.; Tóth-Heyn, P.; Hómlok, J.; Benyó, B.; Chase, J.G. Modelling intestinal glucose absorption in premature infants using continuous glucose monitoring data. Comput. Methods Programs Biomed. 2019, 171, 41–51. [Google Scholar] [CrossRef]
- Dias, C.C.; Kamath, S.; Vidyasagar, S. Design of dual hormone blood glucose therapy and comparison with single hormone using MPC algorithm. IET Syst. Biol. 2020, 14, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Goyal, M.; Aydas, B.; Ghazaleh, H.; Rajasekharan, S. CarbMetSim: A discrete-event simulator for carbohydrate metabolism in humans. PLoS ONE 2020, 15, e0209725. [Google Scholar] [CrossRef]
- Montaser, E.; Diez, J.E.; Rossetti, P.; Rashid, M.; Cinar, A.; Bondia, J. Seasonal Local Models for Glucose Prediction in Type 1 Diabetes. IEEE J. Biomed. Health Inform. 2020, 24, 2064–2072. [Google Scholar] [CrossRef] [PubMed]
- Adelberger, D.; Reiterer, F.; Schrangl, P.; Ringemann, C.H.; Huschto, T.; del Re, L. Prediction of postprandial glucose excursions in type 1 diabetes using control-oriented process models. IFAC-Pap. 2021, 54, 466–471. [Google Scholar] [CrossRef]
- Cervigon, C.; Velasco, J.M.; Burgos-Simon, C.; Villanueva, R.J.; Hidalgo, J.I. Probabilistic Fitting of Glucose Models with Real-Coded Genetic Algorithms. In Proceedings of the 2021 IEEE Congress on Evolutionary Computation (CEC), Kraków, Poland, 28 June–1 July 2021; pp. 736–743. [Google Scholar] [CrossRef]
- Tavarez, J.R.; Sanchez, I.Y.; Maldonado, V.A.; Montes, M.; Ortiz, R.A. Stochastic Identification and Kalman Filter for Blood Glucose Estimation. In Recent Trends in Sustainable Engineering; Flores Rodríguez, K.L., Ramos Alvarado, R., Barati, M., Segovia Tagle, V., Velázquez González, R.S., Eds.; Springer International Publishing: Cham, Switzerland, 2022; Volume 297, pp. 117–130. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lubasinski, N.; Thabit, H.; Nutter, P.W.; Harper, S. Blood Glucose Prediction from Nutrition Analytics in Type 1 Diabetes: A Review. Nutrients 2024, 16, 2214. https://doi.org/10.3390/nu16142214
Lubasinski N, Thabit H, Nutter PW, Harper S. Blood Glucose Prediction from Nutrition Analytics in Type 1 Diabetes: A Review. Nutrients. 2024; 16(14):2214. https://doi.org/10.3390/nu16142214
Chicago/Turabian StyleLubasinski, Nicole, Hood Thabit, Paul W. Nutter, and Simon Harper. 2024. "Blood Glucose Prediction from Nutrition Analytics in Type 1 Diabetes: A Review" Nutrients 16, no. 14: 2214. https://doi.org/10.3390/nu16142214
APA StyleLubasinski, N., Thabit, H., Nutter, P. W., & Harper, S. (2024). Blood Glucose Prediction from Nutrition Analytics in Type 1 Diabetes: A Review. Nutrients, 16(14), 2214. https://doi.org/10.3390/nu16142214





