Lactobacillus plantarum CCFM405 against Rotenone-Induced Parkinson’s Disease Mice via Regulating Gut Microbiota and Branched-Chain Amino Acids Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Experimental Design and Rotenone Treatment
2.3. Measurement of Colon Motility and Stool Collection
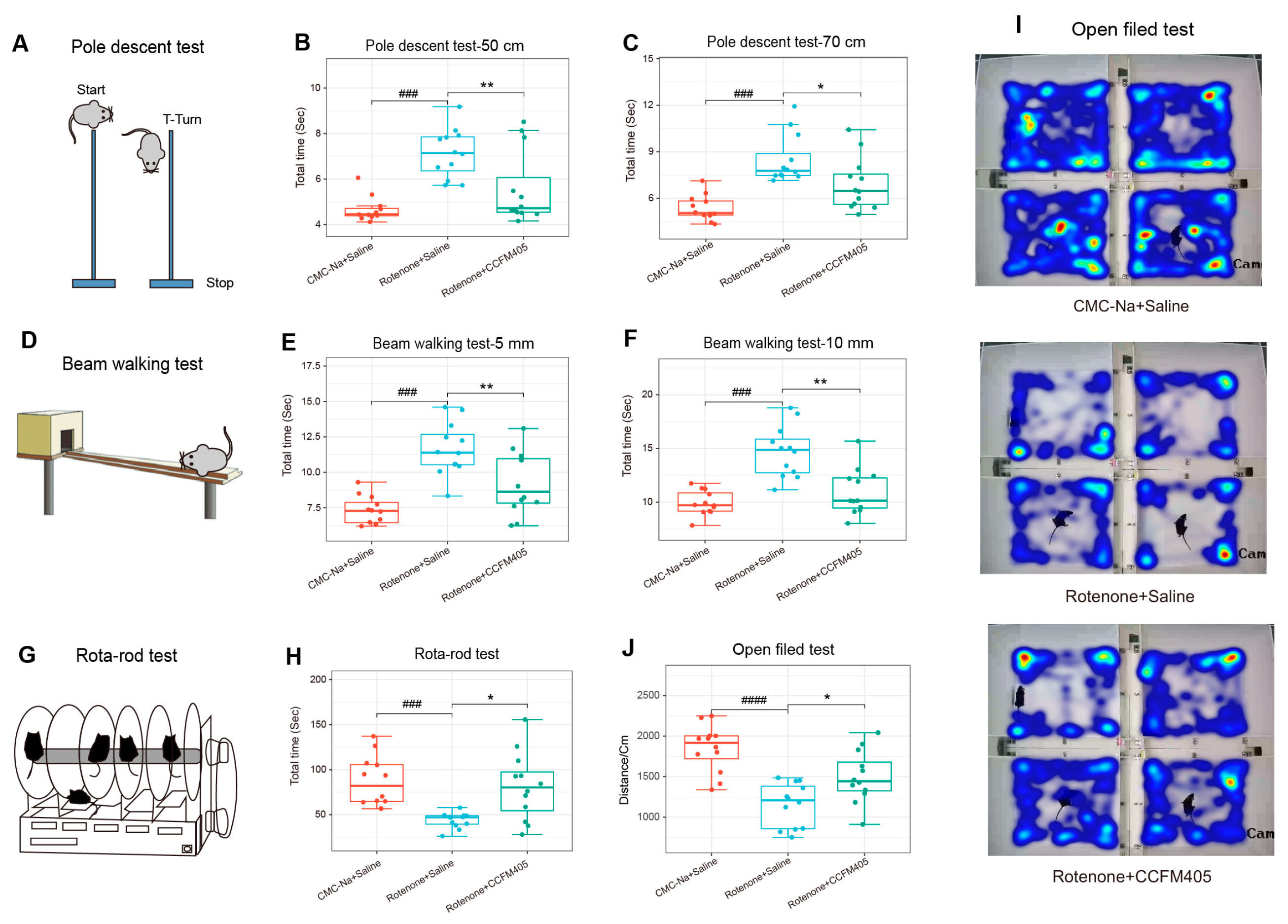
2.4. Behavioral Tests for Motor Functions
2.5. Measurement of Neurotransmitters
2.6. Immunohistochemistry and Image Analysis
2.7. Gut Microbiota Profiling and Functional Prediction Analysis
2.8. Metabolomic Data Analysis
2.9. Sample Collection and Tissue Preparation
2.10. Statistical Analysis
3. Results
3.1. Effects of L. plantarum CCFM405 on Body Weight and Gastrointestinal Function in Rotenone-Induced PD Mice
3.2. Effects of L. plantarum CCFM405 on Motor Function in Rotenone-Induced PD Mice
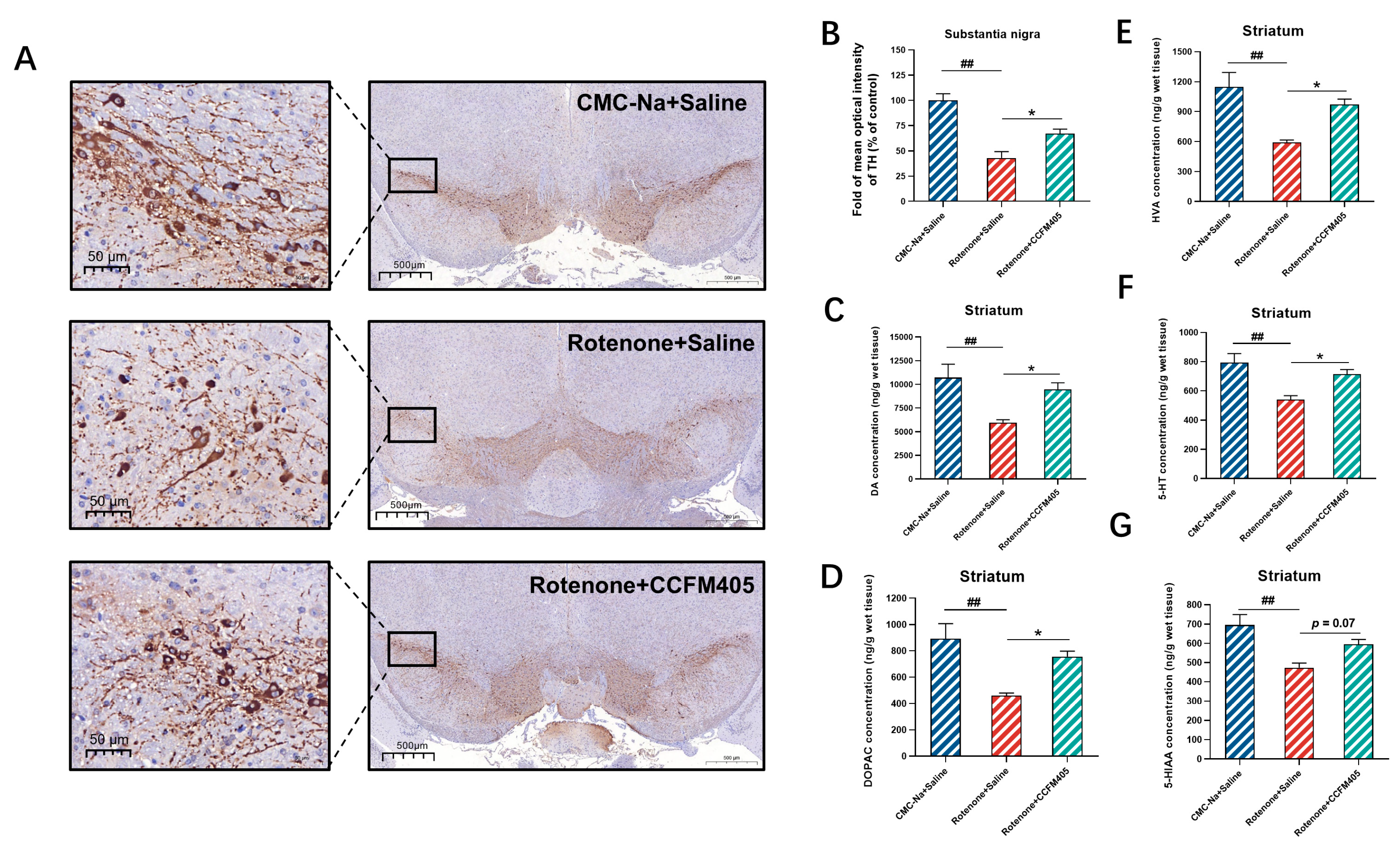
3.3. Effects of L. plantarum CCFM405 on Dopaminergic Neurons and Neurotransmitters in the Striatum of Rotenone-Induced PD Mice
3.4. Effects of L. plantarum CCFM405 on Microglia and Astrocytes in the Nigra Region of the Brain of Rotenone-Induced PD Mice
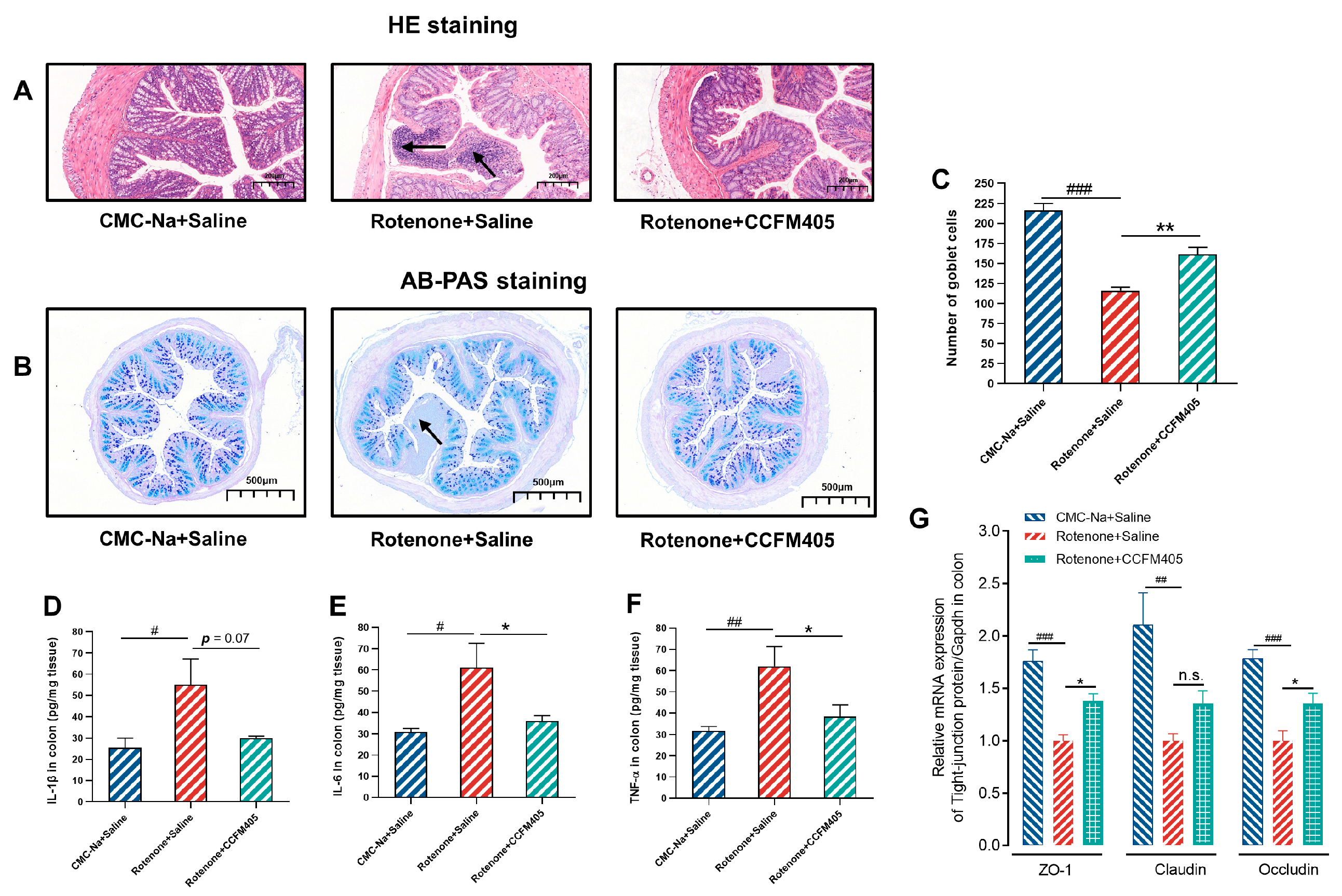
3.5. Effects of L. plantarum CCFM405 on Intestinal Inflammation and Goblet Cell Count in the Colon in Rotenone-Induced PD Mice
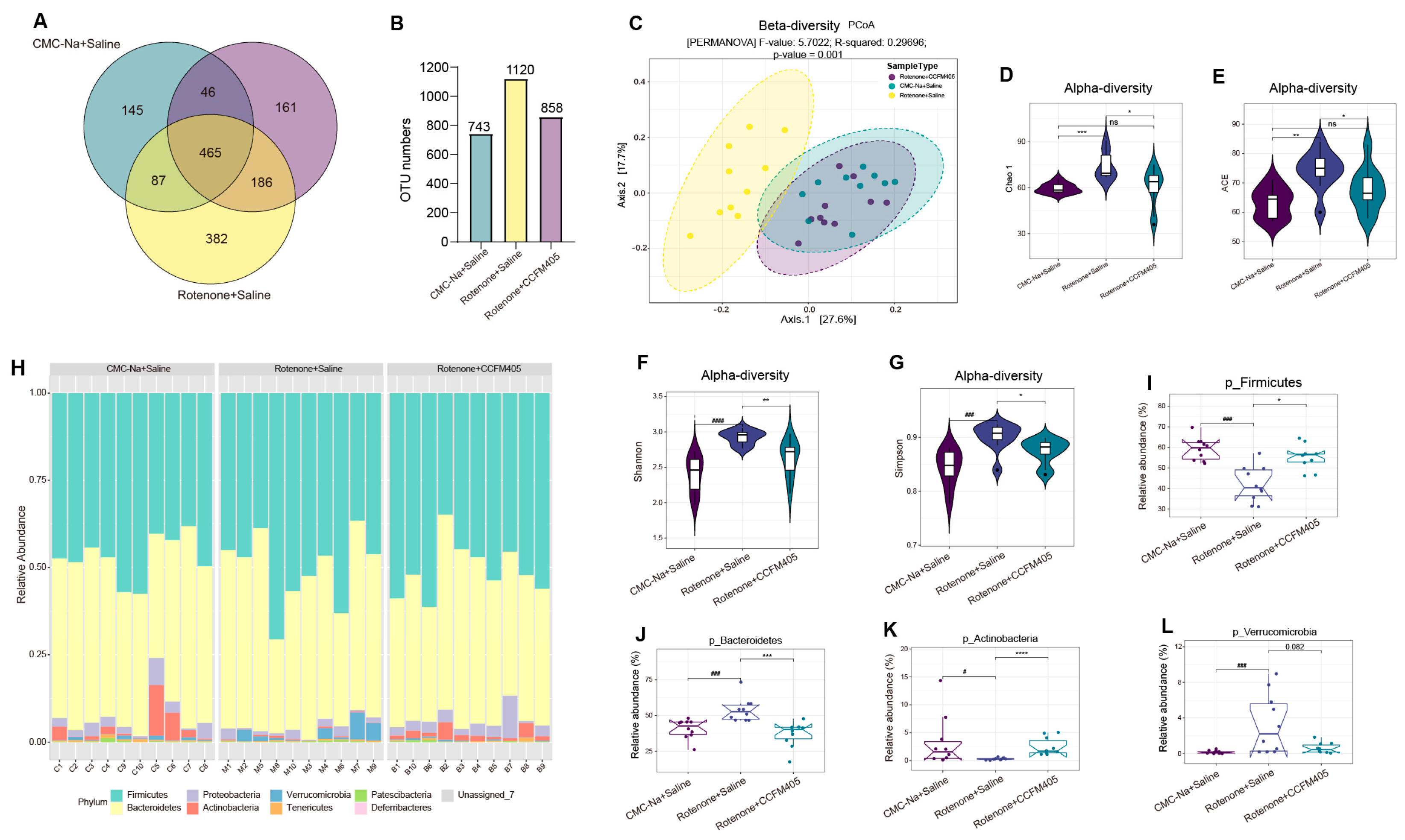
3.6. Effects of L. plantarum CCFM405 on Gut Microbiota Diversity and Structure
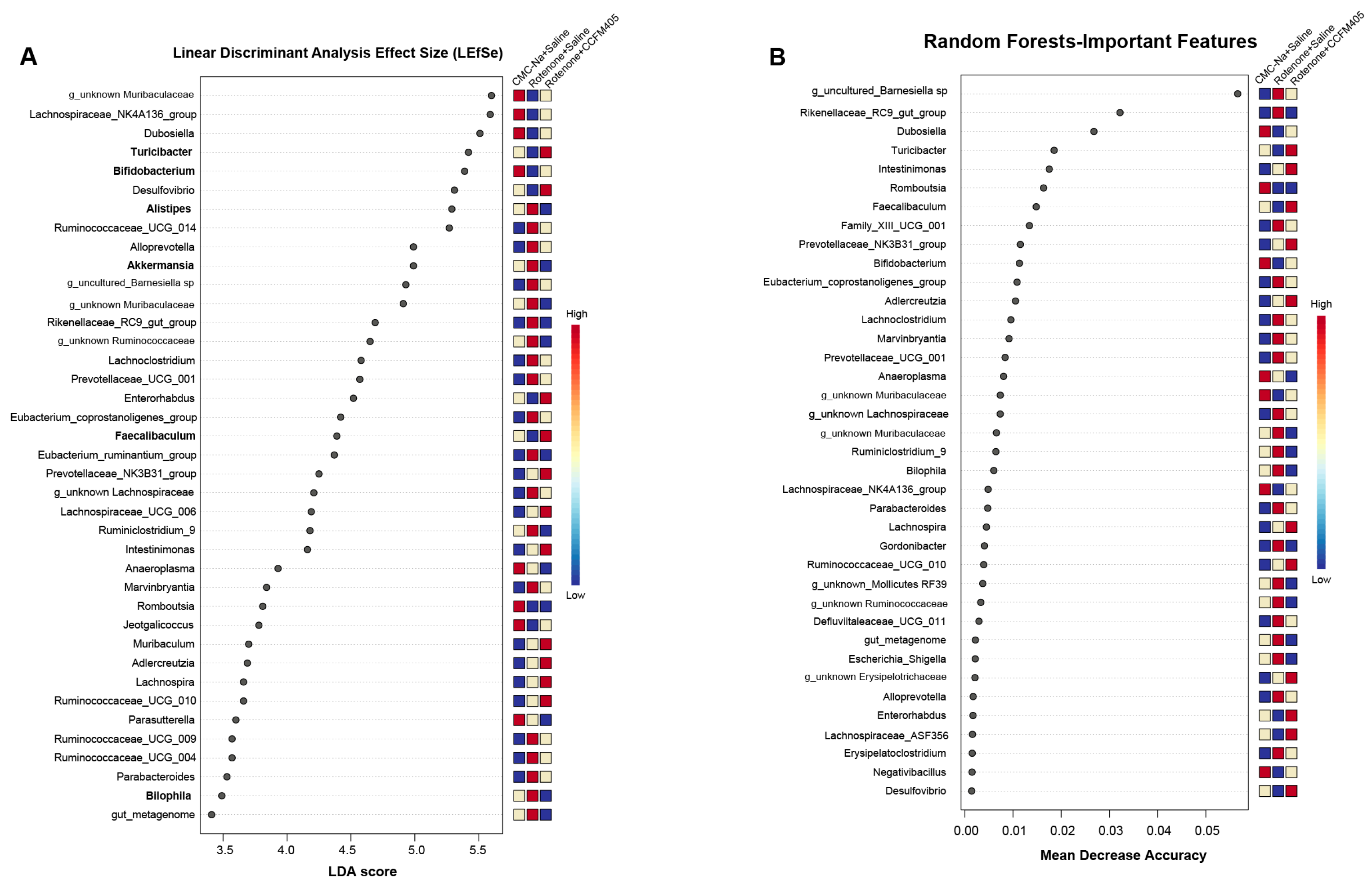
3.7. Effect of L. plantarum CCFM405 on Rotenone-Induced Specific Differential Bacteria in PD Mice
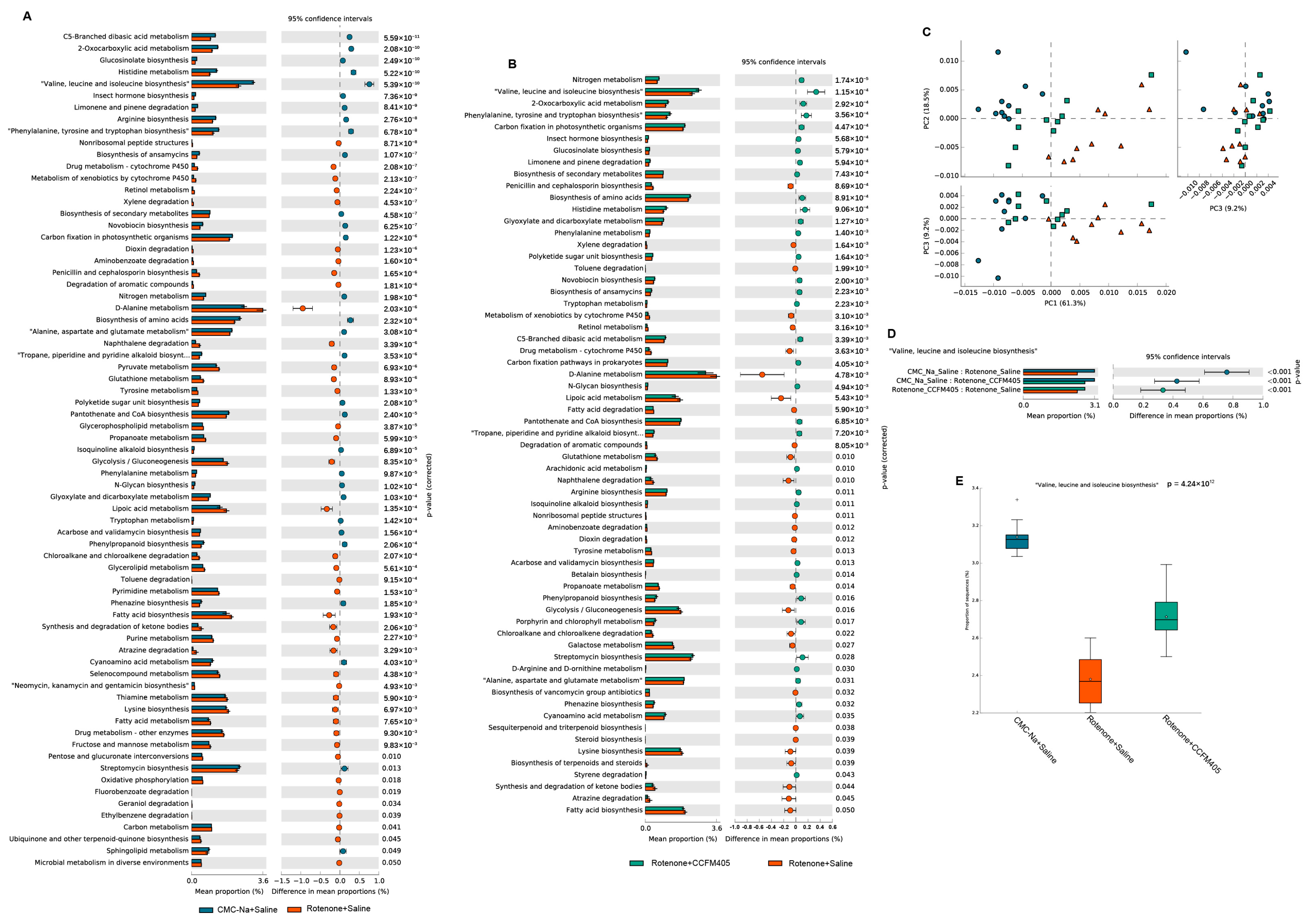
3.8. Predicted Functional Changes in the Gut Microbiota
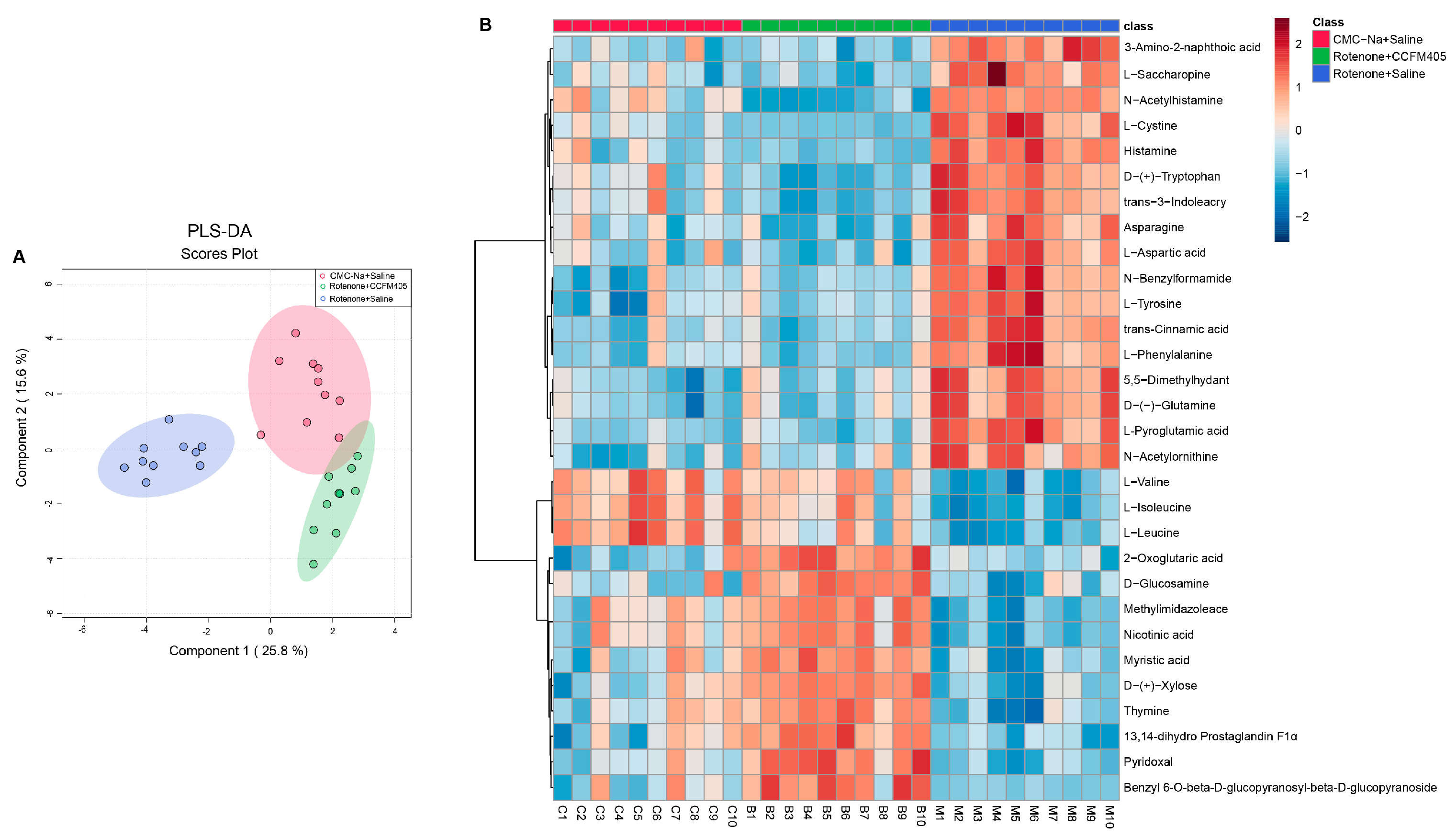
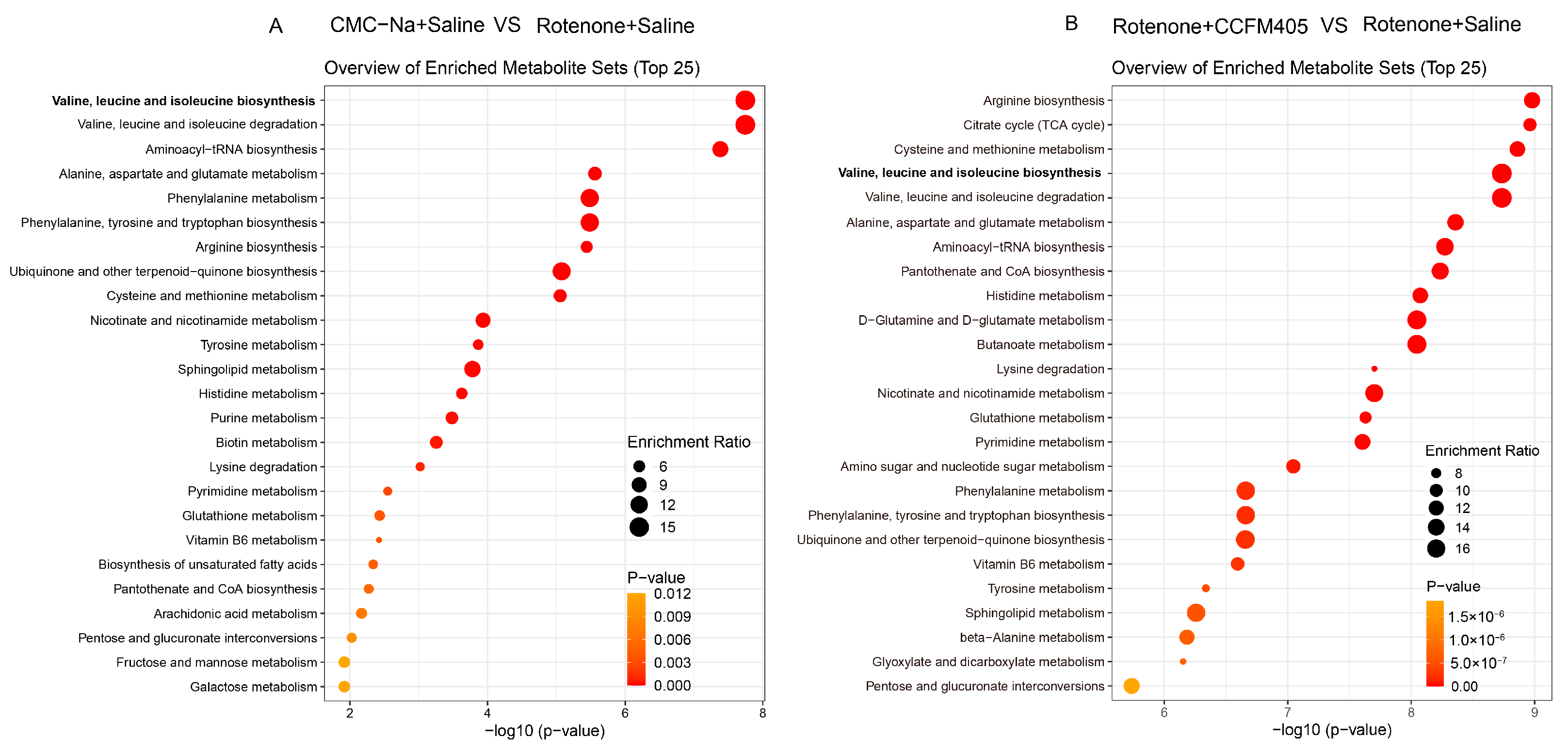
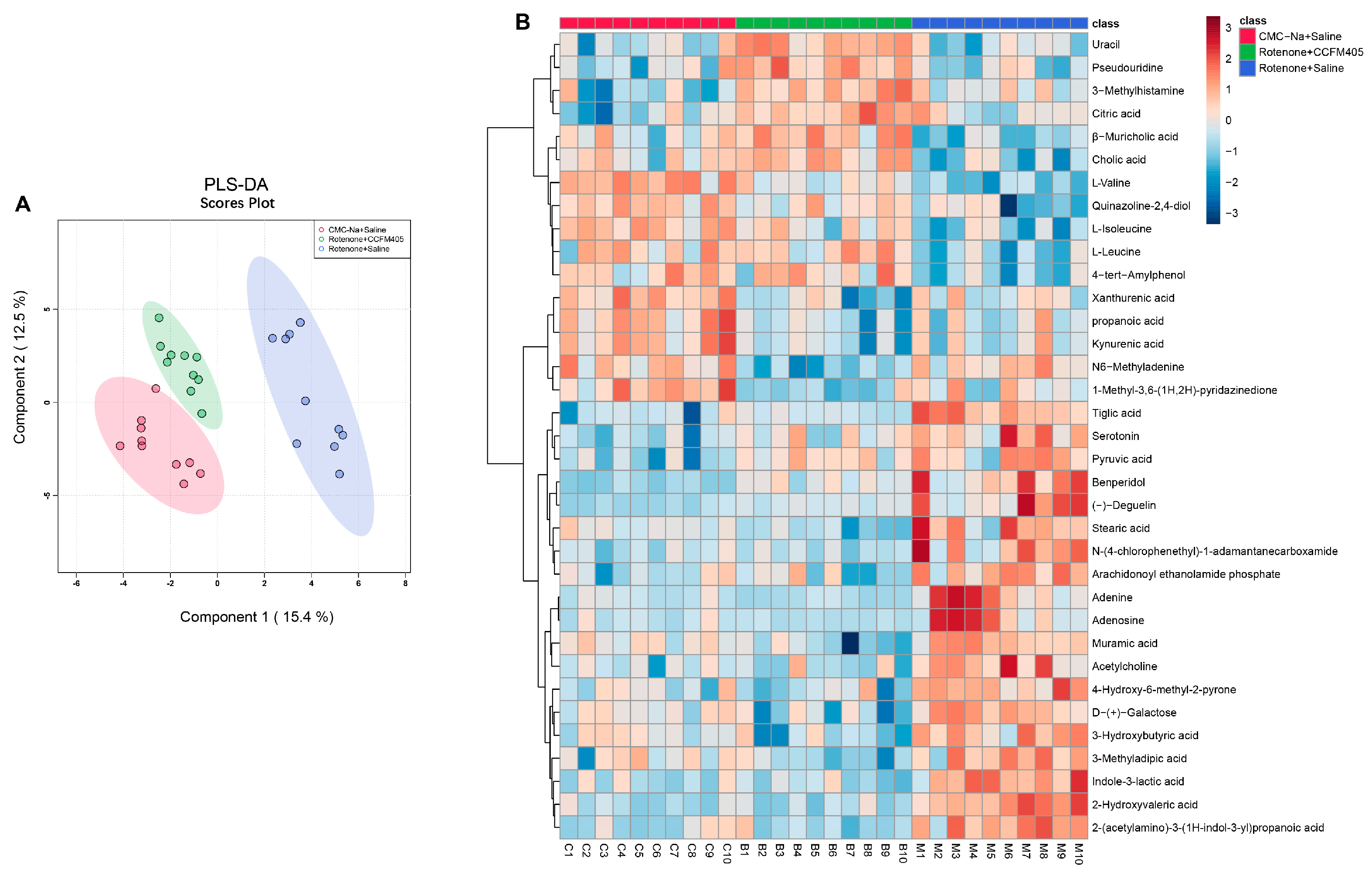
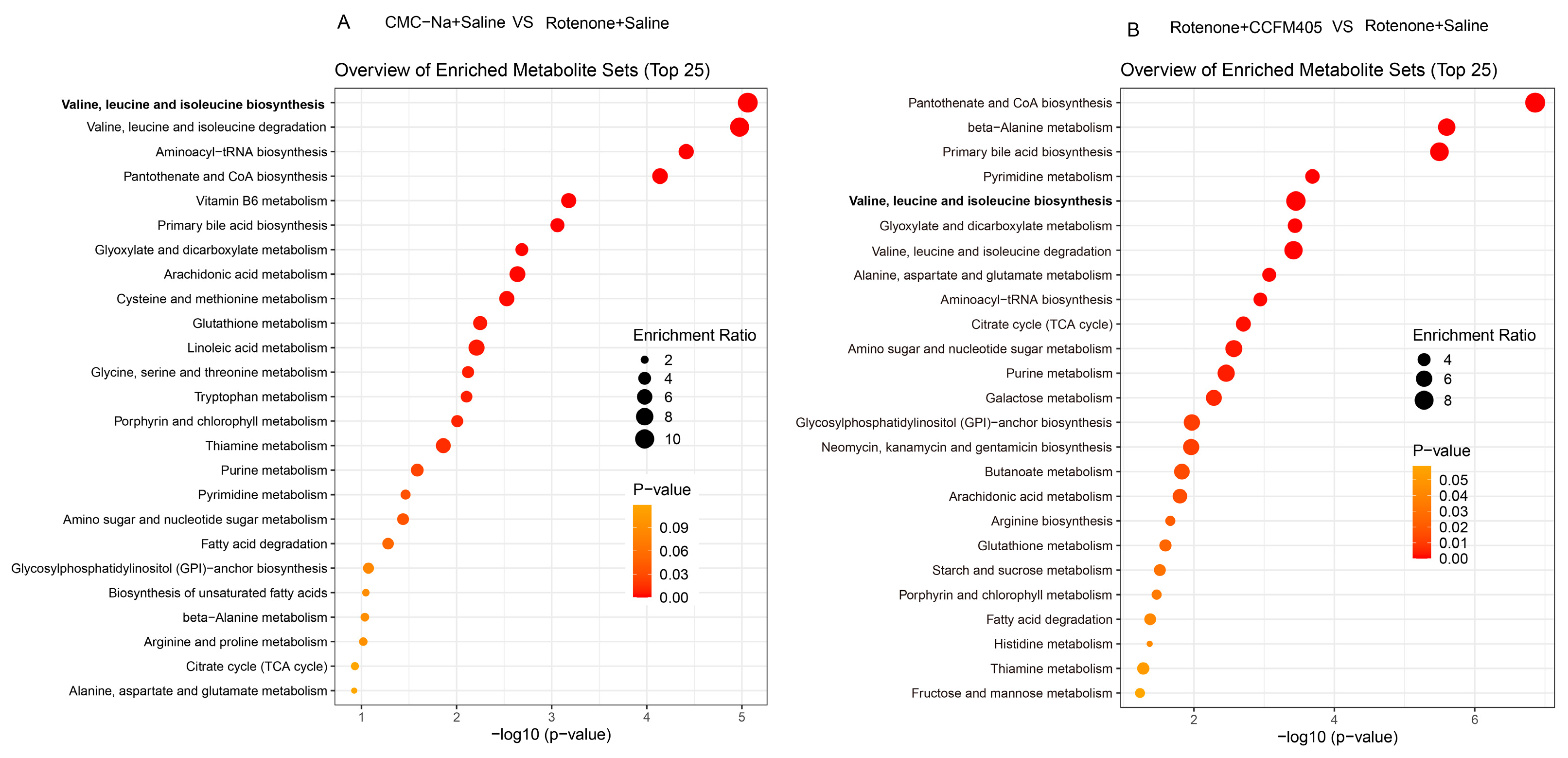
3.9. L. plantarum CCFM405 Altered the Fecal Metabolite Profiles in Rotenone-Induced PD Mice
3.10. L. plantarum CCFM405 Altered the Serum Metabolite Profiles of Rotenone-Induced PD Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feigin, V.L.; Abajobir, A.A.; Abate, K.H.; Abd-Allah, F.; Abdulle, A.M.; Abera, S.F.; Nguyen, G. Global, regional, and national burden of neurological disorders during 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Neurol. 2017, 16, 877–897. [Google Scholar] [CrossRef] [PubMed]
- Dorsey, E.R.; Bloem, B.R. The Parkinson Pandemic—A Call to Action. JAMA Neurol. 2018, 75, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Lotharius, J.; Brundin, P. Pathogenesis of Parkinson’s disease: Dopamine, vesicles and alpha-synuclein. Nat. Rev. Neurosci. 2002, 3, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Bloem, B.R.; Okun, M.S.; Klein, C. Parkinson’s disease. Lancet 2021, 397, 2284–2303. [Google Scholar] [CrossRef] [PubMed]
- Leszek, J.; Barreto, G.E.; Gąsiorowski, K.; Koutsouraki, E.; Ávila-Rodrigues, M.; Aliev, G. Inflammatory Mechanisms and Oxidative Stress as Key Factors Responsible for Progression of Neurodegeneration: Role of Brain Innate Immune System. CNS Neurol. Disord. Drug Targets 2016, 15, 329–336. [Google Scholar] [CrossRef]
- Cao, J.J.; Li, K.S.; Shen, Y.Q. Activated immune cells in Parkinson’s disease. J. Neuroimmune Pharm. 2011, 6, 323–329. [Google Scholar] [CrossRef]
- Calabrese, V.; Cornelius, C.; Dinkova-Kostova, A.T.; Calabrese, E.J.; Mattson, M.P. Cellular stress responses, the hormesis paradigm, and vitagenes: Novel targets for therapeutic intervention in neurodegenerative disorders. Antioxid. Redox Signal. 2010, 13, 1763–1811. [Google Scholar] [CrossRef]
- Calabrese, V.; Cornelius, C.; Dinkova-Kostova, A.T.; Calabrese, E.J. Vitagenes, cellular stress response, and acetylcarnitine: Relevance to hormesis. Biofactors 2009, 35, 146–160. [Google Scholar] [CrossRef]
- Calabrese, V.; Mancuso, C.; Calvani, M.; Rizzarelli, E.; Butterfield, D.A.; Giuffrida Stella, A.M. Nitric oxide in the central nervous system: Neuroprotection versus neurotoxicity. Nat. Rev. Neurosci. 2007, 8, 766–775. [Google Scholar] [CrossRef]
- Sung, H.Y.; Park, J.W.; Kim, J.S. The frequency and severity of gastrointestinal symptoms in patients with early Parkinson’s disease. J. Mov. Disord. 2014, 7, 7–12. [Google Scholar] [CrossRef]
- Margolis, K.G.; Cryan, J.F.; Mayer, E.A. The microbiota-gut-brain axis: From motility to mood. Gastroenterology 2021, 160, 1486–1501. [Google Scholar] [CrossRef] [PubMed]
- Agirman, G.; Hsiao, E.Y. SnapShot: The microbiota-gut-brain axis. Cell 2021, 184, 2524. [Google Scholar] [CrossRef] [PubMed]
- Scheperjans, F.; Aho, V.; Pereira, P.A.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M.; et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2015, 30, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.H.; Lim, S.Y.; Lang, A.E. The microbiome-gut-brain axis in Parkinson disease—From basic research to the clinic. Nat. Rev. Neurol. 2022, 18, 476–495. [Google Scholar] [CrossRef]
- Tan, A.H.; Chong, C.W.; Lim, S.Y.; Yap, I.K.S.; Teh, C.S.J.; Loke, M.F.; Song, S.L.; Tan, J.Y.; Ang, B.H.; Tan, Y.Q. Gut microbial ecosystem in Parkinson disease: New clinicobiological insights from multi-omics. Ann. Neurol. 2021, 89, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Wang, J.; Deng, Y.; Ye, Z.; Ge, Y. Inflammatory microbes and genes as potential biomarkers of Parkinson’s disease. NPJ Biofilms Microbiomes 2022, 8, 101. [Google Scholar] [CrossRef]
- Zhao, Z.; Ning, J.; Bao, X.Q.; Shang, M.; Ma, J.; Li, G.; Zhang, D. Fecal microbiota transplantation protects rotenone-induced Parkinson’s disease mice via suppressing inflammation mediated by the lipopolysaccharide-TLR4 signaling pathway through the microbiota-gut-brain axis. Microbiome 2021, 9, 226. [Google Scholar] [CrossRef]
- Sun, M.-F.; Zhu, Y.-L.; Zhou, Z.-L.; Jia, X.-B.; Xu, Y.-D.; Yang, Q.; Cui, C.; Shen, Y.-Q. Neuroprotective effects of fecal microbiota transplantation on MPTP-induced Parkinson’s disease mice: Gut microbiota, glial reaction and TLR4/TNF-α signaling pathway. Brain Behav. Immun. 2018, 70, 48–60. [Google Scholar] [CrossRef]
- Liao, J.-F.; Cheng, Y.-F.; Li, S.-W.; Lee, W.-T.; Hsu, C.-C.; Wu, C.-C.; Jeng, O.-J.; Wang, S.; Tsai, Y.-C. Lactobacillus plantarum PS128 ameliorates 2, 5-Dimethoxy-4-iodoamphetamine-induced tic-like behaviors via its influences on the microbiota–gut-brain-axis. Brain Res. Bull. 2019, 153, 59–73. [Google Scholar] [CrossRef]
- Liao, J.F.; Cheng, Y.F.; You, S.T.; Kuo, W.C.; Huang, C.W.; Chiou, J.J.; Hsu, C.C.; Hsieh-Li, H.M.; Wang, S.; Tsai, Y.C. Lactobacillus plantarum PS128 alleviates neurodegenerative progression in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced mouse models of Parkinson’s disease. Brain Behav. Immun. 2020, 90, 26–46. [Google Scholar] [CrossRef]
- Lu, C.S.; Chang, H.C.; Weng, Y.H.; Chen, C.C.; Kuo, Y.S.; Tsai, Y.C. The Add-On Effect of Lactobacillus plantarum PS128 in Patients with Parkinson’s Disease: A Pilot Study. Front. Nutr. 2021, 8, 650053. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, Z.; Zhao, L.; Zhao, Y.; Yang, G.; Wang, C.; Gao, L.; Niu, C.; Li, S. Lactobacillus plantarum DP189 reduces α-SYN aggravation in MPTP-induced parkinson’s disease mice via regulating oxidative damage, inflammation, and gut microbiota disorder. J. Agric. Food Chem. 2022, 70, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Srivastav, S.; Neupane, S.; Bhurtel, S.; Katila, N.; Maharjan, S.; Choi, H.; Hong, J.T.; Choi, D.-Y. Probiotics mixture increases butyrate, and subsequently rescues the nigral dopaminergic neurons from MPTP and rotenone-induced neurotoxicity. J. Nutr. Biochem. 2019, 69, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Inden, M.; Kitamura, Y.; Abe, M.; Tamaki, A.; Takata, K.; Taniguchi, T. Parkinsonian rotenone mouse model: Reevaluation of long-term administration of rotenone in C57BL/6 mice. Biol. Pharm. Bull. 2011, 34, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Qian, Y.; Xu, S.; Song, Y.; Xiao, Q. Longitudinal Analysis of Fecal Microbiome and Pathologic Processes in a Rotenone Induced Mice Model of Parkinson’s Disease. Front. Aging Neurosci. 2017, 9, 441. [Google Scholar] [CrossRef]
- Perez-Pardo, P.; Dodiya, H.B.; Engen, P.A.; Forsyth, C.B.; Huschens, A.M.; Shaikh, M.; Voigt, R.M.; Naqib, A.; Green, S.J.; Kordower, J.H.; et al. Role of TLR4 in the gut-brain axis in Parkinson’s disease: A translational study from men to mice. Gut 2019, 68, 829–843. [Google Scholar] [CrossRef]
- Johnson, M.E.; Bobrovskaya, L. An update on the rotenone models of Parkinson’s disease: Their ability to reproduce the features of clinical disease and model gene–environment interactions. Neurotoxicology 2015, 46, 101–116. [Google Scholar] [CrossRef]
- Tong, Y.; Wang, G.; Zhang, Q.; Tian, F.; Liu, X.; Zhao, J.; Zhang, H.; Chen, W. Systematic understanding of the potential manganese-adsorption components of a screened Lactobacillus plantarum CCFM436. RSC Adv. 2016, 6, 102804–102813. [Google Scholar] [CrossRef]
- Zhou, Z.L.; Jia, X.B.; Sun, M.F.; Zhu, Y.L.; Qiao, C.M.; Zhang, B.P.; Zhao, L.P.; Yang, Q.; Cui, C.; Chen, X.; et al. Neuroprotection of Fasting Mimicking Diet on MPTP-Induced Parkinson’s Disease Mice via Gut Microbiota and Metabolites. Neurotherapeutics 2019, 16, 741–760. [Google Scholar] [CrossRef]
- Luong, T.N.; Carlisle, H.J.; Southwell, A.; Patterson, P.H. Assessment of motor balance and coordination in mice using the balance beam. J. Vis. Exp. 2011, 49, 2376. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using Microbiome Analyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef] [PubMed]
- Paulson, J.N.; Stine, O.C.; Bravo, H.C.; Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 2013, 10, 1200–1202. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Travagli, R.A.; Browning, K.N.; Camilleri, M. Parkinson disease and the gut: New insights into pathogenesis and clinical relevance. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 673–685. [Google Scholar] [CrossRef]
- Liu, T.W.; Chen, C.M.; Chang, K.H. Biomarker of Neuroinflammation in Parkinson’s Disease. Int. J. Mol. Sci. 2022, 23, 4148. [Google Scholar] [CrossRef]
- Kang, X.; Ploner, A.; Roelstraete, B.; Khalili, H.; Williams, D.M.; Pedersen, N.L.; Ludvigsson, J.F.; Wirdefeldt, K. Association Between Microscopic Colitis and Parkinson’s Disease in a Swedish Population. Mov. Disord. 2021, 36, 1919–1926. [Google Scholar] [CrossRef]
- Zhang, Y.; He, X.; Qian, Y.; Xu, S.; Mo, C.; Yan, Z.; Yang, X.; Xiao, Q. Plasma branched-chain and aromatic amino acids correlate with the gut microbiota and severity of Parkinson’s disease. NPJ Park. Dis. 2022, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Needham, B.D.; Kaddurah-Daouk, R.; Mazmanian, S.K. Gut microbial molecules in behavioural and neurodegenerative conditions. Nat. Rev. Neurosci. 2020, 21, 717–731. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.A.B.; Trivedi, D.K.; Silverman, J.; Duru, I.C.; Paulin, L.; Auvinen, P.; Scheperjans, F. Multiomics implicate gut microbiota in altered lipid and energy metabolism in Parkinson’s disease. NPJ Park. Dis. 2022, 8, 39. [Google Scholar] [CrossRef]
- Anselmi, L.; Toti, L.; Bove, C.; Hampton, J.; Travagli, R.A. A Nigro−Vagal pathway controls gastric motility and is affected in a rat model of parkinsonism. Gastroenterology 2017, 153, 1581–1593. [Google Scholar] [CrossRef]
- Lai, F.; Jiang, R.; Xie, W.; Liu, X.; Tang, Y.; Xiao, H.; Gao, J.; Jia, Y.; Bai, Q. Intestinal pathology and gut microbiota alterations in a methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine (MPTP) mouse model of Parkinson’s disease. Neurochem. Res. 2018, 43, 1986–1999. [Google Scholar] [CrossRef]
- Gries, M.; Christmann, A.; Schulte, S.; Weyland, M.; Rommel, S.; Martin, M.; Baller, M.; Röth, R.; Schmitteckert, S.; Unger, M. Parkinson mice show functional and molecular changes in the gut long before motoric disease onset. Mol. Neurodegener. 2021, 16, 34. [Google Scholar] [CrossRef]
- Siracusa, R.; Scuto, M.; Fusco, R.; Trovato, A.; Ontario, M.L.; Crea, R.; Di Paola, R.; Cuzzocrea, S.; Calabrese, V. Anti-inflammatory and Anti-oxidant Activity of Hidrox(®) in Rotenone-Induced Parkinson’s Disease in Mice. Antioxidants 2020, 9, 824. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, V.; Cornelius, C.; Mancuso, C.; Barone, E.; Calafato, S.; Bates, T.; Rizzarelli, E.; Kostova, A.T. Vitagenes, dietary antioxidants and neuroprotection in neurodegenerative diseases. Front. Biosci. (Landmark Ed.) 2009, 14, 376–397. [Google Scholar] [CrossRef]
- Leta, V.; Chaudhuri, K.R.; Milner, O.; Chung-Faye, G.; Metta, V.; Pariante, C.M.; Borsini, A. Neurogenic and anti-inflammatory effects of probiotics in Parkinson’s disease: A systematic review of preclinical and clinical evidence. Brain Behav. Immun. 2021, 98, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Savva, G.M.; Bedarf, J.R.; Charles, I.G.; Hildebrand, F.; Narbad, A. Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. NPJ Park. Dis. 2021, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Toh, T.S.; Chong, C.W.; Lim, S.Y.; Bowman, J.; Cirstea, M.; Lin, C.H.; Chen, C.C.; Appel-Cresswell, S.; Finlay, B.B.; Tan, A.H. Gut microbiome in Parkinson’s disease: New insights from meta-analysis. Parkinsonism Relat. Disord. 2021, 94, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.-F.; Shen, Y.-Q. Dysbiosis of gut microbiota and microbial metabolites in Parkinson’s Disease. Ageing Res. Rev. 2018, 45, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Nuzum, N.D.; Loughman, A.; Szymlek-Gay, E.A.; Hendy, A.; Teo, W.P.; Macpherson, H. Gut microbiota differences between healthy older adults and individuals with Parkinson’s disease: A systematic review. Neurosci. Biobehav. Rev. 2020, 112, 227–241. [Google Scholar] [CrossRef]
- Li, T.; Chu, C.; Yu, L.; Zhai, Q.; Wang, S.; Zhao, J.; Zhang, H.; Chen, W.; Tian, F. Neuroprotective Effects of Bifidobacterium breve CCFM1067 in MPTP-Induced Mouse Models of Parkinson’s Disease. Nutrients 2022, 14, 4678. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kinoshita, T.; Matsumoto, A.; Yoshino, K.; Saito, I.; Xiao, J.-Z. Bifidobacterium breve A1 supplementation improved cognitive decline in older adults with mild cognitive impairment: An open-label, single-arm study. J. Prev. Alzheimer’s Dis. 2019, 6, 70–75. [Google Scholar] [CrossRef]
- Tian, P.; O’Riordan, K.J.; Lee, Y.-k.; Wang, G.; Zhao, J.; Zhang, H.; Cryan, J.F.; Chen, W. Towards a psychobiotic therapy for depression: Bifidobacterium breve CCFM1025 reverses chronic stress-induced depressive symptoms and gut microbial abnormalities in mice. Neurobiol. Stress 2020, 12, 100216. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Chu, C.; Wu, C.; Wang, C.; Zhang, C.; Li, T.; Zhai, Q.; Yu, L.; Tian, F.; Chen, W. Efficacy of probiotics in multiple sclerosis: A systematic review of preclinical trials and meta-analysis of randomized controlled trials. Food Funct. 2021, 12, 2354–2377. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Furuoka, H.; Kaya, M.; Kuhara, T. Oral administration of probiotic Bifidobacterium breve improves facilitation of hippocampal memory extinction via restoration of aberrant higher induction of neuropsin in an MPTP-induced mouse model of Parkinson’s disease. Biomedicines 2021, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Shin, J.; Kim, S.; Kim, S.; Cho, B.; Park, S.-j.; Park, G.; Shin, H.; Park, M.S.; Kim, J. Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI promotes neuronal rejuvenation in aged mice. Biochem. Biophys. Res. Commun. 2022, 603, 41–48. [Google Scholar] [CrossRef]
- Kong, Q.; Chen, Q.; Mao, X.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Bifidobacterium longum CCFM1077 Ameliorated Neurotransmitter Disorder and Neuroinflammation Closely Linked to Regulation in the Kynurenine Pathway of Autistic-like Rats. Nutrients 2022, 14, 1615. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhao, F.; Liu, Y.; Ma, T.; Jin, H.; Quan, K.; Leng, B.; Zhao, J.; Yuan, X.; Li, Z. Probiotics synergized with conventional regimen in managing Parkinson’s disease. NPJ Park. Dis. 2022, 8, 62. [Google Scholar] [CrossRef]
- D’Amato, A.; Di Cesare Mannelli, L.; Lucarini, E.; Man, A.L.; Le Gall, G.; Branca, J.J.V.; Ghelardini, C.; Amedei, A.; Bertelli, E.; Regoli, M.; et al. Faecal microbiota transplant from aged donor mice affects spatial learning and memory via modulating hippocampal synaptic plasticity- and neurotransmission-related proteins in young recipients. Microbiome 2020, 8, 140. [Google Scholar] [CrossRef]
- Chen, S.J.; Chen, C.C.; Liao, H.Y.; Lin, Y.T.; Wu, Y.W.; Liou, J.M.; Wu, M.S.; Kuo, C.H.; Lin, C.H. Association of Fecal and Plasma Levels of Short-Chain Fatty Acids with Gut Microbiota and Clinical Severity in Patients with Parkinson Disease. Neurology 2022, 98, e848–e858. [Google Scholar] [CrossRef]
- Baldini, F.; Hertel, J.; Sandt, E.; Thinnes, C.C.; Neuberger-Castillo, L.; Pavelka, L.; Betsou, F.; Krüger, R.; Thiele, I. Parkinson’s disease-associated alterations of the gut microbiome predict disease-relevant changes in metabolic functions. BMC Biol. 2020, 18, 62. [Google Scholar] [CrossRef]
- Gao, H.; Jiang, Q.; Ji, H.; Ning, J.; Li, C.; Zheng, H. Type 1 diabetes induces cognitive dysfunction in rats associated with alterations of the gut microbiome and metabolomes in serum and hippocampus. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2019, 1865, 165541. [Google Scholar] [CrossRef]
- Gao, X.; Chang, S.; Liu, S.; Peng, L.; Xie, J.; Dong, W.; Tian, Y.; Sheng, J. Correlations between α-linolenic acid-improved multitissue homeostasis and gut microbiota in mice fed a high-fat diet. mSystems 2020, 5, e00391-20. [Google Scholar] [CrossRef] [PubMed]
- Hertel, J.; Harms, A.C.; Heinken, A.; Baldini, F.; Thinnes, C.C.; Glaab, E.; Vasco, D.A.; Pietzner, M.; Stewart, I.D.; Wareham, N.J. Integrated analyses of microbiome and longitudinal metabolome data reveal microbial-host interactions on sulfur metabolism in Parkinson’s disease. Cell Rep. 2019, 29, 1767–1777. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, A.; Cattane, N.; Galluzzi, S.; Provasi, S.; Lopizzo, N.; Festari, C.; Ferrari, C.; Guerra, U.P.; Paghera, B.; Muscio, C.; et al. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol. Aging 2017, 49, 60–68. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria with Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Yang, X.; Xu, S.; Huang, P.; Li, B.; Du, J.; He, Y.; Su, B.; Xu, L.-M.; Wang, L. Gut metagenomics-derived genes as potential biomarkers of Parkinson’s disease. Brain 2020, 143, 2474–2489. [Google Scholar] [CrossRef]
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 2015, 6, 81. [Google Scholar] [CrossRef]
- Reunanen, J.; Kainulainen, V.; Huuskonen, L.; Ottman, N.; Belzer, C.; Huhtinen, H.; de Vos, W.M.; Satokari, R. Akkermansia muciniphila Adheres to Enterocytes and Strengthens the Integrity of the Epithelial Cell Layer. Appl. Env. Microbiol. 2015, 81, 3655–3662. [Google Scholar] [CrossRef]
- Corazziari, E.S. Intestinal mucus barrier in normal and inflamed colon. J. Pediatr. Gastroenterol. Nutr. 2009, 48, S54–S55. [Google Scholar] [CrossRef]
- Yan, Z.; Yang, F.; Sun, L.; Yu, J.; Sun, L.; Si, Y.; Yao, L. Role of gut microbiota-derived branched-chain amino acids in the pathogenesis of Parkinson’s disease: An animal study. Brain Behav. Immun. 2022, 106, 307–321. [Google Scholar] [CrossRef]
- Cui, C.; Han, Y.; Li, H.; Yu, H.; Zhang, B.; Li, G. Curcumin-driven reprogramming of the gut microbiota and metabolome ameliorates motor deficits and neuroinflammation in a mouse model of Parkinson’s disease. Front. Cell Infect. Microbiol. 2022, 12, 887407. [Google Scholar] [CrossRef] [PubMed]
- Gentile, C.L.; Weir, T.L. The gut microbiota at the intersection of diet and human health. Science 2018, 362, 776–780. [Google Scholar] [CrossRef]
- Neis, E.P.; Dejong, C.H.; Rensen, S.S. The role of microbial amino acid metabolism in host metabolism. Nutrients 2015, 7, 2930–2946. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Nishiwaki, H.; Ito, M.; Ishida, T.; Hamaguchi, T.; Maeda, T.; Kashihara, K.; Tsuboi, Y.; Ueyama, J.; Shimamura, T.; Mori, H.; et al. Meta-Analysis of Gut Dysbiosis in Parkinson’s Disease. Mov. Disord. 2020, 35, 1626–1635. [Google Scholar] [CrossRef]
- Yudkoff, M. Interactions in the Metabolism of Glutamate and the Branched-Chain Amino Acids and Ketoacids in the CNS. Neurochem. Res. 2017, 42, 10–18. [Google Scholar] [CrossRef]
- Yielding, K.L.; Tomkins, G.M. An effect of L-leucine and other essential amino acids on the structure and activity of glutamic dehydrogenase. Proc. Natl. Acad. Sci. USA 1961, 47, 983–989. [Google Scholar] [CrossRef]
- Wang, J.; Wang, F.; Mai, D.; Qu, S. Molecular Mechanisms of Glutamate Toxicity in Parkinson’s Disease. Front. Neurosci. 2020, 14, 585584. [Google Scholar] [CrossRef]
- Lynch, C.J.; Adams, S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 2014, 10, 723–736. [Google Scholar] [CrossRef]
- De Simone, R.; Vissicchio, F.; Mingarelli, C.; De Nuccio, C.; Visentin, S.; Ajmone-Cat, M.A.; Minghetti, L. Branched-chain amino acids influence the immune properties of microglial cells and their responsiveness to pro-inflammatory signals. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2013, 1832, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Scarnà, A.; McTavish, S.F.; Cowen, P.J.; Goodwin, G.M.; Rogers, R.D. The effects of a branched chain amino acid mixture supplemented with tryptophan on biochemical indices of neurotransmitter function and decision-making. Psychopharmacology 2005, 179, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Tărlungeanu, D.C.; Deliu, E.; Dotter, C.P.; Kara, M.; Janiesch, P.C.; Scalise, M.; Galluccio, M.; Tesulov, M.; Morelli, E.; Sonmez, F.M.; et al. Impaired Amino Acid Transport at the Blood Brain Barrier Is a Cause of Autism Spectrum Disorder. Cell 2016, 167, 1481–1494. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, C.; Yu, L.; Li, Y.; Guo, H.; Zhai, Q.; Chen, W.; Tian, F. Lactobacillus plantarum CCFM405 against Rotenone-Induced Parkinson’s Disease Mice via Regulating Gut Microbiota and Branched-Chain Amino Acids Biosynthesis. Nutrients 2023, 15, 1737. https://doi.org/10.3390/nu15071737
Chu C, Yu L, Li Y, Guo H, Zhai Q, Chen W, Tian F. Lactobacillus plantarum CCFM405 against Rotenone-Induced Parkinson’s Disease Mice via Regulating Gut Microbiota and Branched-Chain Amino Acids Biosynthesis. Nutrients. 2023; 15(7):1737. https://doi.org/10.3390/nu15071737
Chicago/Turabian StyleChu, Chuanqi, Leilei Yu, Yiwen Li, Hang Guo, Qixiao Zhai, Wei Chen, and Fengwei Tian. 2023. "Lactobacillus plantarum CCFM405 against Rotenone-Induced Parkinson’s Disease Mice via Regulating Gut Microbiota and Branched-Chain Amino Acids Biosynthesis" Nutrients 15, no. 7: 1737. https://doi.org/10.3390/nu15071737
APA StyleChu, C., Yu, L., Li, Y., Guo, H., Zhai, Q., Chen, W., & Tian, F. (2023). Lactobacillus plantarum CCFM405 against Rotenone-Induced Parkinson’s Disease Mice via Regulating Gut Microbiota and Branched-Chain Amino Acids Biosynthesis. Nutrients, 15(7), 1737. https://doi.org/10.3390/nu15071737






