In Vitro Effect of Enzymes and Human Milk Oligosaccharides on FODMAP Digestion and Fecal Microbiota Composition
Abstract
1. Introduction
2. Materials and Methods
2.1. Upper Gastrointestinal Tract Simulation
2.2. Carbohydrate Quantification and FOS Profiling
2.3. Short-Term Colonic Incubations
2.4. QuintSHIME® Model
2.5. Microbial Community Activity
2.6. Microbial Community Composition Analysis by qPCR
2.7. Microbial Community Composition Analysis by Quantitative 16S-Targeted Sequencing and Flow Cytometry
2.8. Statistical Analysis
3. Results
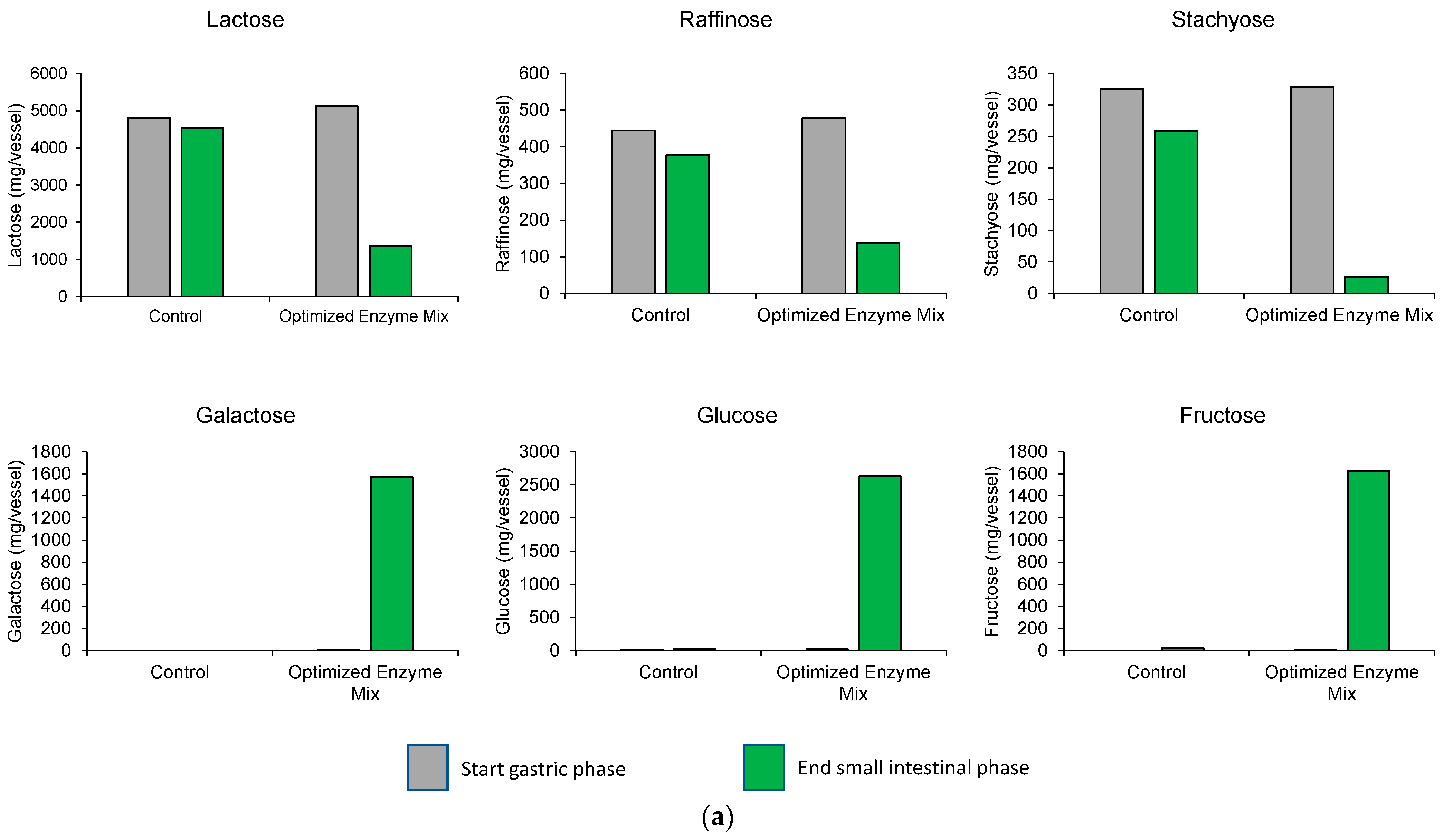
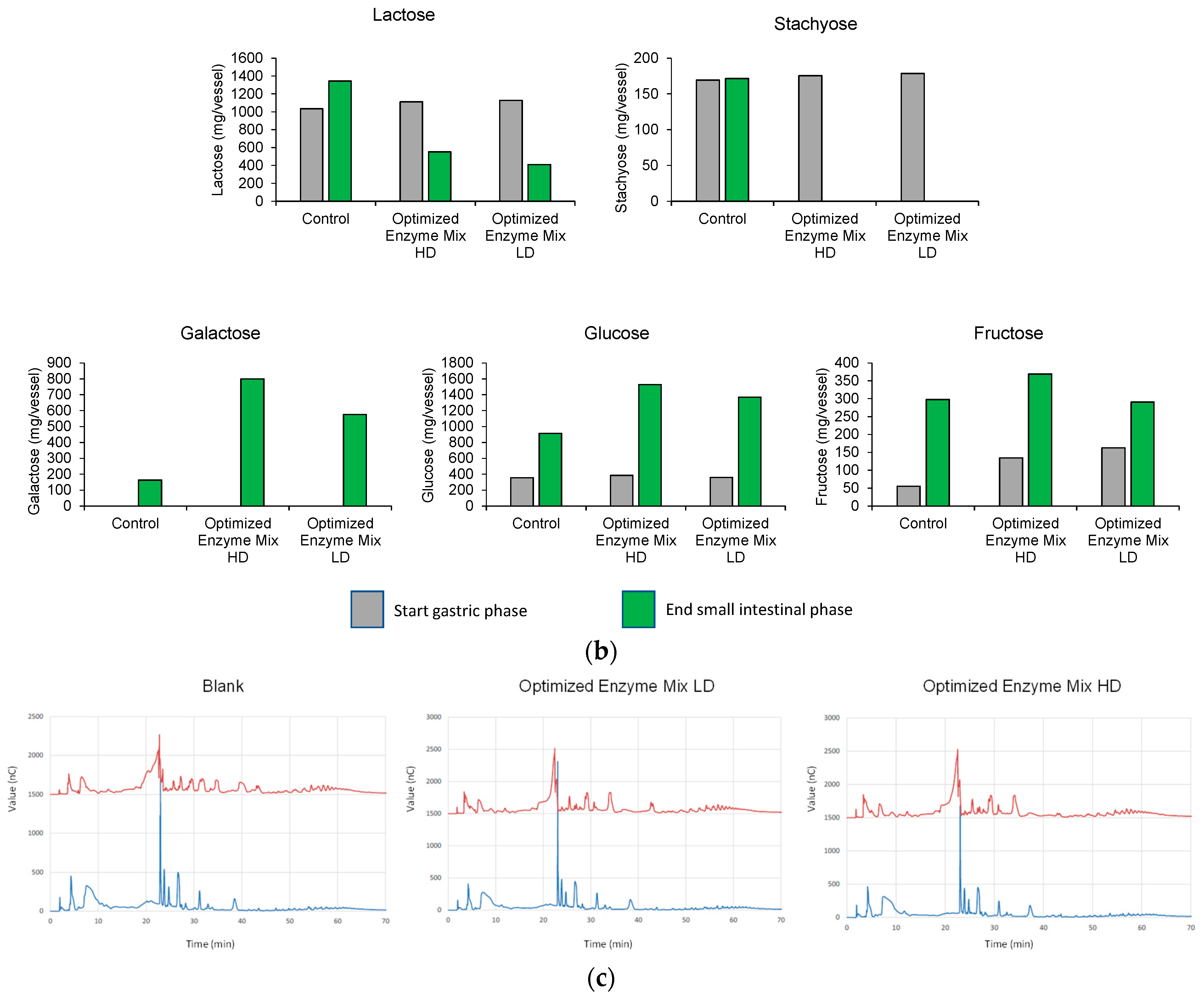
3.1. Upper GIT Simulation
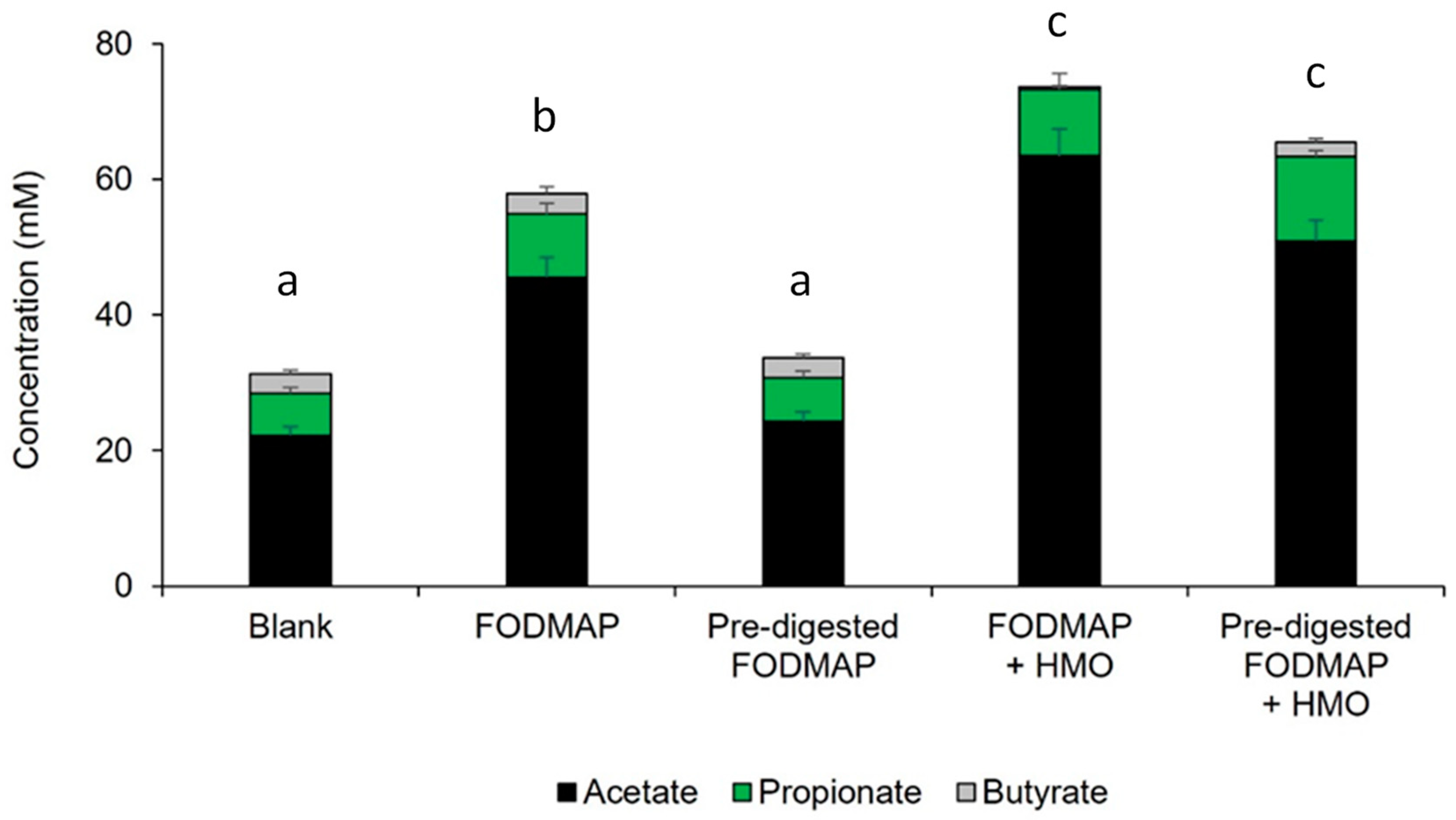
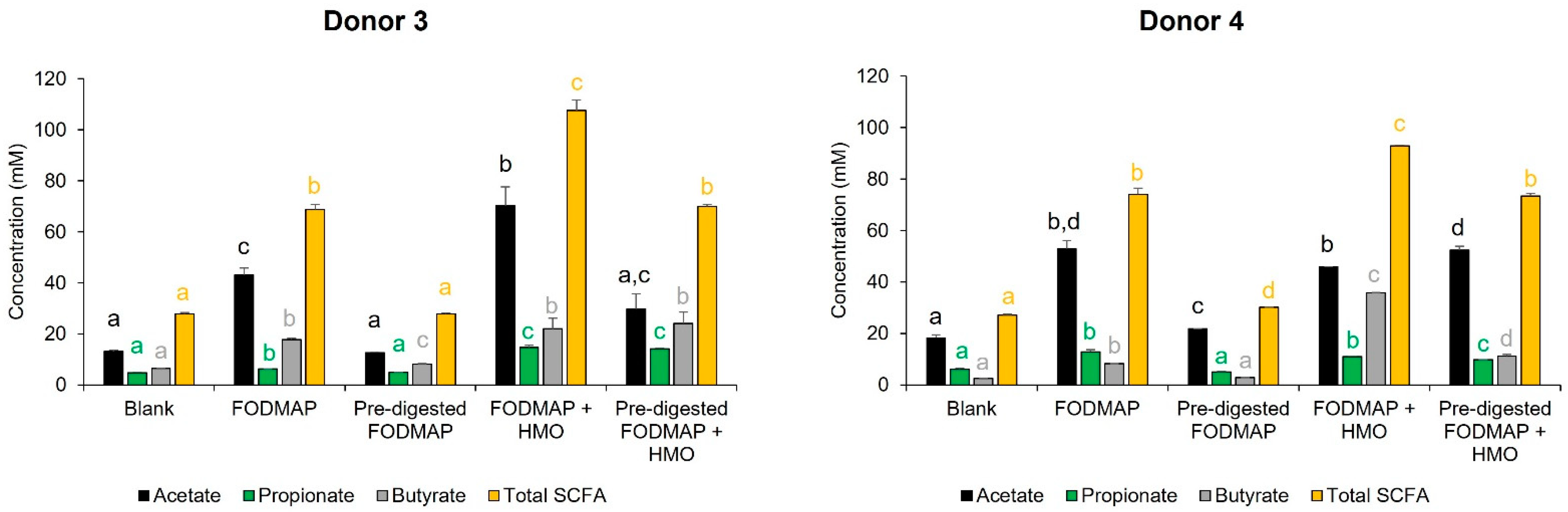
3.2. Short-Term Colonic Incubations
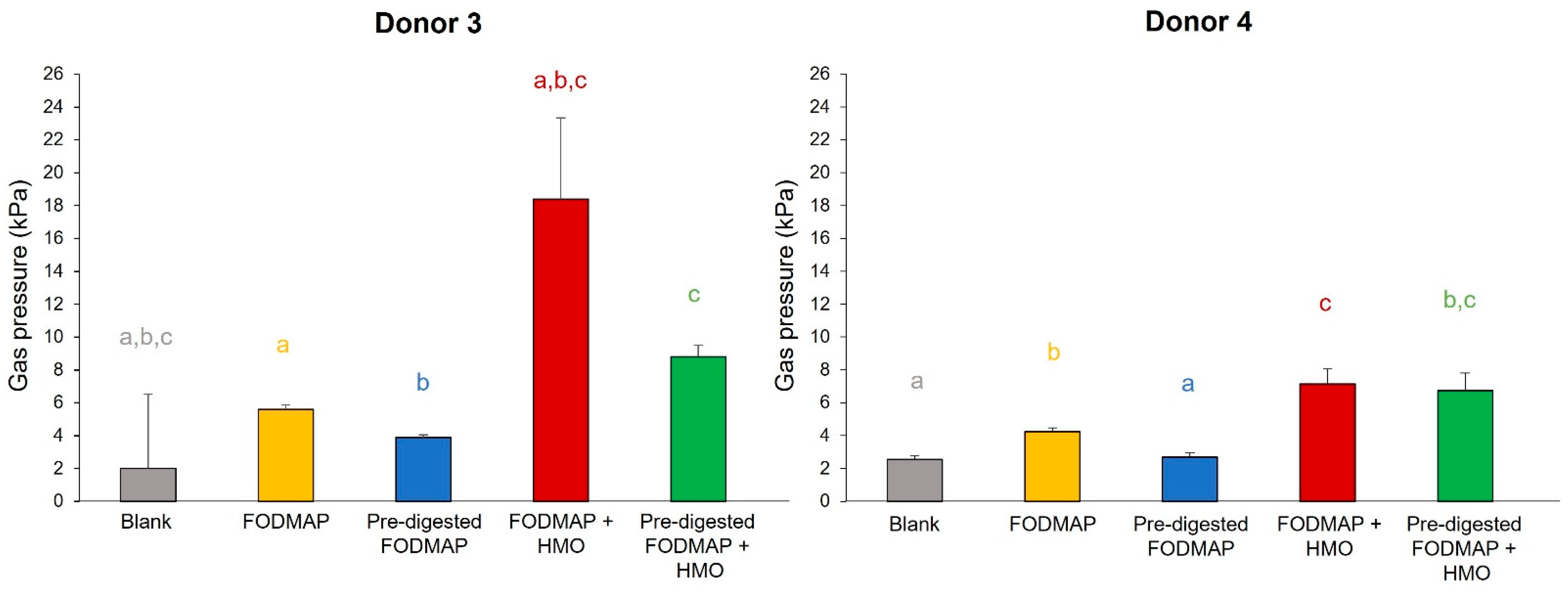
3.3. QuintSHIME® Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gibson, P.R.; Shepherd, S.J. Personal view: Food for thought--western lifestyle and susceptibility to Crohn’s disease. The FODMAP hypothesis. Aliment. Pharmacol. Ther. 2005, 21, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- Gibson, P.R.; Shepherd, S.J. Evidence-based dietary management of functional gastrointestinal symptoms: The FODMAP approach. J. Gastroenterol. Hepatol. 2010, 25, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Murray, K.; Wilkinson-Smith, V.; Hoad, C.; Costigan, C.; Cox, E.; Lam, C.; Marciani, L.; Gowland, P.; Spiller, R.C. Differential effects of FODMAPs (fermentable oligo-, di-, mono-saccharides and polyols) on small and large intestinal contents in healthy subjects shown by MRI. Am. J. Gastroenterol. 2014, 109, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.S.; Gearry, R.B.; Muir, J.G.; Irving, P.M.; Rose, R.; Rosella, O.; Haines, M.L.; Shepherd, S.J.; Gibson, P.R. Dietary poorly absorbed, short-chain carbohydrates increase delivery of water and fermentable substrates to the proximal colon. Aliment. Pharmacol. Ther. 2010, 31, 874–882. [Google Scholar] [CrossRef]
- Sperber, A.D.; Dumitrascu, D.; Fukudo, S.; Gerson, C.; Ghoshal, U.C.; Gwee, K.A.; Hungin, A.P.S.; Kang, J.Y.; Minhu, C.; Schmulson, M.; et al. The global prevalence of IBS in adults remains elusive due to the heterogeneity of studies: A Rome Foundation working team literature review. Gut 2017, 66, 1075–1082. [Google Scholar] [CrossRef]
- Major, G.; Pritchard, S.; Murray, K.; Alappadan, J.P.; Hoad, C.L.; Marciani, L.; Gowland, P.; Spiller, R. Colon hypersensitivity to distension, rather than excessive gas production, produces carbohydrate-related symptoms in individuals with irritable bowel syndrome. Gastroenterology 2017, 152, 124–133.e2. [Google Scholar] [CrossRef]
- Gibson, P.R.; Halmos, E.P.; Muir, J.G. Review article: FODMAPS, prebiotics and gut health-the FODMAP hypothesis revisited. Aliment. Pharmacol. Ther. 2020, 52, 233–246. [Google Scholar] [CrossRef]
- Bellini, M.; Tonarelli, S.; Nagy, A.G.; Pancetti, A.; Costa, F.; Ricchiuti, A.; de Bortoli, N.; Mosca, M.; Marchi, S.; Rossi, A. Low FODMAP diet: Evidence, doubts, and hopes. Nutrients 2020, 12, 148. [Google Scholar] [CrossRef]
- Lacy, B.E.; Pimentel, M.; Brenner, D.M.; Chey, W.D.; Keefer, L.A.; Long, M.D.; Moshiree, B. ACG clinical guideline: Management of irritable bowel syndrome. Am. J. Gastroenterol. 2021, 116, 17–44. [Google Scholar] [CrossRef]
- McKenzie, Y.A.; Bowyer, R.K.; Leach, H.; Gulia, P.; Horobin, J.; O’Sullivan, N.A.; Pettitt, C.; Reeves, L.B.; Seamark, L.; Williams, M.; et al. British Dietetic Association systematic review and evidence-based practice guidelines for the dietary management of irritable bowel syndrome in adults (2016 update). J. Hum. Nutr. Diet. 2016, 29, 549–575. [Google Scholar] [CrossRef]
- O’Keeffe, M.; Lomer, M.C. Who should deliver the low FODMAP diet and what educational methods are optimal: A review. J. Gastroenterol. Hepatol. 2017, 32 (Suppl. 1), 23–26. [Google Scholar] [CrossRef]
- Barrett, J.S. How to institute the low-FODMAP diet. J. Gastroenterol. Hepatol. 2017, 32 (Suppl. 1), 8–10. [Google Scholar] [CrossRef]
- Maagaard, L.; Ankersen, D.V.; Vegh, Z.; Burisch, J.; Jensen, L.; Pedersen, N.; Munkholm, P. Follow-up of patients with functional bowel symptoms treated with a low FODMAP diet. World J. Gastroenterol. 2016, 22, 4009–4019. [Google Scholar] [CrossRef]
- O’Keeffe, M.; Jansen, C.; Martin, L.; Williams, M.; Seamark, L.; Staudacher, H.M.; Irving, P.M.; Whelan, K.; Lomer, M.C. Long-term impact of the low-FODMAP diet on gastrointestinal symptoms, dietary intake, patient acceptability, and healthcare utilization in irritable bowel syndrome. Neurogastroenterol. Motil. 2018, 30, e13154. [Google Scholar] [CrossRef]
- Gearry, R.B.; Irving, P.M.; Barrett, J.S.; Nathan, D.M.; Shepherd, S.J.; Gibson, P.R. Reduction of dietary poorly absorbed short-chain carbohydrates (FODMAPs) improves abdominal symptoms in patients with inflammatory bowel disease-a pilot study. J. Crohns. Colitis. 2009, 3, 8–14. [Google Scholar] [CrossRef]
- Halmos, E.P.; Christophersen, C.T.; Bird, A.R.; Shepherd, S.J.; Gibson, P.R.; Muir, J.G. Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut 2015, 64, 93–100. [Google Scholar] [CrossRef]
- Dieterich, W.; Schuppan, D.; Schink, M.; Schwappacher, R.; Wirtz, S.; Agaimy, A.; Neurath, M.F.; Zopf, Y. Influence of low FODMAP and gluten-free diets on disease activity and intestinal microbiota in patients with non-celiac gluten sensitivity. Clin. Nutr. 2019, 38, 697–707. [Google Scholar] [CrossRef]
- Ianiro, G.; Pecere, S.; Giorgio, V.; Gasbarrini, A.; Cammarota, G. Digestive enzyme supplementation in gastrointestinal diseases. Curr. Drug. Metab. 2016, 17, 187–193. [Google Scholar] [CrossRef]
- Musilova, S.; Rada, V.; Vlkova, E.; Bunesova, V. Beneficial effects of human milk oligosaccharides on gut microbiota. Benef. Microbes. 2014, 5, 273–283. [Google Scholar] [CrossRef]
- Suligoj, T.; Vigsnaes, L.K.; Abbeele, P.V.D.; Apostolou, A.; Karalis, K.; Savva, G.M.; McConnell, B.; Juge, N. Effects of human milk oligosaccharides on the adult gut microbiota and barrier function. Nutrients 2020, 12, 2808. [Google Scholar] [CrossRef]
- Palsson, O.S.; Peery, A.; Seitzberg, D.; Amundsen, I.D.; McConnell, B.; Simren, M. Human milk oligosaccharides support normal bowel function and improve symptoms of irritable bowel syndrome: A multicenter, open-label trial. Clin. Transl. Gastroenterol. 2020, 11, e00276. [Google Scholar] [CrossRef] [PubMed]
- Iribarren, C.; Tornblom, H.; Aziz, I.; Magnusson, M.K.; Sundin, J.; Vigsnaes, L.K.; Amundsen, I.D.; McConnell, B.; Seitzberg, D.; Ohman, L.; et al. Human milk oligosaccharide supplementation in irritable bowel syndrome patients: A parallel, randomized, double-blind, placebo-controlled study. Neurogastroenterol. Motil. 2020, 32, e13920. [Google Scholar] [CrossRef] [PubMed]
- Mackie, A.; Rigby, N. InfoGest consensus method. In The Impact of Food Bioactives on Health; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 13–22. [Google Scholar]
- Van den Abbeele, P.; Kamil, A.; Fleige, L.; Chung, Y.; De Chavez, P.; Marzorati, M. Different oat ingredients stimulate specific microbial metabolites in the gut microbiome of three human individuals in vitro. ACS Omega 2018, 3, 12446–12456. [Google Scholar] [CrossRef] [PubMed]
- Molly, K.; Vande Woestyne, M.; Verstraete, W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 1993, 39, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Possemiers, S.; Verthe, K.; Uyttendaele, S.; Verstraete, W. PCR-DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol. Ecol. 2004, 49, 495–507. [Google Scholar] [CrossRef]
- De Weirdt, R.; Possemiers, S.; Vermeulen, G.; Moerdijk-Poortvliet, T.C.; Boschker, H.T.; Verstraete, W.; Van de Wiele, T. Human faecal microbiota display variable patterns of glycerol metabolism. FEMS Microbiol. Ecol. 2010, 74, 601–611. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Verstrepen, L.; Ghyselinck, J.; Albers, R.; Marzorati, M.; Mercenier, A. A novel non-digestible, carrot-derived polysaccharide (cRG-I) selectively modulates the human gut microbiota while promoting gut barrier integrity: An integrated in vitro approach. Nutrients 2020, 12, 1917. [Google Scholar] [CrossRef]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Dore, J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel. Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic. Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- De Paepe, K.; Kerckhof, F.M.; Verspreet, J.; Courtin, C.M.; Van de Wiele, T. Inter-individual differences determine the outcome of wheat bran colonization by the human gut microbiome. Environ. Microbiol. 2017, 19, 3251–3267. [Google Scholar] [CrossRef]
- Van Nevel, S.; Koetzsch, S.; Weilenmann, H.U.; Boon, N.; Hammes, F. Routine bacterial analysis with automated flow cytometry. J. Microbiol. Methods 2013, 94, 73–76. [Google Scholar] [CrossRef]
- Shepherd, S.J.; Lomer, M.C.; Gibson, P.R. Short-chain carbohydrates and functional gastrointestinal disorders. Am. J. Gastroenterol. 2013, 108, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Ong, D.K.; Mitchell, S.B.; Barrett, J.S.; Shepherd, S.J.; Irving, P.M.; Biesiekierski, J.R.; Smith, S.; Gibson, P.R.; Muir, J.G. Manipulation of dietary short chain carbohydrates alters the pattern of gas production and genesis of symptoms in irritable bowel syndrome. J. Gastroenterol. Hepatol. 2010, 25, 1366–1373. [Google Scholar] [CrossRef]
- Gill, P.A.; van Zelm, M.C.; Muir, J.G.; Gibson, P.R. Review article: Short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment. Pharmacol. Ther. 2018, 48, 15–34. [Google Scholar] [CrossRef]
- Staudacher, H.M.; Lomer, M.C.; Anderson, J.L.; Barrett, J.S.; Muir, J.G.; Irving, P.M.; Whelan, K. Fermentable carbohydrate restriction reduces luminal bifidobacteria and gastrointestinal symptoms in patients with irritable bowel syndrome. J. Nutr. 2012, 142, 1510–1518. [Google Scholar] [CrossRef]
- Halmos, E.P.; Christophersen, C.T.; Bird, A.R.; Shepherd, S.J.; Muir, J.G.; Gibson, P.R. Consistent prebiotic effect on gut microbiota with altered FODMAP intake in patients with Crohn’s disease: A randomised, controlled cross-over trial of well-defined diets. Clin. Transl. Gastroenterol. 2016, 7, e164. [Google Scholar] [CrossRef]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Shen, J.; Ran, Z.H. Association between Faecalibacterium prausnitzii reduction and inflammatory bowel disease: A meta-analysis and systematic review of the literature. Gatroenterol. Res. Pract. 2014, 2014, 872725. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Strukelj, B. The influence of probiotics on the Firmicutes/Bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The controversial role of human gut Lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef]
- Sasaki, K.; Inoue, J.; Sasaki, D.; Hoshi, N.; Shirai, T.; Fukuda, I.; Azuma, T.; Kondo, A.; Osawa, R. Construction of a model culture system of human colonic microbiota to detect decreased Lachnospiraceae abundance and butyrogenesis in the feces of ulcerative colitis patients. Biotechnol. J. 2019, 14, e1800555. [Google Scholar] [CrossRef]
- Zhang, C.; Yin, A.; Li, H.; Wang, R.; Wu, G.; Shen, J.; Zhang, M.; Wang, L.; Hou, Y.; Ouyang, H.; et al. Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. eBioMedicine 2015, 2, 968–984. [Google Scholar] [CrossRef]




| Digestion Rate | ||||
|---|---|---|---|---|
| FODMAP Enzyme | FOS | Stachyose | Raffinose | Lactose |
| Maxinvert | ||||
| 500 SU | 46% | – | – | – |
| 2500 SU | 100% | – | – | – |
| 10,000 SU | 100% | – | – | – |
| Fibractase | ||||
| 600 GalU | – | 55% | 24% | – |
| 1200 GalU | – | 63% | 31% | – |
| 2400 GalU | – | 76% | 52% | – |
| Tolerase L | ||||
| 1000 ALU | – | – | – | 56% |
| 2500 ALU | – | – | – | 76% |
| 10,000 ALU | – | – | – | 94% |
| Reciprocal Simpson Diversity Index | |||||
|---|---|---|---|---|---|
| Donor | Blank | FODMAP | Pre-Digested FODMAP | FODMAP + HMO | Pre-Digested FODMAP + HMO |
| Donor 3 | 13.0 | 3.2 * | 11.5 | 5.3 * | 10.2 |
| Donor 4 | 6.5 | 3.7 * | 6.0 | 4.0 * | 5.4 |
| Average | 9.7 | 3.5 | 8.7 | 4.7 * | 7.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, V.T.; Steinert, R.E.; Duysburgh, C.; Ghyselinck, J.; Marzorati, M.; Dekker, P.J.T. In Vitro Effect of Enzymes and Human Milk Oligosaccharides on FODMAP Digestion and Fecal Microbiota Composition. Nutrients 2023, 15, 1637. https://doi.org/10.3390/nu15071637
Pham VT, Steinert RE, Duysburgh C, Ghyselinck J, Marzorati M, Dekker PJT. In Vitro Effect of Enzymes and Human Milk Oligosaccharides on FODMAP Digestion and Fecal Microbiota Composition. Nutrients. 2023; 15(7):1637. https://doi.org/10.3390/nu15071637
Chicago/Turabian StylePham, Van T., Robert E. Steinert, Cindy Duysburgh, Jonas Ghyselinck, Massimo Marzorati, and Peter J. T. Dekker. 2023. "In Vitro Effect of Enzymes and Human Milk Oligosaccharides on FODMAP Digestion and Fecal Microbiota Composition" Nutrients 15, no. 7: 1637. https://doi.org/10.3390/nu15071637
APA StylePham, V. T., Steinert, R. E., Duysburgh, C., Ghyselinck, J., Marzorati, M., & Dekker, P. J. T. (2023). In Vitro Effect of Enzymes and Human Milk Oligosaccharides on FODMAP Digestion and Fecal Microbiota Composition. Nutrients, 15(7), 1637. https://doi.org/10.3390/nu15071637





