Identification of Natural Compounds of the Apple as Inhibitors against Cholinesterase for the Treatment of Alzheimer’s Disease: An In Silico Molecular Docking Simulation and ADMET Study
Abstract
1. Introduction
2. Material and Methods
2.1. Preparation of Ligand Structures
2.2. Preparation of Enzyme Structures
2.3. Virtual Screening
2.4. Molecular Interaction Analysis
2.5. Drug-Likeness and ADMET
2.6. Molecular Dynamics Simulation
3. Results and Discussions
3.1. Docking Results
3.2. Drug-Likeness and ADMET Analysis
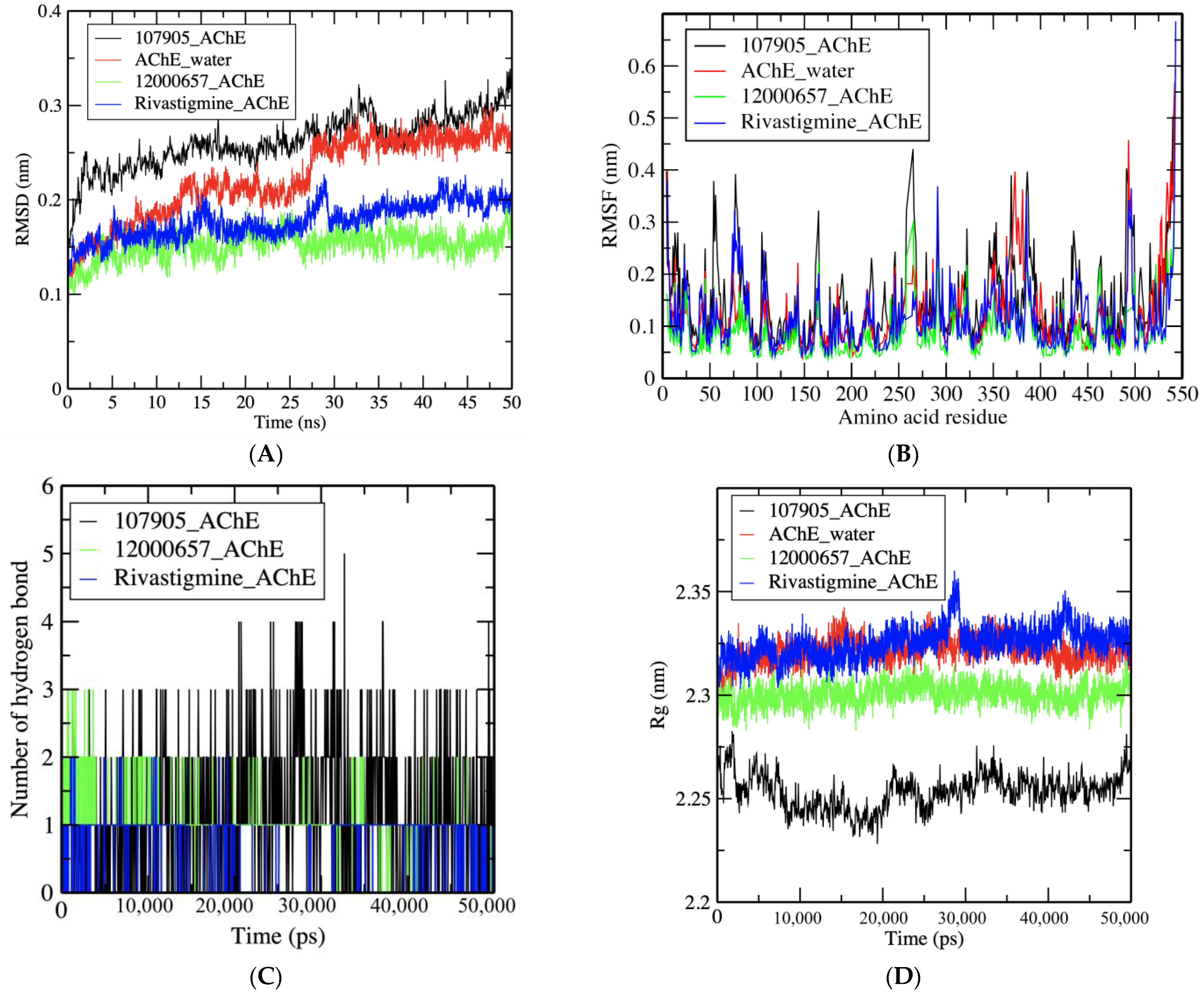
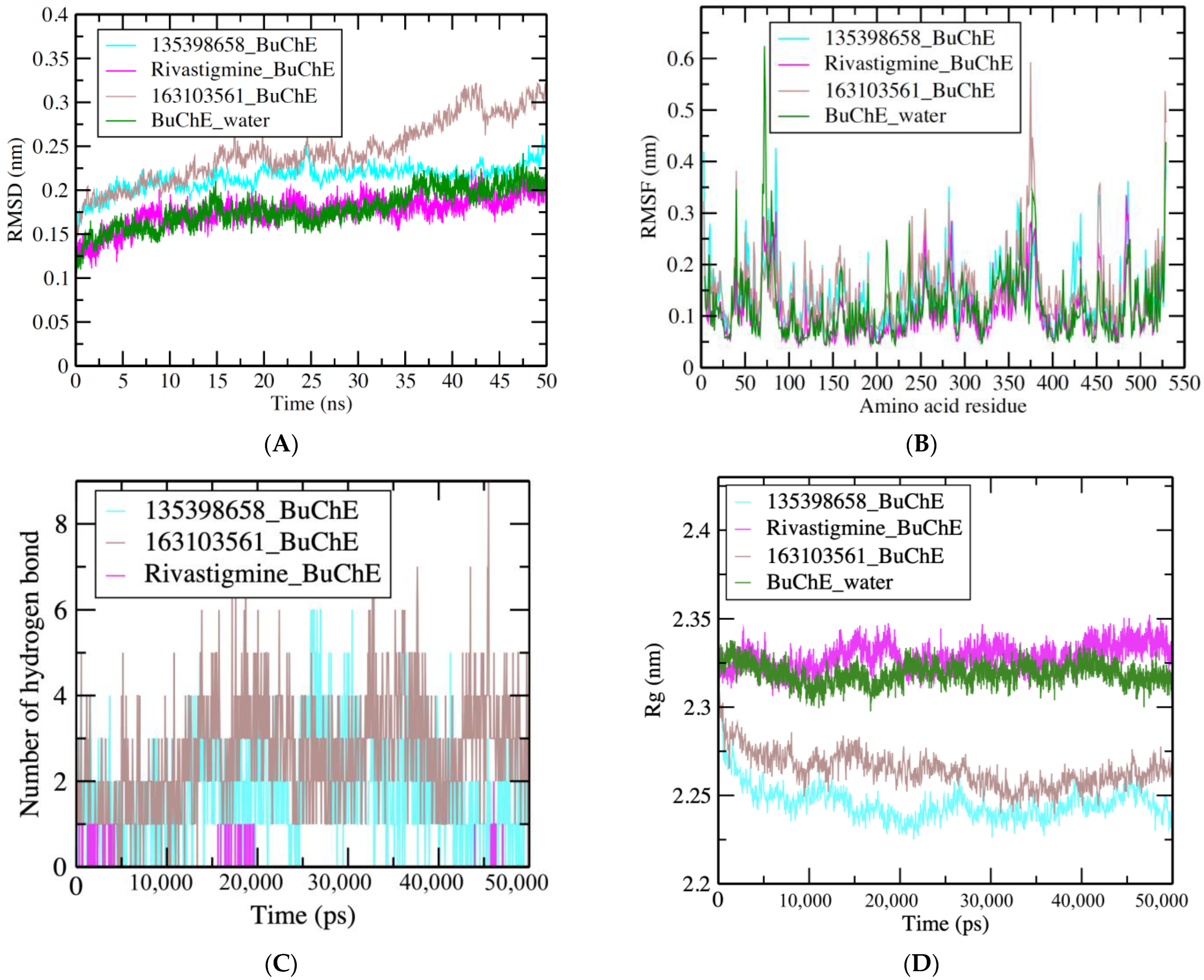
3.3. MDS Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hashimoto, M.; Rockenstein, E.; Crews, L.; Masliah, E. Role of protein aggregation in mitochondrial dysfunction and neurodegeneration in Alzheimer’s and Parkinson’s diseases. Neuromol. Med. 2003, 4, 21–36. [Google Scholar] [CrossRef]
- Salawu, F.K.; Umar, J.T.; Olokoba, A.B. Alzheimer′s disease: A review of recent developments. Ann. Afr. Med. 2011, 10, 73–79. [Google Scholar] [CrossRef]
- Rutten, B.P.F.; Steinbusch, H.W.M.; Korr, H.; Schmitz, C. Antioxidants and Alzheimer’s disease: From bench to bedside (and back again). Curr. Opin. Clin. Nutr. Metab. Care 2002, 5, 645–651. [Google Scholar] [CrossRef]
- Ansari, N.; Khodagholi, F. Natural products as promising drug candidates for the treatment of Alzheimer’s disease: Molecular mechanism aspect. Curr. Neuropharmacol. 2013, 11, 414–429. [Google Scholar] [CrossRef] [PubMed]
- Greig, N.H.; Lahiri, D.K.; Sambamurti, K. Butyrylcholinesterase: An important new target in Alzheimer’s disease therapy. Int. Psychogeriatr. 2002, 14, 77–91. [Google Scholar] [CrossRef]
- Orhan, I.; Kartal, M.; Tosun, F.; Şener, B. Screening of various phenolic acids and flavonoid derivatives for their anticholinesterase potential. Z. Für Nat. C 2007, 62, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, T.; Rao, P.P.N. Design, synthesis and evaluation of 2,4-disubstituted pyrimidines as cholinesterase inhibitors. Bioorganic Med. Chem. Lett. 2010, 20, 3606–3609. [Google Scholar] [CrossRef] [PubMed]
- Mesulam, M. The cholinergic lesion of Alzheimer’s disease: Pivotal factor or side show? Learn. Mem. 2004, 11, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Osborn, G.G.; Saunders, A.V. Current treatments for patients with Alzheimer disease. J. Am. Osteopath. Assoc. 2010, 110 (Suppl. S8), S16–S26. [Google Scholar]
- Suh, Y.-H. Amyloid precursor protein, presenilins, and alpha-synuclein: Molecular pathogenesis and pharmacological applications in Alzheimer’s disease. Pharmacol. Rev. 2002, 54, 469–525. [Google Scholar] [CrossRef] [PubMed]
- Mohandas, E.; Rajmohan, V.; Raghunath, B. Neurobiology of Alzheimer’s disease. Indian J. Psychiatry 2009, 51, 55–61. [Google Scholar] [CrossRef]
- Hegde, M.L.; Hegde, P.M.; Rao, K.S.; Mitra, S. Oxidative genome damage and its repair in neurodegenerative diseases: Function of transition metals as a double-edged sword. J. Alzheimer’s Dis. 2011, 24, 183–198. [Google Scholar] [CrossRef]
- Racchi, M.; Mazzucchelli, M.; Lenzken, S.C.; Porrello, E.; Lanni, C.; Govoni, S. Role of acetylcholinesterase inhibitors in the regulation of amyloid β precursor protein (AβPP) metabolism. Chem. Biol. Interact. 2005, 157–158, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, S.; Takano, K. Amyloid fibrils from the viewpoint of protein folding. Cell. Mol. Life Sci. (CMLS) 2004, 61, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Inestrosa, N.C.; Alvarez, A.; Pérez, C.A.; Moreno, R.D.; Vicente, M.; Linker, C.; Casanueva, O.I.; Soto, C.; Garrido, J. Acetylcholinesterase accelerates assembly of amyloid-β-peptides into Alzheimer’s fibrils: Possible role of the peripheral site of the enzyme. Neuron 1996, 16, 881–891. [Google Scholar] [CrossRef]
- Talesa, V.N. Acetylcholinesterase in Alzheimer’s disease. Mech. Ageing Dev. 2001, 122, 1961–1969. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Su, T.; Li, X. Natural products as sources of new lead compounds for the treatment of Alzheimer’s disease. Curr. Top. Med. Chem. 2013, 13, 1864–1878. [Google Scholar] [CrossRef]
- Ved, H.S.; Best, J.M.; Dave, J.R.; Doctor, B.P. Comparative inhibition of acetylcholinesterase by tacrine, physostigmine and Huperzine in the adult Rat Brain. In Enzymes of the Cholinesterase Family; Quinn, D.M., Balasubramanian, A.S., Doctor, B.P., Taylor, P., Eds.; Springer: Boston, MA, USA, 1995; pp. 477–478. [Google Scholar]
- Howes, M.-J.R.; Perry, N.S.; Houghton, P.J. Plants with traditional uses and activities, relevant to the management of Alzheimer’s disease and other cognitive disorders. Phytother. Res. 2003, 17, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Sramek, J.J.; Frackiewicz, E.J.; Cutler, N.R. Review of the acetylcholinesterase inhibitor galanthamine. Expert Opin. Investig. Drugs 2000, 9, 2393–2402. [Google Scholar] [CrossRef]
- Okello, E.J.; Savelev, S.U.; Perry, E.K. In Vitro anti-β-secretase and dual anti-cholinesterase activities of Camellia sinensis L. (TEA) relevant to treatment of dementia. Phytother. Res. 2004, 18, 624–627. [Google Scholar] [CrossRef]
- Ellman, G.L.; Courtney, K.D.; Andres, V.; Featherstone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Kumar, V.; Mal, M.; Houghton, P.J. Acetylcholinesterase inhibitors from plants. Phytomedicine 2007, 14, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Roche, M.; Dufour, C.; Loonis, M.; Reist, M.; Carrupt, P.-A.; Dangles, O. Olive phenols efficiently inhibit the oxidation of serum albumin-bound linoleic acid and butyrylcholine esterase. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2009, 1790, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Gilani, A.-H. Inhibitory effect of curcuminoids on acetylcholinesterase activity and attenuation of scopolamine-induced amnesia may explain medicinal use of turmeric in Alzheimer’s disease. Pharmacol. Biochem. Behav. 2009, 91, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.-H.; Ku, C.-Y.; Ho, C.-T.; Chen, C.-S.; Huang, C.-S.; Lee, C.-H.; Chen, L.-C.; Pan, M.-H.; Chang, H.-W.; Chang, C.-H.; et al. Tea Polyphenol (−)-epigallocatechin-3-gallate inhibits nicotine- and estrogen-induced α9-nicotinic acetylcholine receptor upregulation in human breast cancer cells. Mol. Nutr. Food Res. 2010, 55, 455–466. [Google Scholar] [CrossRef]
- Zhang, L.; Cao, H.; Wen, J.; Xu, M. Green tea polyphenol (–)-epigallocatechin-3-gallate enhances the inhibitory effect of Huperzine A on acetylcholinesterase by increasing the affinity with serum albumin. Nutr. Neurosci. 2009, 12, 142–148. [Google Scholar] [CrossRef]
- Aboul Ezz, H.S.; Khadrawy, Y.A.; Mourad, I.M. The effect of bisphenol A on some oxidative stress parameters and acetylcholinesterase activity in the heart of male Albino Rats. Cytotechnology 2013, 67, 145–155. [Google Scholar] [CrossRef]
- Akintunde, J.K.; Akintola, T.E.; Hammed, M.O.; Amoo, C.O.; Adegoke, A.M.; Ajisafe, L.O. Naringin protects against bisphenol-a induced oculopathy as implication of cataract in hypertensive rat model. Biomed. Pharmacother. 2020, 126, 110043. [Google Scholar] [CrossRef]
- Flieger, J.; Śniegocki, T.; Dolar-Szczasny, J.; Załuska, W.; Rejdak, R. The First Evidence on the Occurrence of Bisphenol Analogues in the Aqueous Humor of Patients Undergoing Cataract Surgery. J. Clin. Med. 2022, 11, 6402. [Google Scholar] [CrossRef]
- Orhan, I.; Şener, B.; Choudhary, M.I.; Khalid, A. Acetylcholinesterase and butyrylcholinesterase inhibitory activity of some Turkish medicinal plants. J. Ethnopharmacol. 2004, 91, 57–60. [Google Scholar] [CrossRef]
- Chan, A.; Graves, V.; Shea, T.B. Apple juice concentrate maintains acetylcholine levels following dietary compromise. J. Alzheimer’s Dis. 2006, 9, 287–291. [Google Scholar] [CrossRef]
- Ali, B.; Jamal, Q.M.S.; Shams, S.; Al-Wabel, N.A.; Siddiqui, M.U.; Alzohairy, M.A.; Al Karaawi, M.A.; Kumar Kesari, K.; Mushtaq, G.; Kamal, M.A. In Silico analysis of green tea polyphenols as inhibitors of Ache and BCHE enzymes in Alzheimer’s disease treatment. CNS Neurol. Disord.-Drug Targets 2016, 15, 624–628. [Google Scholar] [CrossRef]
- Kennedy, D.O.; Wightman, E.L.; Reay, J.L.; Lietz, G.; Okello, E.J.; Wilde, A.; Haskell, C.F. Effects of resveratrol on cerebral blood flow variables and cognitive performance in humans: A double-blind, placebo-controlled, crossover investigation. Am. J. Clin. Nutr. 2010, 91, 1590–1597. [Google Scholar] [CrossRef]
- Kennedy, D.O.; Wightman, E.L. Herbal extracts and phytochemicals: Plant secondary metabolites and the enhancement of human brain function. Adv. Nutr. 2011, 2, 32–50. [Google Scholar] [CrossRef]
- Kim, H.K.; Kim, M.; Kim, S.; Kim, M.; Chung, J.H. Effects of green tea polyphenol on cognitive and acetylcholinesterase activities. Biosci. Biotechnol. Biochem. 2004, 68, 1977–1979. [Google Scholar] [CrossRef]
- Heo, H.J.; Kim, M.-J.; Lee, J.-M.; Choi, S.J.; Cho, H.-Y.; Hong, B.; Kim, H.-K.; Kim, E.; Shin, D.-H. Naringenin from citrus junos has an inhibitory effect on acetylcholinesterase and a mitigating effect on amnesia. Dement. Geriatr. Cogn. Disord. 2004, 17, 151–157. [Google Scholar] [CrossRef]
- Hertog, M.G.; Hollman, P.C.; Katan, M.B.; Kromhout, D. Intake of potentially anticarcinogenic flavonoids and their determinants in adults in the Netherlands. Nutr. Cancer 1993, 20, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Tota, S.; Awasthi, H.; Kamat, P.K.; Nath, C.; Hanif, K. Protective effect of quercetin against intracerebral streptozotocin induced reduction in cerebral blood flow and impairment of memory in mice. Behav. Brain Res. 2010, 209, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Hansen, R.A.; Gartlehner, G.; Webb, A.P.; Morgan, L.C.; Moore, C.G.; Jonas, D.E. Efficacy and safety of donepezil, galantamine, and rivastigmine for the treatment of Alzheimer’s disease: A systematic review and meta-analysis. Clin. Interv. Aging 2008, 3, 211–225. [Google Scholar] [PubMed]
- Ceymann, M.; Arrigoni, E.; Schärer, H.; Nising, A.B.; Hurrell, R.F. Identification of apples rich in health-promoting flavan-3-ols and phenolic acids by measuring the polyphenol profile. J. Food Compos. Anal 2012, 26, 128–135. [Google Scholar] [CrossRef]
- Boyer, J.; Liu, H.R. Antioxidants of Apples. N. Y. Fruit Q. 2004, 11, 11–15. [Google Scholar]
- Wojdyło, A.; Oszmiański, J.; Laskowski, P. Polyphenolic compounds and antioxidant activity of new and Old Apple varieties. J. Agric. Food Chem. 2008, 56, 6520–6530. [Google Scholar] [CrossRef]
- Escarpa, A.; González, M.C. High-performance liquid chromatography with diode-array detection for the determination of phenolic compounds in peel and pulp from different Apple varieties. J. Chromatogr. A 1998, 823, 331–337. [Google Scholar] [CrossRef]
- Hyson, D.A. A comprehensive review of apples and Apple components and their relationship to human health. Adv. Nutr. 2011, 2, 408–420. [Google Scholar] [CrossRef]
- Wimo, A.; Jönsson, L.; Gustavsson, A.; McDaid, D.; Ersek, K.; Georges, J.; Gulácsi, L.; Karpati, K.; Kenigsberg, P.; Valtonen, H. The economic impact of dementia in Europe in 2008-cost estimates from the Eurocode Project. Int. J. Geriatr. Psychiatry 2010, 26, 825–832. [Google Scholar] [CrossRef]
- Joseph, J.A.; Shukitt-Hale, B.; Denisova, N.A.; Bielinski, D.; Martin, A.; McEwen, J.J.; Bickford, P.C. Reversals of age-related declines in neuronal signal transduction, cognitive, and motor behavioral deficits with blueberry, spinach, or strawberry dietary supplementation. J. Neurosci. 1999, 19, 8114–8121. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; McGeer, E.G.; McGeer, P.L. Quercetin, not caffeine, is a major neuroprotective component in coffee. Neurobiol. Aging 2016, 46, 113–123. [Google Scholar] [CrossRef]
- Protein Data Bank (PDB). Available online: www.rcsb.org (accessed on 23 December 2022).
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov (accessed on 23 December 2022).
- Dassault Systèmes. Discovery BIOVIA, Dassault Systèmes, [Discovery Studio Visualizer], Software version 2021; Dassault Systèmes: San Diego, CA, USA, 2021. [Google Scholar]
- DrugBank Database. Available online: https://go.drugbank.com/drugs/DB00989 (accessed on 23 December 2022).
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with pyrx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AUTODOCK4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Haneef, M.; Lohani, M.; Dhasmana, A.; Jamal, Q.M.S.; Shahid, S.M.A.; Firdaus, S. Molecular docking of known carcinogen 4-(methyl-nitrosamino)-1-(3-pyridyl)-1-butanone (NNK) with cyclin dependent kinases towards its potential role in cell cycle perturbation. Bioinformation 2014, 10, 526–532. [Google Scholar] [CrossRef]
- Malik, M.S.; Farooq Adil, S.; Moussa, Z.; Altass, H.M.; Althagafi, I.I.; Morad, M.; Ansari, M.A.; Sajid Jamal, Q.M.; Obaid, R.J.; Al-Warthan, A.A.; et al. Rational design and synthesis of naphthalene Diimide linked bis-naphthalimides as DNA interactive agents. Front. Chem. 2021, 9, 630357. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. Gromacs: Fast, flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Zoete, V.; Cuendet, M.A.; Grosdidier, A.; Michielin, O. SwissParam: A fast force field generation tool for small organic molecules. J. Comput. Chem. 2011, 32, 2359–2368. [Google Scholar] [CrossRef]
- Kufareva, I.; Abagyan, R. Methods of protein structure comparison. Methods Mol. Biol. 2011, 857, 231–257. [Google Scholar]
- Kuzmanic, A.; Zagrovic, B. Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys. J. 2010, 98, 861–871. [Google Scholar] [CrossRef]
- Turner, P.J. XMGRACE, Version 5.1. 19; Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology: Beaverton, OR, USA, 2005.
- Colovic, M.B.; Krstic, D.Z.; Lazarevic-Pasti, T.D.; Bondzic, A.M.; Vasic, V.M. Acetylcholinesterase inhibitors: Pharmacology and toxicology. Curr. Neuropharmacol. 2013, 11, 315–335. [Google Scholar] [CrossRef] [PubMed]
- Haux, J.E.; Quistad, G.B.; Casida, J.E. Phosphobutyrylcholinesterase: phosphorylation of the esteratic site of butyrylcholinesterase by ethephon [(2-chloroethyl)phosphonic acid] dianion. Chem. Res. Toxicol. 2000, 13, 646–651. [Google Scholar] [CrossRef]
- Macdonald, I.R.; Martin, E.; Rosenberry, T.L.; Darvesh, S. Probing the peripheral site of human butyrylcholinesterase. Biochemistry 2012, 51, 7046–7053. [Google Scholar] [CrossRef]
- Jamal, Q.M.; Alharbi, A.H. Molecular docking and Dynamics Studies of cigarette smoke carcinogens interacting with acetylcholinesterase and butyrylcholinesterase enzymes of the central nervous system. Environ. Sci. Pollut. Res. 2021, 29, 61972–61992. [Google Scholar] [CrossRef] [PubMed]
- Dvir, H.; Silman, I.; Harel, M.; Rosenberry, T.L.; Sussman, J.L. Acetylcholinesterase: From 3D structure to function. Chem.-Biol. Interact. 2010, 187, 10–22. [Google Scholar] [CrossRef]
- Mallender, W.D.; Szegletes, T.; Rosenberry, T.L. Acetylthiocholine binds to ASP74 at the peripheral site of human acetylcholinesterase as the first step in the catalytic pathway. Biochemistry 2000, 39, 7753–7763. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Luo, S.; Sun, X.; Huang, M.; Ma, Q.; Du, L.; Cui, Y. Enhanced neuroprotective effects of epicatechin gallate encapsulated by bovine milk-derived exosomes against Parkinson’s disease through antiapoptosis and antimitophagy. J. Agric. Food Chem. 2021, 69, 5134–5143. [Google Scholar] [CrossRef]
- Herges, K.; Millward, J.M.; Hentschel, N.; Infante-Duarte, C.; Aktas, O.; Zipp, F. Neuroprotective effect of combination therapy of glatiramer acetate and epigallocatechin-3-gallate in neuroinflammation. PLoS ONE 2011, 6, e25456. [Google Scholar] [CrossRef]
- Ryan, S.N.; Laing, W.A.; McManus, M.T. A cysteine proteinase inhibitor purified from Apple Fruit. Phytochemistry 1998, 49, 957–963. [Google Scholar] [CrossRef]
- Hook, G.; Hook, V.Y.H.; Kindy, M. Cysteine protease inhibitors reduce brain β-amyloid and β-secretase activity in vivo and are potential Alzheimer’s disease therapeutics. Biol. Chem. 2007, 388, 979–983. [Google Scholar] [CrossRef]
- Siklos, M.; BenAissa, M.; Thatcher, G.R.J. Cysteine proteases as therapeutic targets: Does selectivity matter? A systematic review of calpain and cathepsin inhibitors. Acta Pharm. Sin. B 2015, 5, 506–519. [Google Scholar] [CrossRef]
- Roemmelt, S.; Zimmermann, N.; Rademacher, W.; Treutter, D. Formation of novel flavonoids in Apple (malus×domestica) treated with the 2-oxoglutarate-dependent dioxygenase inhibitor prohexadione-CA. Phytochemistry 2003, 64, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Patocka, J.; Bhardwaj, K.; Klimova, B.; Nepovimova, E.; Wu, Q.; Landi, M.; Kuca, K.; Valis, M.; Wu, W. Malus domestica: A Review on Nutritional Features, Chemical Composition, Traditional and Medicinal Value. Plants 2020, 9, 1408. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.ayurtimes.com/apple-fruit-malus-domestica/ (accessed on 6 March 2023).
- Available online: https://www.oecd-ilibrary.org/sites/832cfaee-en/index.html?itemId=/content/component/832cfaee-en (accessed on 6 March 2023).
- Zammit, S.; Lewis, S.; Gunnell, D.; Smith, G.D. Schizophrenia and neural tube defects: Comparisons from an epidemiological perspective. Schizophr. Bull. 2007, 33, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Ichi, S.; Nakazaki, H.; Boshnjaku, V.; Singh, R.M.; Mania-Farnell, B.; Xi, G.; McLone, D.G.; Tomita, T.; Mayanil, C.S. Fetal neural tube stem cells from pax3 mutant mice proliferate, differentiate, and form synaptic connections when stimulated with folic acid. Stem Cells Dev. 2012, 21, 321–330. [Google Scholar] [CrossRef]
- Li, W.; Liu, H.; Yu, M.; Zhang, X.; Zhang, Y.; Liu, H.; Wilson, J.X.; Huang, G. Folic acid alters methylation profile of jak-stat and long-term depression signaling pathways in Alzheimer’s disease models. Mol. Neurobiol. 2015, 53, 6548–6556. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, E. Vitamin B12, folic acid, and the nervous system. Lancet Neurol. 2006, 5, 949–960. [Google Scholar] [CrossRef] [PubMed]
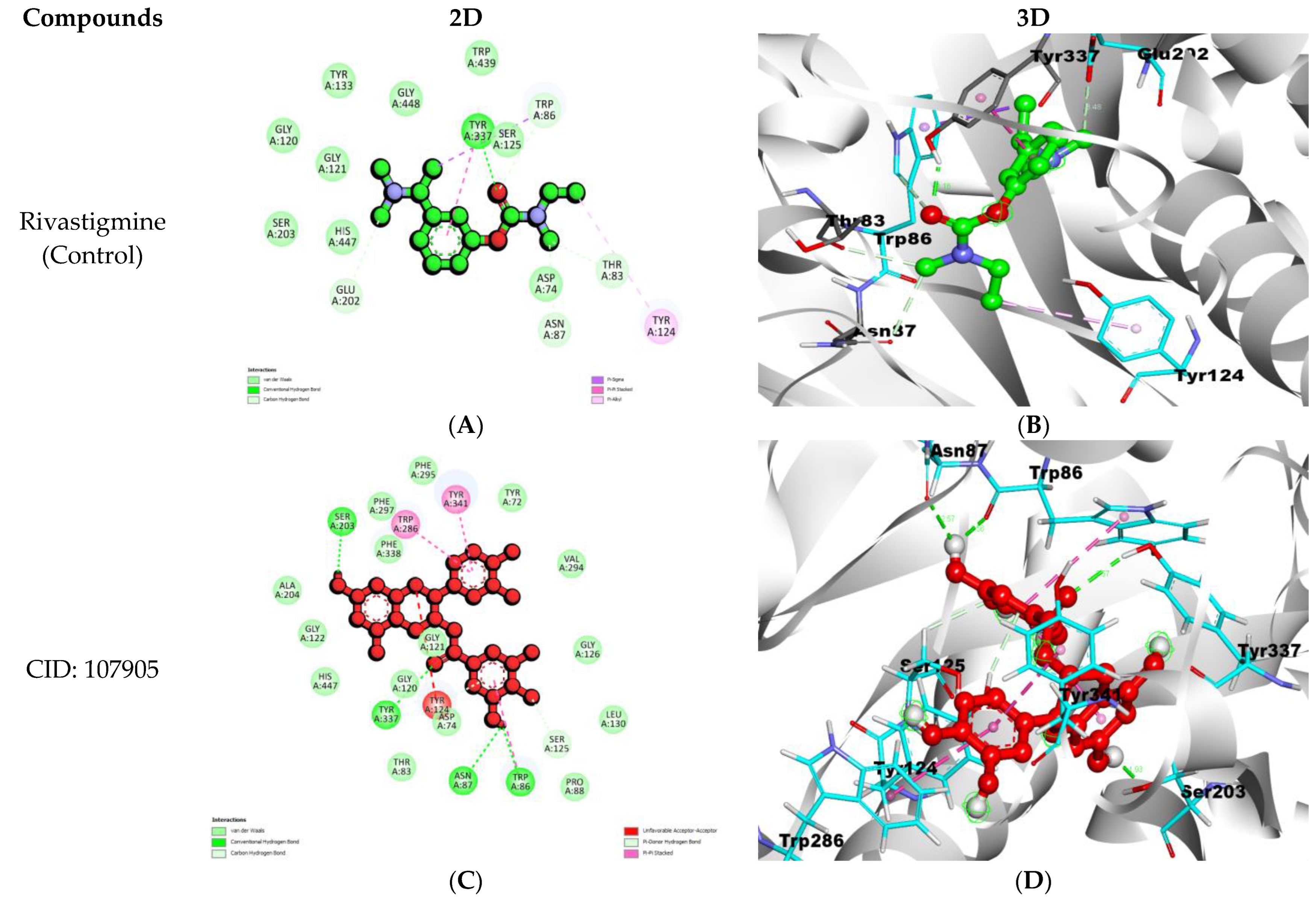
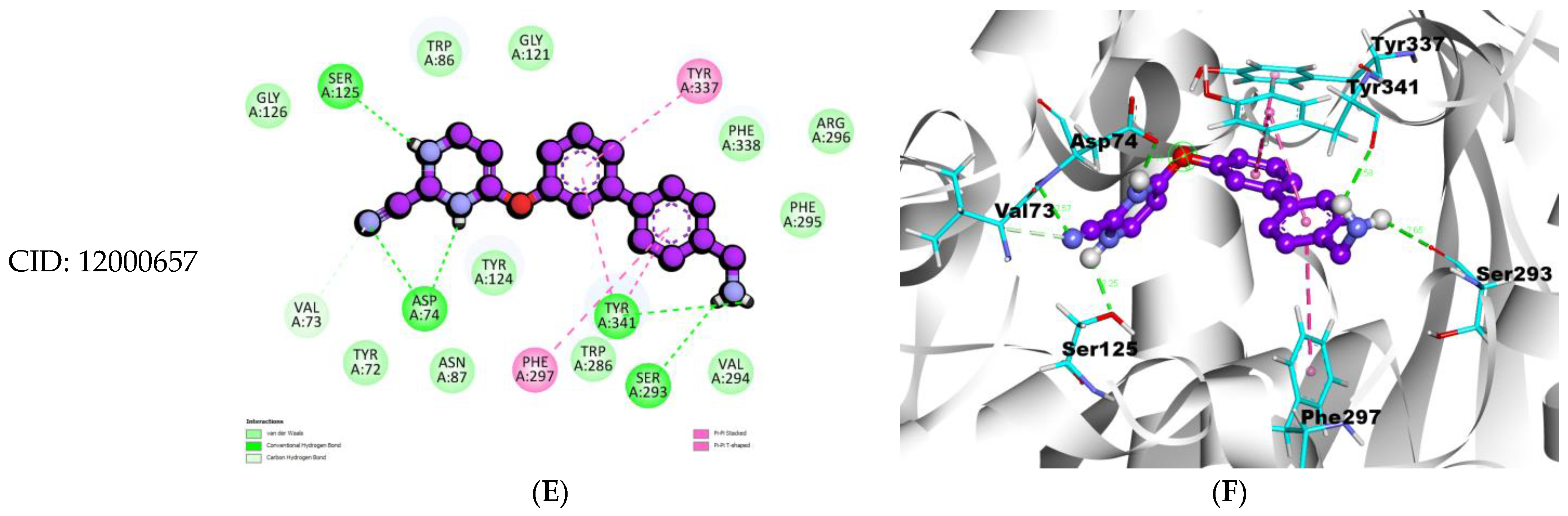

| Compounds | Binding Affinity (Kcal/mol) | Hydrogen Bond Names | Hydrogen Bond Lengths (Angstrom) | Van Der Waals Interactions | Other Types of Bond Formation |
|---|---|---|---|---|---|
| Rivastigmine (Control) | −7.8 | A:TYR337:HH-UNL1:O1 | 2.15667 | GLY120, GLY121, SER203, HIS447, TYR133, GLY448, TRP439, SER125, | PI–PI TYR124 PI–ALKYL TYR337 PI–SIGMA TRP86 |
| A:TRP86:CD1-:UNL1:O1 | 3.46816 | ||||
| :UNL1:C3-A:THR83:O | 3.0719 | ||||
| :UNL1:C3-A:ASN87:OD1 | 3.32779 | ||||
| :UNL1:C13-A:GLU202:OE1 | 3.47924 | ||||
| (-)-Epicatechin gallate CID: 107905 | −12.2 | A:TYR337:HH-N:UNK1:O | 2.57161 | ALA204, GLY122, HIS447, THR83, ASP74, GLY120, GLY121, PRO88, LEU130, GLY126, VAL294, TYR72, PHE295, PHE297, PHE338 | PI–PI TRP286, TYR341 |
| N:UNK1:H-A:TRP86:O | 2.56415 | ||||
| N:UNK1:H-A:ASN87:OD1 | 2.57352 | ||||
| N:UNK1:H-A:SER203:OG | 1.92587 | ||||
| A:SER125:HB1-N:UNK1:O | 2.81437 | ||||
| A:TYR124:HH-N:UNK1 | 2.80141 | ||||
| 4-((4′-(Aminomethyl)-[1,1′-biphenyl]-3-yl)oxy)pyrimidine-2-carbonitrile CID: 12000657 | −11.6 | A:ASP74:HN-N:UNK1:N | 2.56919 | TYR72, VAL73, TRP86, ASN87, TYR124, GLY121, SER125, GLY126, TRP286, VAL294, PHE295, ARG296, PHE338 | PI–PI PHE297, TYR337, TYR341 |
| N:UNK1:HN-A:ASP74:OD2 | 2.10567 | ||||
| N:UNK1:HN-A:SER125:OG | 2.25043 | ||||
| N:UNK1:H-A:TYR341:O | 2.58107 | ||||
| N:UNK1:H-A:SER293:O | 2.6484 | ||||
| A:VAL73:HA-N:UNK1:N | 3.01081 |
| Compounds | Binding Affinity (Kcal/mol) | Hydrogen Bond Names | Hydrogen Bond Lengths (Angstrom) | Van Der Waals Interactions | Other Types of Bond Formation |
|---|---|---|---|---|---|
| Rivastigmine (Control) | −6.8 | :UNL1:C3-A:TRP82:O | 3.42913 | ALA328, TRP430, TYR440, GLY439, SER79, TYR332, TYR128, GLY115, THR120, GLY121, LEU125, | PI–PI STACKING HIS438 |
| :UNL1:C13-A:ASP70:OD1 | 3.52033 | ||||
| [(2R,3S,4S,5S,6S)-6-[5-[(2S)-5,7-dihydroxy-4-oxo-2,3-dihydrochromen-2-yl]-2-hydroxyphenoxy]-3,4,5-trihydroxyoxan-2-yl]methyl (E)-3-(4-hydroxyphenyl)prop-2-enoate CID: 163103561 | −11.2 | N:UNK1:H-A:GLU197:OE1 | 2.48466 | PRO84, TYR332, GLN119, ASN83, PHE398, VAL288, GLY116, GLY117, SER287, SER198, GLY115, GLY439, TRP112 | PI–ANINON GLU197 PI–ALKYL LEU286 PI–PI T SHAPED PHE329,TRP231, TRP82 |
| N:UNK1:H-A:LEU286:O | 1.8801 | ||||
| N:UNK1:H-A:PRO285:O | 2.54319 | ||||
| N:UNK1:H-A:GLN67:OE1 | 2.29008 | ||||
| A:GLY121:HA1-N:UNK1:O | 2.84111 | ||||
| A:LEU286:HA-N:UNK1:O | 2.85756 | ||||
| N:UNK1:C-A:HIS438:NE2 | 3.13846 | ||||
| Folic acid CID: 135398658 | −10.0 | N:UNK1:HN-A:ASP70:OD1 | 2.30031 | TRP231, ALA199, VAL288, SER287, PRO285, GLN119, GLY116, ALA328, PHE398, HIS438, TYR332, TRP430, TRP82, ILE69, GLN67, PRO84, GLY121, THR120 | PI–PI T SHAPED PHE329 |
| N:UNK1:HN-A:SER79:O | 2.58808 | ||||
| N:UNK1:HN-A:ASN83:OD1 | 2.26671 | ||||
| N:UNK1:H-A:THR120:OG1 | 2.52606 | ||||
| N:UNK1:H-A:ASN83:OD1 | 2.12821 | ||||
| N:UNK1:H-A:SER198:OG | 2.55222 | ||||
| N:UNK1:H-A:LEU286:O | 2.84045 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamal, Q.M.S.; Khan, M.I.; Alharbi, A.H.; Ahmad, V.; Yadav, B.S. Identification of Natural Compounds of the Apple as Inhibitors against Cholinesterase for the Treatment of Alzheimer’s Disease: An In Silico Molecular Docking Simulation and ADMET Study. Nutrients 2023, 15, 1579. https://doi.org/10.3390/nu15071579
Jamal QMS, Khan MI, Alharbi AH, Ahmad V, Yadav BS. Identification of Natural Compounds of the Apple as Inhibitors against Cholinesterase for the Treatment of Alzheimer’s Disease: An In Silico Molecular Docking Simulation and ADMET Study. Nutrients. 2023; 15(7):1579. https://doi.org/10.3390/nu15071579
Chicago/Turabian StyleJamal, Qazi Mohammad Sajid, Mohammad Imran Khan, Ali H. Alharbi, Varish Ahmad, and Brijesh Singh Yadav. 2023. "Identification of Natural Compounds of the Apple as Inhibitors against Cholinesterase for the Treatment of Alzheimer’s Disease: An In Silico Molecular Docking Simulation and ADMET Study" Nutrients 15, no. 7: 1579. https://doi.org/10.3390/nu15071579
APA StyleJamal, Q. M. S., Khan, M. I., Alharbi, A. H., Ahmad, V., & Yadav, B. S. (2023). Identification of Natural Compounds of the Apple as Inhibitors against Cholinesterase for the Treatment of Alzheimer’s Disease: An In Silico Molecular Docking Simulation and ADMET Study. Nutrients, 15(7), 1579. https://doi.org/10.3390/nu15071579







