Siraitia grosvenorii Residual Extract Inhibits Inflammation in RAW264.7 Macrophages and Attenuates Osteoarthritis Progression in a Rat Model
Abstract
1. Introduction
2. Materials and Methods
2.1. SGRE Preparation and Chemical Profiling of SGRE
2.2. Ultra-High-Performance Liquid Chromatography-Charged Aerosol Detector (UHPLC-CAD) Analysis of SGRE
2.3. Cell Culture and Lipopolysaccharide-Induced Inflammation
2.4. Cell Viability Assay
2.5. Measurement of the Pro-Inflammatory Cytokine and Mediator Levels
2.6. Western Blotting
2.7. Induction of MIA-Induced OA in Rats
2.8. Joint Pain Assessment
2.9. Serum Analysis
2.10. Real-Time Polymerase Chain Reaction (RT-PCR) Analysis
2.11. Histopathology Analysis
2.12. Statistical Analysis
3. Results
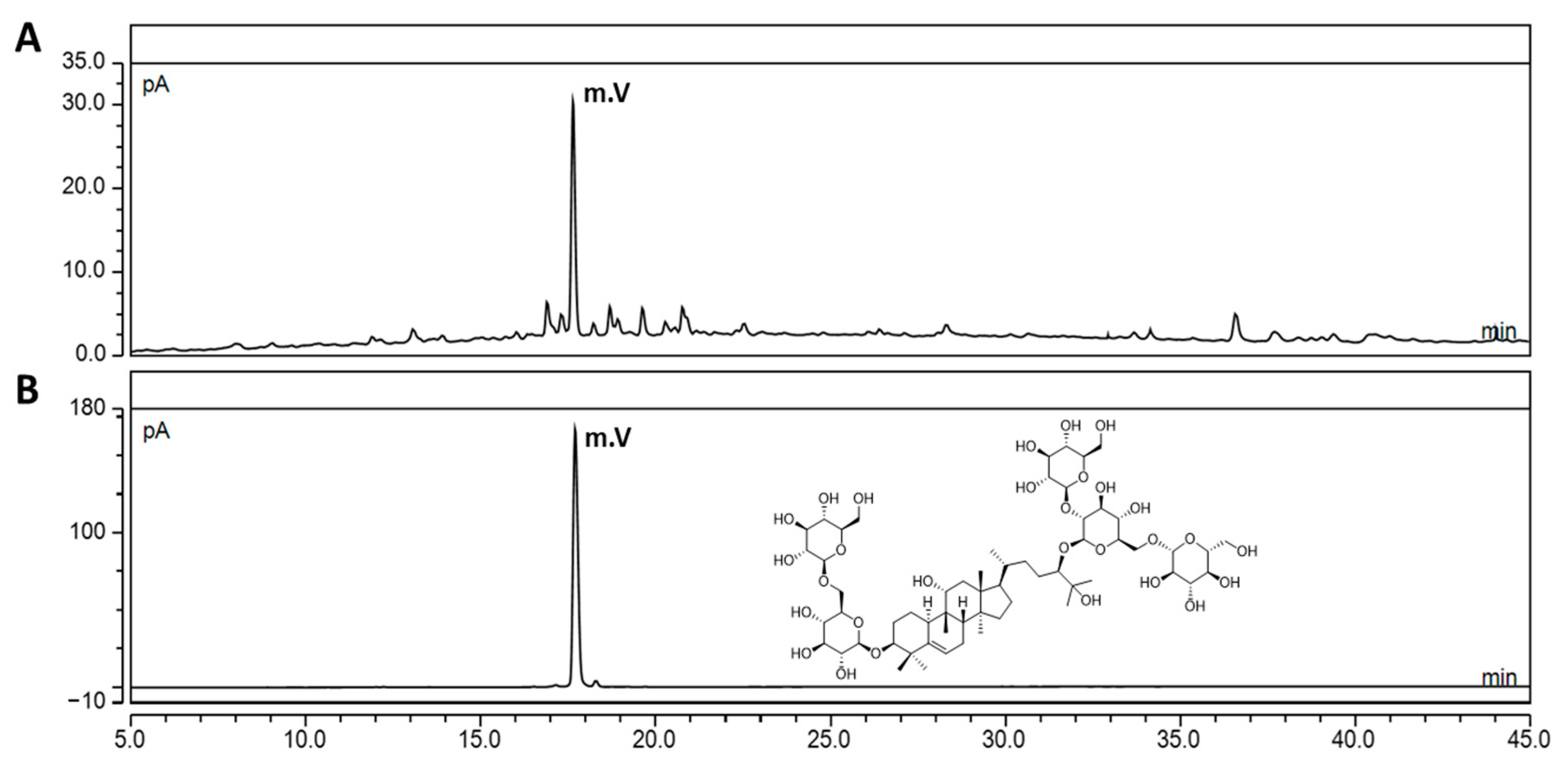
3.1. Quantification of the Mogroside V Level in SGRE Using UHPLC-CAD
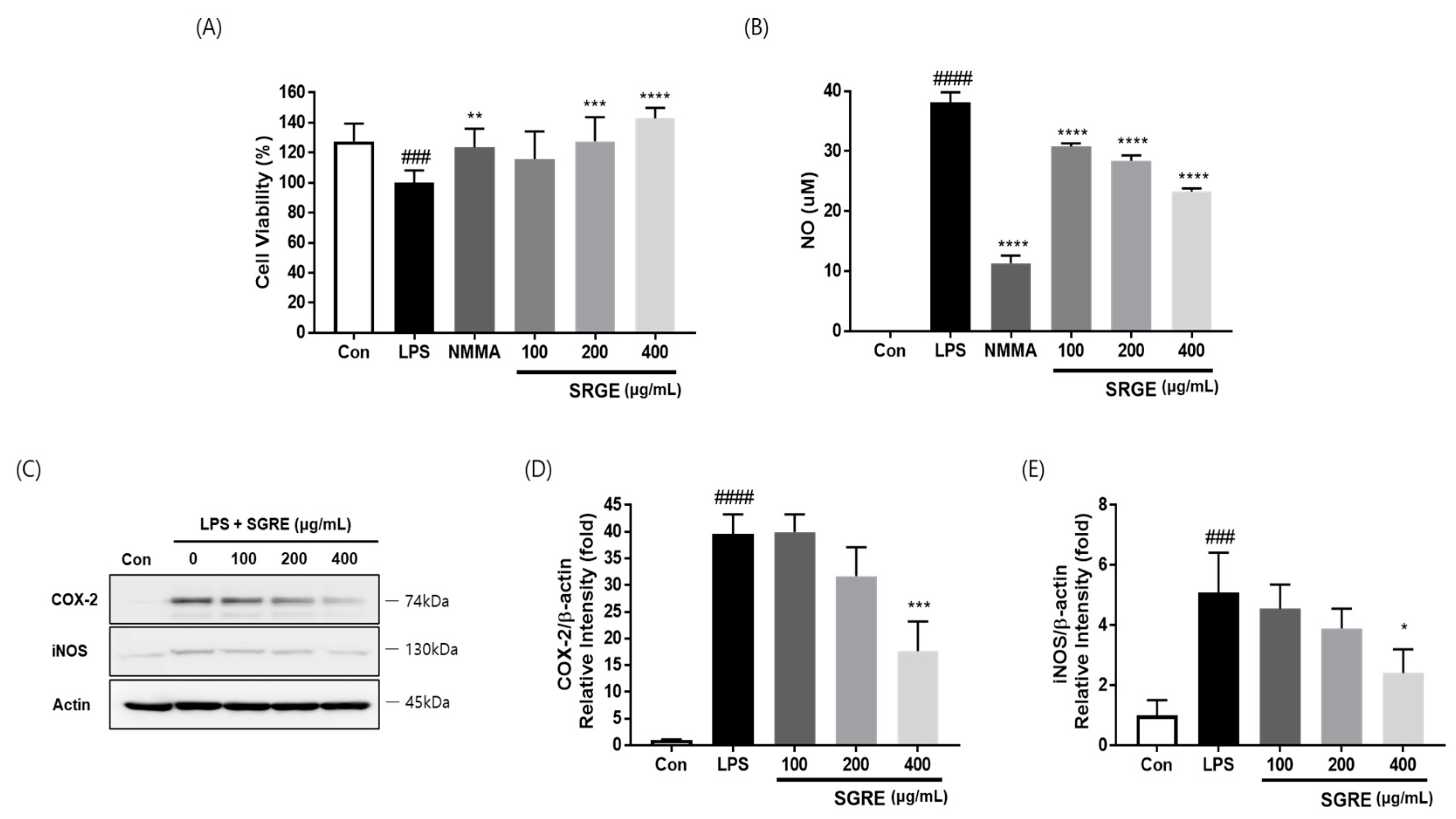
3.2. Effects of SGRE on the Viability of LPS-Induced RAW264.7 Macrophages
3.3. Effect of SGRE on NO Production and the Expression of COX-2 and iNOS in LPS-Induced RAW264.7 Macrophages
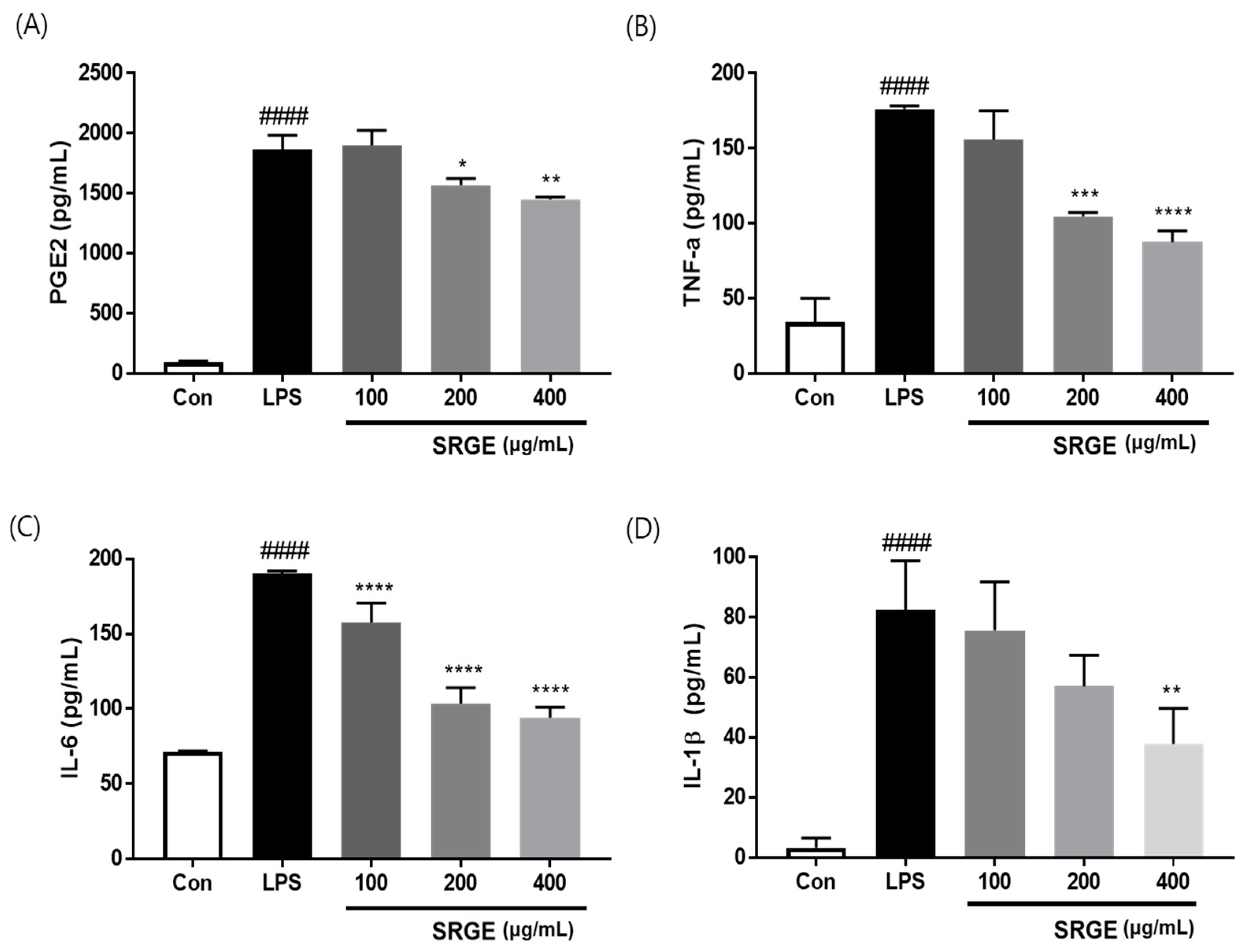
3.4. Effect of SGRE on the Levels of Pro-Inflammatory Factors in LPS-Induced RAW264.7 Macrophages
3.5. Effect of SGRE on MAPK Activation in LPS-Induced RAW264.7 Macrophages
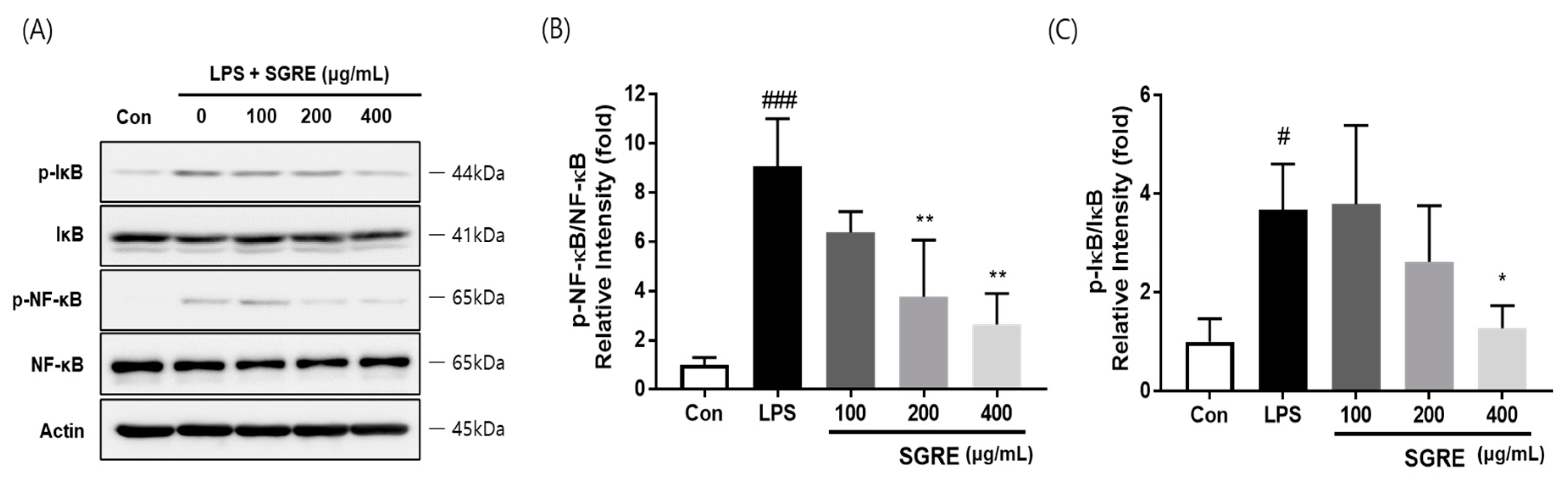
3.6. Effect of SGRE on IκB Degradation and NF-κB p65 Phosphorylation in LPS-Induced RAW264.7 Macrophages
3.7. Effects of SGRE on the Hind Paw Weight-Bearing Ratio
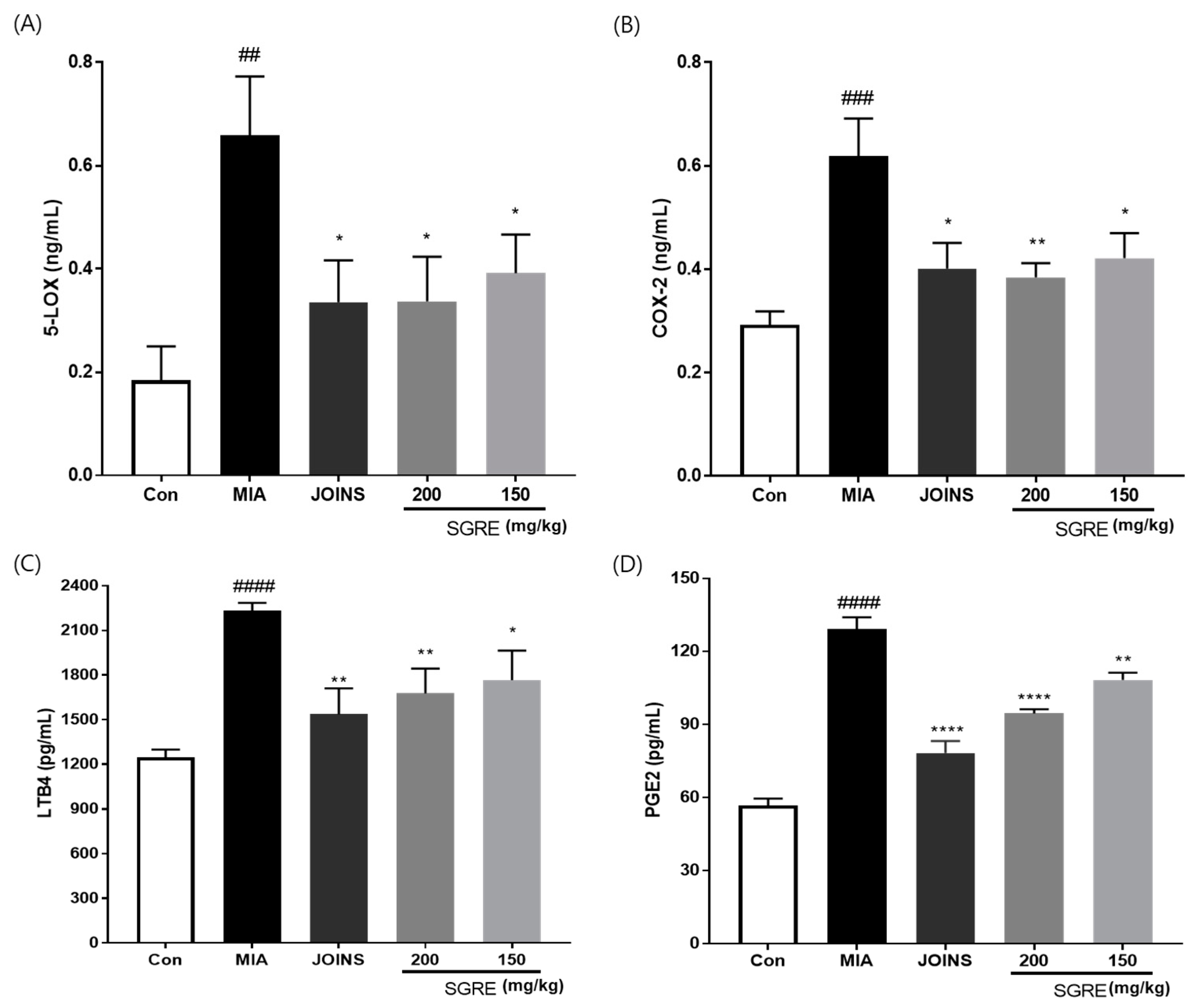
3.8. Effects of SGRE on the Mediators and Cytokines Involved in the Inflammatory Response
3.9. Effects of SGRE on Cartilage Degradation in MIA-Induced Rats
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Klein, J.C.; Keith, A.; Rice, S.J.; Shepherd, C.; Agarwal, V.; Loughlin, J.; Shendure, J. Functional testing of thousands of osteoarthritis-associated variants for regulatory activity. Nat. Commun. 2019, 10, 2434. [Google Scholar] [CrossRef] [PubMed]
- Ju, L.; Hu, P.; Chen, P.; Xue, X.; Li, Z.; He, F.; Qiu, Z.; Cheng, J.; Huang, F. Huoxuezhitong capsule ameliorates MIA-induced osteoarthritis of rats through suppressing PI3K/Akt/NF-kappaB pathway. Biomed. Pharmacother. 2020, 129, 110471. [Google Scholar] [CrossRef]
- Camacho-Encina, M.; Balboa-Barreiro, V.; Rego-Perez, I.; Picchi, F.; VanDuin, J.; Qiu, J.; Fuentes, M.; Oreiro, N.; LaBaer, J.; Ruiz-Romero, C.; et al. Discovery of an autoantibody signature for the early diagnosis of knee osteoarthritis: Data from the Osteoarthritis Initiative. Ann. Rheum. Dis. 2019, 78, 1699–1705. [Google Scholar] [CrossRef] [PubMed]
- Orlowsky, E.W.; Kraus, V.B. The role of innate immunity in osteoarthritis: When our first line of defense goes on the offensive. J. Rheumatol. 2015, 42, 363–371. [Google Scholar] [CrossRef]
- Robinson, W.H.; Lepus, C.M.; Wang, Q.; Raghu, H.; Mao, R.; Lindstrom, T.M.; Sokolove, J. Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2016, 12, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Li, X.; Lei, S.; Feng, S.; Zhou, C.; Tong, X.; Han, R. Protective effects of Pudilan Tablets against osteoarthritis in mice induced by monosodium iodoacetate. Sci. Rep. 2023, 13, 2760. [Google Scholar] [CrossRef]
- Nees, T.A.; Rosshirt, N.; Reiner, T.; Schiltenwolf, M.; Moradi, B. Inflammation and osteoarthritis-related pain. Schmerz 2019, 33, 4–12. [Google Scholar] [CrossRef]
- Wojdasiewicz, P.; Poniatowski, L.A.; Szukiewicz, D. The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis. Mediat. Inflamm. 2014, 2014, 561459. [Google Scholar] [CrossRef]
- Chevalier, X.; Eymard, F.; Richette, P. Biologic agents in osteoarthritis: Hopes and disappointments. Nat. Rev. Rheumatol. 2013, 9, 400–410. [Google Scholar] [CrossRef]
- Poulet, B.; Staines, K.A. New developments in osteoarthritis and cartilage biology. Curr. Opin. Pharmacol. 2016, 28, 8–13. [Google Scholar] [CrossRef]
- Yabas, M.; Orhan, C.; Er, B.; Tuzcu, M.; Durmus, A.S.; Ozercan, I.H.; Sahin, N.; Bhanuse, P.; Morde, A.A.; Padigaru, M.; et al. A Next Generation Formulation of Curcumin Ameliorates Experimentally Induced Osteoarthritis in Rats via Regulation of Inflammatory Mediators. Front. Immunol. 2021, 12, 609629. [Google Scholar] [CrossRef]
- Kapoor, M.; Martel-Pelletier, J.; Lajeunesse, D.; Pelletier, J.P.; Fahmi, H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 2011, 7, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.E.; Sulistyowati, E.; Chao, Y.Y.; Wu, B.N.; Dai, Z.K.; Hsu, J.H.; Yeh, J.L. In Vitro Evaluation of the Anti-Inflammatory Effect of KMUP-1 and In Vivo Analysis of Its Therapeutic Potential in Osteoarthritis. Biomedicines 2021, 9, 615. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.C.; Jo, J.; Park, J.; Kang, H.K.; Park, Y. NF-kappaB Signaling Pathways in Osteoarthritic Cartilage Destruction. Cells 2019, 8, 734. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Jeong, S.; Kim, H.; Kang, D.; Lee, J.; Kang, S.B.; Kim, J.H. Disease-modifying therapeutic strategies in osteoarthritis: Current status and future directions. Exp. Mol. Med. 2021, 53, 1689–1696. [Google Scholar] [CrossRef]
- Lee, D.; Ju, M.K.; Kim, H. Commiphora Extract Mixture Ameliorates Monosodium Iodoacetate-Induced Osteoarthritis. Nutrients 2020, 12, 1477. [Google Scholar] [CrossRef]
- Rahmati, M.; Mobasheri, A.; Mozafari, M. Inflammatory mediators in osteoarthritis: A critical review of the state-of-the-art, current prospects, and future challenges. Bone 2016, 85, 81–90. [Google Scholar] [CrossRef]
- Sharma, L. Osteoarthritis of the Knee. N. Engl. J. Med. 2021, 384, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, C.I.; Perez, M.J.; Manautou, J.E.; Mottino, A.D. Acetaminophen from liver to brain: New insights into drug pharmacological action and toxicity. Pharmacol. Res. 2016, 109, 119–131. [Google Scholar] [CrossRef]
- Deligiannidou, G.E.; Papadopoulos, R.E.; Kontogiorgis, C.; Detsi, A.; Bezirtzoglou, E.; Constantinides, T. Unraveling Natural Products’ Role in Osteoarthritis Management-An Overview. Antioxidants 2020, 9, 348. [Google Scholar] [CrossRef]
- Marques, R.V.; Sestito, S.E.; Bourgaud, F.; Miguel, S.; Cailotto, F.; Reboul, P.; Jouzeau, J.Y.; Rahuel-Clermont, S.; Boschi-Muller, S.; Simonsen, H.T.; et al. Anti-Inflammatory Activity of Bryophytes Extracts in LPS-Stimulated RAW264.7 Murine Macrophages. Molecules 2022, 27, 1940. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Chen, N.; Ren, K.; Jia, J.; Wei, K.; Zhang, L.; Lv, Y.; Wang, J.; Li, M. The Fruits of Siraitia grosvenorii: A Review of a Chinese Food-Medicine. Front. Pharmacol. 2019, 10, 1400. [Google Scholar] [CrossRef] [PubMed]
- Soejarto, D.D.; Addo, E.M.; Kinghorn, A.D. Highly sweet compounds of plant origin: From ethnobotanical observations to wide utilization. J. Ethnopharmacol. 2019, 243, 112056. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.Y.; Kim, S.H.; Yuk, H.J.; Yang, W.K.; Lee, Y.M.; Son, E.; Kim, D.S. Siraitia grosvenorii residual extract attenuates ovalbumin-induced lung inflammation by down-regulating IL-4, IL-5, IL-13, IL-17, and MUC5AC expression in mice. Phytomedicine 2019, 61, 152835. [Google Scholar] [CrossRef]
- Sung, Y.Y.; Yuk, H.J.; Yang, W.K.; Kim, S.H.; Kim, D.S. Siraitia grosvenorii Residual Extract Attenuates Atopic Dermatitis by Regulating Immune Dysfunction and Skin Barrier Abnormality. Nutrients 2020, 12, 3638. [Google Scholar] [CrossRef]
- Lee, Y.M.; Son, E.; Kim, S.H.; Kim, D.S. Effect of Alpinia oxyphylla extract in vitro and in a monosodium iodoacetate-induced osteoarthritis rat model. Phytomedicine 2019, 65, 153095. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Wang, G.; Hu, Z.; Fu, Q.; Song, X.; Cui, Q.; Jia, R.; Zou, Y.; He, C.; Li, L.; Yin, Z. Resveratrol mitigates lipopolysaccharide-mediated acute inflammation in rats by inhibiting the TLR4/NF-kappaBp65/MAPKs signaling cascade. Sci. Rep. 2017, 7, 45006. [Google Scholar] [CrossRef]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-kappaB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Troeberg, L.; Nagase, H. Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim. Biophys. Acta 2012, 1824, 133–145. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, W.; Yong, H.; He, M.; Yang, Y.; Deng, Z.; Li, Y. Macrophages in osteoarthritis: Pathophysiology and therapeutics. Am. J. Transl. Res. 2020, 12, 261–268. [Google Scholar] [PubMed]
- Yuan, Q.; Wang, J.; Guo, L.; Xu, Y.; Hu, L.; Mao, H.; Miao, L.; Zhang, H.; Chai, L. Neobavaisoflavone ameliorates LPS-induced RAW264.7 cell inflammations by suppressing the activation of NF-kappaB and MAPKs signaling pathways. Iran. J. Basic. Med. Sci. 2022, 25, 1021–1027. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Kaminska, B. MAPK signalling pathways as molecular targets for anti-inflammatory therapy--from molecular mechanisms to therapeutic benefits. Biochim. Biophys. Acta 2005, 1754, 253–262. [Google Scholar] [CrossRef]
- Gottschalk, R.A.; Martins, A.J.; Angermann, B.R.; Dutta, B.; Ng, C.E.; Uderhardt, S.; Tsang, J.S.; Fraser, I.D.; Meier-Schellersheim, M.; Germain, R.N. Distinct NF-kappaB and MAPK Activation Thresholds Uncouple Steady-State Microbe Sensing from Anti-pathogen Inflammatory Responses. Cell Syst. 2016, 2, 378–390. [Google Scholar] [CrossRef]
- Asthana, C.; Peterson, G.M.; Shastri, M.; Patel, R.P. Development and validation of a novel high performance liquid chromatography-coupled with Corona charged aerosol detector method for quantification of glucosamine in dietary supplements. PLoS ONE 2019, 14, e0216039. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Son, E.; Kim, D.S. Treatment with Peanut Sprout Root Extract Alleviates Inflammation in a Lipopolysaccharide-Stimulated Mouse Macrophage Cell Line by Inhibiting the MAPK Signaling Pathway. Int. J. Mol. Sci. 2019, 20, 5907. [Google Scholar] [CrossRef] [PubMed]
- Bonizzi, G.; Karin, M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004, 25, 280–288. [Google Scholar] [CrossRef]
- Shou, J.; Kong, X.; Wang, X.; Tang, Y.; Wang, C.; Wang, M.; Zhang, L.; Liu, Y.; Fei, C.; Xue, F.; et al. Tizoxanide Inhibits Inflammation in LPS-Activated RAW264.7 Macrophages via the Suppression of NF-kappaB and MAPK Activation. Inflammation 2019, 42, 1336–1349. [Google Scholar] [CrossRef]
- Fujii, Y.; Liu, L.; Yagasaki, L.; Inotsume, M.; Chiba, T.; Asahara, H. Cartilage Homeostasis and Osteoarthritis. Int. J. Mol. Sci. 2022, 23, 6316. [Google Scholar] [CrossRef]
- Bauvois, B. New facets of matrix metalloproteinases MMP-2 and MMP-9 as cell surface transducers: Outside-in signaling and relationship to tumor progression. Biochim. Biophys. Acta 2012, 1825, 29–36. [Google Scholar] [CrossRef]
- Chow, Y.Y.; Chin, K.Y. The Role of Inflammation in the Pathogenesis of Osteoarthritis. Mediators Inflamm. 2020, 2020, 8293921. [Google Scholar] [CrossRef] [PubMed]
- Haltmayer, E.; Ribitsch, I.; Gabner, S.; Rosser, J.; Gueltekin, S.; Peham, J.; Giese, U.; Dolezal, M.; Egerbacher, M.; Jenner, F. Co-culture of osteochondral explants and synovial membrane as in vitro model for osteoarthritis. PLoS ONE 2019, 14, e0214709. [Google Scholar] [CrossRef] [PubMed]
- Bove, S.E.; Calcaterra, S.L.; Brooker, R.M.; Huber, C.M.; Guzman, R.E.; Juneau, P.L.; Schrier, D.J.; Kilgore, K.S. Weight bearing as a measure of disease progression and efficacy of anti-inflammatory compounds in a model of monosodium iodoacetate-induced osteoarthritis. Osteoarthr. Cartil. 2003, 11, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jian, Y.; Wang, H.; Huang, H.; Gong, L.; Liu, G.; Yang, Y.; Wang, W. A Review of the Phytochemistry and Pharmacology of the Fruit of Siraitia grosvenorii (Swingle): A Traditional Chinese Medicinal Food. Molecules 2022, 27, 6618. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.W.; Hill, A.P.; Readshaw, S.A.; Hollerton, J.C.; Upton, R.J.; Lynn, S.M.; Besley, S.C.; Boughtflower, B.J. Use of Calculated Physicochemical Properties to Enhance Quantitative Response When Using Charged Aerosol Detection. Anal. Chem. 2017, 89, 1772–1777. [Google Scholar] [CrossRef] [PubMed]





| Gene | Primer Sequence | |
|---|---|---|
| IL-1β | Forward | 5′-ACAAGGCTGCCCCGACTAT-3′ |
| Reverse | 5′-CTCCTGGTATGAAGTGGCAAATC-3′ | |
| IL-6 | Forward | 5′-GCCCTTCAGGAACAGCTATGA-3′ |
| Reverse | 5′-TGTCAACAACATCAGTCCCAAGA-3′ | |
| TNF-α | Forward | 5′-ACAAGG CTGCCCCGACTAT-3′ |
| Reverse | 5′-CTCCTGGTATGAAGTGGCAAATC-3′ | |
| MMP-1 | Forward | 5′-CTCCCTTGGACTCACTCATTCTA-3′ |
| Reverse | 5′-AGAACATCACCTCTCCCCTAAAC-3′ | |
| MMP-9 | Forward | 5′-GCGCCAGCCGACTTATGT-3′ |
| Reverse | 5′-AATCCTCTGCCAGCTGTGTGT-3′ | |
| MMP-13 | Forward | 5′-ACGTTCAAGGAATCCAGTCTCTCT-3′ |
| Reverse | 5′-GGATAGGGCTGGGTCACACTT-3′ | |
| SOX-9 | Forward | 5′-CTGAAGGGCTACGACTGGAC-3′ |
| Reverse | 5′-TACTGGTCTGCCAGCTTCCT-3′ | |
| COL2A1 | Forward | 5′-GCAACAGCAGGTTCACGTACA-3′ |
| Reverse | 5′-TCGGTACTCGATGATGGTCTTG-3′ | |
| ACAN | Forward | 5′-GAAGTGGCGTCCAAACCAA-3′ |
| Reverse | 5′-CGTTCCATTCACCCCTCTCA-3′ | |
| GAPDH | Forward | 5′-TGGCCTCCAAGGAGTAAGAAAC-3′ |
| Reverse | 5′-CAGCAACTGAGGGCCTCTCT-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.M.; Kim, M.; Yuk, H.J.; Kim, S.-H.; Kim, D.-S. Siraitia grosvenorii Residual Extract Inhibits Inflammation in RAW264.7 Macrophages and Attenuates Osteoarthritis Progression in a Rat Model. Nutrients 2023, 15, 1417. https://doi.org/10.3390/nu15061417
Lee YM, Kim M, Yuk HJ, Kim S-H, Kim D-S. Siraitia grosvenorii Residual Extract Inhibits Inflammation in RAW264.7 Macrophages and Attenuates Osteoarthritis Progression in a Rat Model. Nutrients. 2023; 15(6):1417. https://doi.org/10.3390/nu15061417
Chicago/Turabian StyleLee, Yun Mi, Misun Kim, Heung Joo Yuk, Seung-Hyung Kim, and Dong-Seon Kim. 2023. "Siraitia grosvenorii Residual Extract Inhibits Inflammation in RAW264.7 Macrophages and Attenuates Osteoarthritis Progression in a Rat Model" Nutrients 15, no. 6: 1417. https://doi.org/10.3390/nu15061417
APA StyleLee, Y. M., Kim, M., Yuk, H. J., Kim, S.-H., & Kim, D.-S. (2023). Siraitia grosvenorii Residual Extract Inhibits Inflammation in RAW264.7 Macrophages and Attenuates Osteoarthritis Progression in a Rat Model. Nutrients, 15(6), 1417. https://doi.org/10.3390/nu15061417







