Untargeted and Targeted Blood Lipidomic Signature Profile of Gestational Alcohol Exposure
Abstract
1. Introduction
2. Materials and Methods
2.1. Treatment Groups and Alcohol Dosing Paradigm
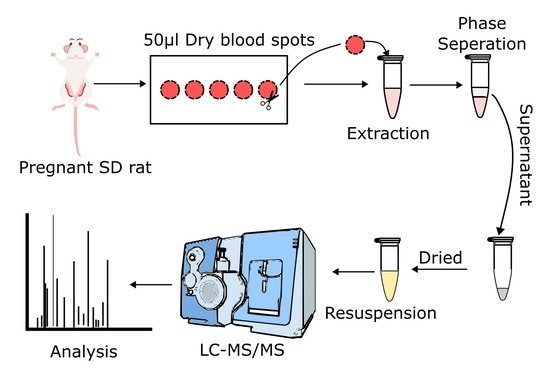
2.2. Blood Collection
2.3. Untargeted and Targeted Lipid Analysis
2.4. Quality Control
2.5. Data Processing
2.6. Statistical Analysis
3. Results
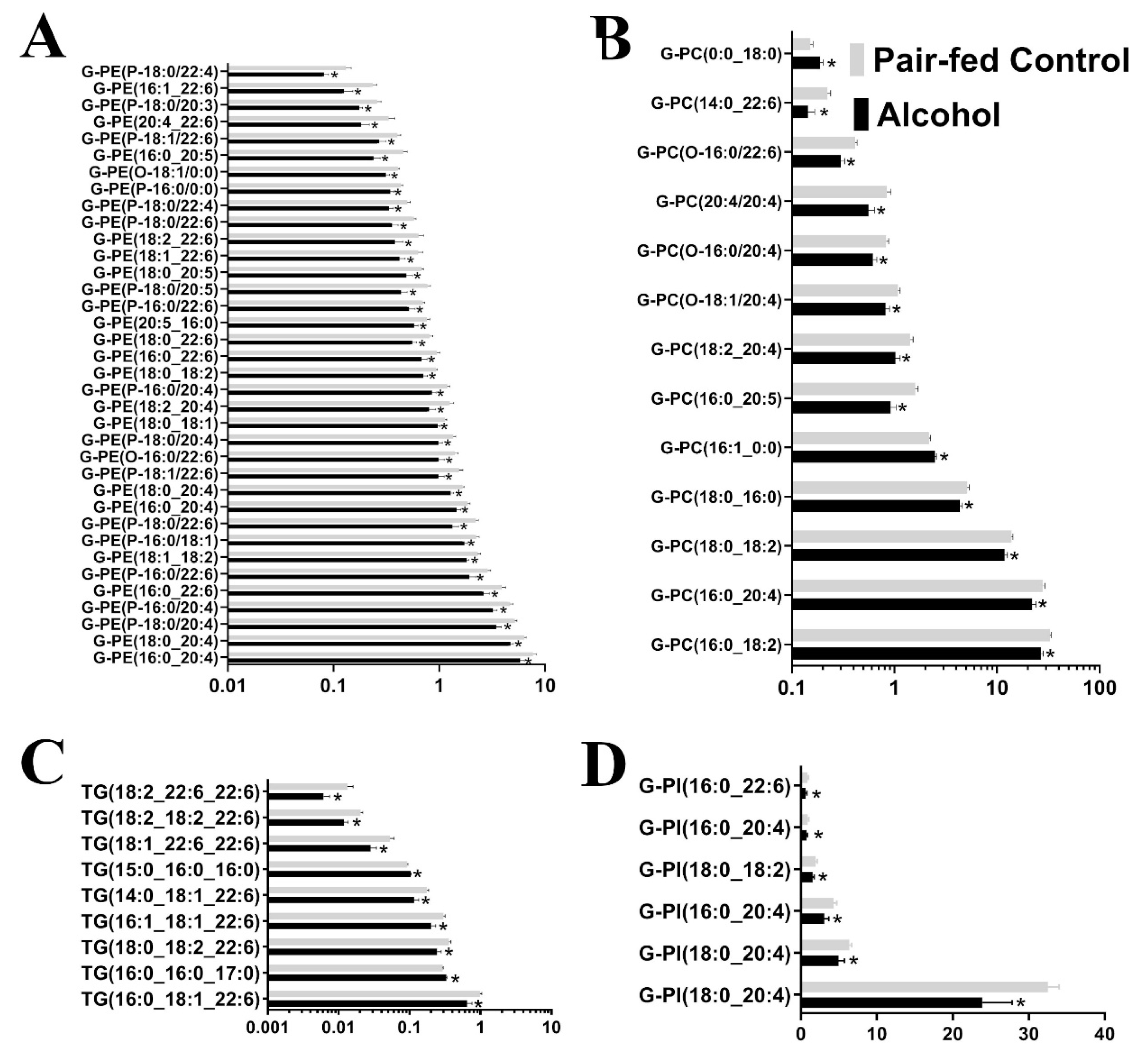
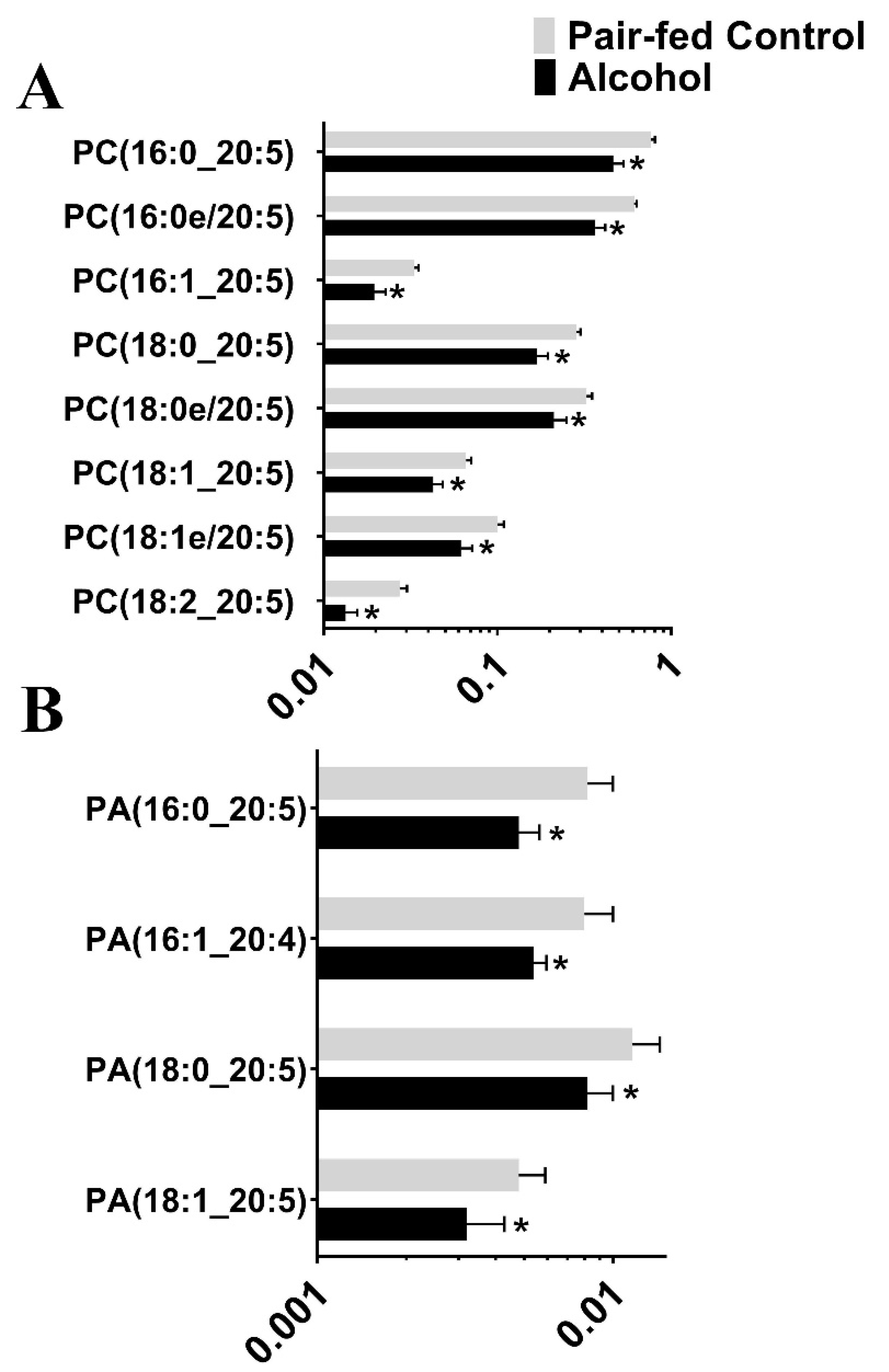
3.1. Untargeted Lipidomic Analysis
3.2. Targeted Lipidomic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, K.; Smith, D. Recognition of the Fetal Alcohol Syndrome in Early Infancy. Lancet 1973, 302, 999–1001. [Google Scholar] [CrossRef] [PubMed]
- Sokol, R.J. Fetal Alcohol Spectrum Disorder. JAMA 2003, 290, 2996. [Google Scholar] [CrossRef]
- Goodlett, C.R.; Horn, K.H. Mechanisms of alcohol-induced damage to the developing nervous system. Alcohol Res. Health 2001, 25, 175–184. [Google Scholar] [PubMed]
- Schenker, S.; Becker, H.C.; Randall, C.L.; Phillips, D.K.; Baskin, G.S.; Henderson, G.I. Fetal Alcohol Syndrome—Current Status of Pathogenesis. Alcohol. Clin. Exp. Res. 1990, 14, 635–647. [Google Scholar] [CrossRef]
- Herrera, E. Lipid metabolism in pregnancy and its consequences in the fetus and newborn. Endocrine 2002, 19, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Linask, K.K.; Han, M. Acute alcohol exposure during mouse gastrulation alters lipid metabolism in placental and heart development: Folate prevention. Birth Defects Res. Part A Clin. Mol. Teratol. 2016, 106, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Herrera, E.; Gomez-Coronado, D.; Lasuncion, M.A. Lipid metabolism in pregnancy. Biol. Neonate 1987, 51, 70–77. [Google Scholar] [CrossRef]
- Lopezluna, P.; Munoz, T.; Herrera, E. Body-Fat in Pregnant Rats at Midgestation and Late-Gestation. Life Sci. 1986, 39, 1389–1393. [Google Scholar] [CrossRef]
- Beblo, S.; Stark, K.D.; Murthy, M.; Janisse, J.; Rockett, H.; Whitty, J.E.; Buda-Abela, M.; Martier, S.S.; Sokol, R.J.; Hannigan, J.H.; et al. Effects of alcohol intake during pregnancy on docosahexaenoic acid and arachidonic acid in umbilical cord vessels of black women. Pediatrics 2005, 115, e194–e203. [Google Scholar] [CrossRef]
- Sowell, K.D.; Holt, R.R.; Uriu-Adams, J.Y.; Chambers, C.D.; Coles, C.D.; Kable, J.A.; Yevtushok, L.; Zymak-Zakutnya, N.; Wertelecki, W.; Keen, C.L.; et al. Altered Maternal Plasma Fatty Acid Composition by Alcohol Consumption and Smoking during Pregnancy and Associations with Fetal Alcohol Spectrum Disorders. J. Am. Coll. Nutr. 2020, 39, 249–260. [Google Scholar] [CrossRef]
- Pawlosky, R.J.; Salem, N. Alcohol consumption in rhesus monkeys depletes tissues of polyunsaturated fatty acids and alters essential fatty acid metabolism. Alcohol Clin. Exp. Res. 1999, 23, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Valimaki, M.; Halmesmaki, E.; Keso, L.; Ylikorkala, O.; Ylikahri, R. Serum-Lipids and Lipoproteins in Alcoholic Women during Pregnancy. Metabolism 1990, 39, 486–493. [Google Scholar] [CrossRef]
- Tsui-Pierchala, B.A.; Encinas, M.; Milbrandt, J.; Johnson, E.M., Jr. Lipid rafts in neuronal signaling and function. Trends Neurosci. 2002, 25, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Farah, B.; He, M.; Fox, S.; Malouf, A.; Littner, Y.; Bearer, C.F. Ethanol causes the redistribution of L1 cell adhesion molecule in lipid rafts. J. Neurochem. 2011, 119, 859–867. [Google Scholar] [CrossRef] [PubMed]
- Ramadoss, J.; Jobe, S.O.; Magness, R.R. Alcohol and maternal uterine vascular adaptations during pregnancy-part I: Effects of chronic in vitro binge-like alcohol on uterine endothelial nitric oxide system and function. Alcohol. Clin. Exp. Res. 2011, 35, 1686–1693. [Google Scholar] [CrossRef]
- Church, M.W.; Gerkin, K.P. Hearing disorders in children with fetal alcohol syndrome: Findings from case reports. Pediatrics 1988, 82, 147–154. [Google Scholar] [CrossRef]
- Cudd, T.A.; Chen, W.-J.A.; West, J.R. Fetal and Maternal Thyroid Hormone Responses to Ethanol Exposure During the Third Trimester Equivalent of Gestation in Sheep. Alcohol. Clin. Exp. Res. 2002, 26, 53–58. [Google Scholar] [CrossRef]
- Thomas, J.D.; Sather, T.M.; Whinery, L.A. Voluntary exercise influences behavioral development in rats exposed to alcohol during the neonatal brain growth spurt. Behav. Neurosci. 2008, 122, 1264–1273. [Google Scholar] [CrossRef]
- Potter, J.M.; Nestel, P.J. The hyperlipidemia of pregnancy in normal and complicated pregnancies. Am. J. Obstet. Gynecol. 1979, 133, 165–170. [Google Scholar] [CrossRef]
- Piechota, W.; Staszewski, A. Reference Ranges of Lipids and Apolipoproteins in Pregnancy. Eur. J. Obstet. Gynecol. Reprod. Biol. 1992, 45, 27–35. [Google Scholar] [CrossRef]
- Fahraeus, L.; Larsson-Cohn, U.; Wallentin, L. Plasma lipoproteins including high density lipoprotein subfractions during normal pregnancy. Obstet. Gynecol. 1985, 66, 468–472. [Google Scholar]
- Sattar, N.; Greer, I.A.; Galloway, P.J.; Packard, C.J.; Shepherd, J.; Kelly, T.; Mathers, A. Lipid and lipoprotein concentrations in pregnancies complicated by intrauterine growth restriction. J. Clin. Endocrinol. Metab. 1999, 84, 128–130. [Google Scholar] [CrossRef]
- Yang, Y.B.; Lee, M.; Fairn, G.D. Phospholipid subcellular localization and dynamics. J. Biol. Chem. 2018, 293, 6230–6240. [Google Scholar] [CrossRef]
- Vance, J.E. Phospholipid Synthesis and Transport in Mammalian Cells. Traffic 2015, 16, 1–18. [Google Scholar] [CrossRef]
- Schick, P.K.; Kurica, K.B.; Chacko, G.K. Location of phosphatidylethanolamine and phosphatidylserine in the human platelet plasma membrane. J. Clin. Investig. 1976, 57, 1221–1226. [Google Scholar] [CrossRef]
- Dowhan, W. Lipid-assisted folding of membrane proteins. FASEB J. 2000, 14, A1312. [Google Scholar]
- Sunshine, H.; Iruela-Arispe, M.L. Membrane lipids and cell signaling. Curr. Opin. Lipidol. 2017, 28, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Ikonen, E. Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 2001, 13, 470–477. [Google Scholar] [CrossRef]
- Dall’Armi, C.; Devereaux, K.A.; Di Paolo, G. The role of lipids in the control of autophagy. Curr. Biol. 2013, 23, R33–R45. [Google Scholar] [CrossRef]
- LaBarre, J.L.; Puttabyatappa, M.; Song, P.X.K.; Goodrich, J.M.; Zhou, L.; Rajendiran, T.M.; Soni, T.; Domino, S.E.; Treadwell, M.C.; Dolinoy, D.C.; et al. Maternal lipid levels across pregnancy impact the umbilical cord blood lipidome and infant birth weight. Sci. Rep. UK 2020, 10, 14209. [Google Scholar] [CrossRef] [PubMed]
- Korkes, H.A.; Sass, N.; Moron, A.F.; Câmara, N.O.; Bonetti, T.; Cerdeira, A.S.; Silva, I.D.; De Oliveira, L.G. [143-POS]: Lipid composition of placental tissue from patients with early-onset preeclampsia. Pregnancy Hypertens. Int. J. Womens Cardiovasc. Health 2015, 5, 74. [Google Scholar] [CrossRef]
- Korkes, H.A.; Sass, N.; Moron, A.F.; Camara, N.O.; Bonetti, T.; Cerdeira, A.S.; Da Silva, I.D.; De Oliveira, L. Lipidomic assessment of plasma and placenta of women with early-onset preeclampsia. PLoS ONE 2014, 9, e110747. [Google Scholar] [CrossRef]
- Knopp, R.H.; Magee, M.S.; Walden, C.E.; Bonet, B.; Benedetti, T.J. Prediction of infant birth weight by GDM screening tests. Importance of plasma triglyceride. Diabetes Care 1992, 15, 1605–1613. [Google Scholar] [CrossRef] [PubMed]
- Nolan, C.J.; Riley, S.F.; Sheedy, M.T.; Walstab, J.E.; Beischer, N.A. Maternal serum triglyceride, glucose tolerance, and neonatal birth weight ratio in pregnancy. Diabetes Care 1995, 18, 1550–1556. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Trewhella, M.A.; Fairclough, R.J.; Jenkin, G. Changes in uterine endometrial phospholipids and fatty acids throughout the oestrous cycle and early pregnancy in the ewe. Prostaglandins Leukot. Essent. Fat. Acids 1997, 57, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Colicino, E.; Ferrari, F.; Cowell, W.; Niedzwiecki, M.M.; Pedretti, N.F.; Joshi, A.; Wright, R.O.; Wright, R.J. Non-linear and non-additive associations between the pregnancy metabolome and birthweight. Environ. Int. 2021, 156, 106750. [Google Scholar] [CrossRef]
- Hallman, M.; Kulovich, M.; Kirkpatrick, E.; Sugarman, R.G.; Gluck, L. Phosphatidylinositol and phosphatidylglycerol in amniotic fluid: Indices of lung maturity. Am. J. Obstet. Gynecol. 1976, 125, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Houtkooper, R.H.; Vaz, F.M. Cardiolipin, the heart of mitochondrial metabolism. Cell. Mol. Life Sci. 2008, 65, 2493–2506. [Google Scholar] [CrossRef]
- Reddy, V.D.; Padmavathi, P.; Kavitha, G.; Saradamma, B.; Varadacharyulu, N. Alcohol-induced oxidative/nitrosative stress alters brain mitochondrial membrane properties. Mol. Cell. Biochem. 2013, 375, 39–47. [Google Scholar] [CrossRef]
- Torkhovskaya, T.I.; Zakharova, T.S.; Korotkevich, E.I.; Kasum-zade, N.K.; Shalina, R.I.; Markin, S.S. Blood Plasma Lipidome: Opportunities in the Early Diagnostics of Preeclampsia. Russ. J. Bioorganic Chem. 2020, 46, 280–286. [Google Scholar] [CrossRef]



| Lipid Class | %CV |
|---|---|
| Diacylglycerols | 13% |
| Sphingomyelins | 3% |
| Sterols | 7% |
| Triacylglycerols | 8% |
| Glycerophosphocholines | 5% |
| Glycerophosphoethanolamines | 8% |
| Glycerophosphoinositols | 11% |
| Glycerophosphoserines | 10% |
| Lipid Class | Number of Lipid Subclasses Affected by Alcohol | Effect |
|---|---|---|
| Glycerophosphoethanolamines (G-PE) | 37 | Downregulated |
| Glycerophosphocholines (G-PC) | 13 | 11 Downregulated, 2 upregulated |
| Triacylglycerols (TG) | 9 | 7 Downregulated, 2 upregulated |
| Phosphosphingolipids (SL) | 6 | 5 Downregulated, 1 upregulated |
| Glycerophosphoinositols (G-PI) | 6 | Downregulated |
| Glycerophosphoserines (G-PS) | 1 | Downregulated |
| Sterols | 1 | Upregulated |
| Total | 73 |
| Lipid Species | Lipid Subspecies Altered by Alcohol | Effect |
|---|---|---|
| Phosphatidic Acid (PA) | 4 | Downregulated |
| Phosphatidylcholine (PC) | 8 | Downregulated |
| Phosphatidylethanolamine (PE) | 17 | 16 Downregulated, 1 upregulated |
| Phosphatidylglycerol (PG) | 19 | Upregulated |
| Phosphatidylinositol (PI) | 3 | 2 Downregulated, 1 upregulated |
| Phosphatidylserine (PS) | 6 | Downregulated |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naik, V.D.; Ramadoss, J. Untargeted and Targeted Blood Lipidomic Signature Profile of Gestational Alcohol Exposure. Nutrients 2023, 15, 1411. https://doi.org/10.3390/nu15061411
Naik VD, Ramadoss J. Untargeted and Targeted Blood Lipidomic Signature Profile of Gestational Alcohol Exposure. Nutrients. 2023; 15(6):1411. https://doi.org/10.3390/nu15061411
Chicago/Turabian StyleNaik, Vishal D., and Jayanth Ramadoss. 2023. "Untargeted and Targeted Blood Lipidomic Signature Profile of Gestational Alcohol Exposure" Nutrients 15, no. 6: 1411. https://doi.org/10.3390/nu15061411
APA StyleNaik, V. D., & Ramadoss, J. (2023). Untargeted and Targeted Blood Lipidomic Signature Profile of Gestational Alcohol Exposure. Nutrients, 15(6), 1411. https://doi.org/10.3390/nu15061411