Transcriptomics Dissection of Calorie Restriction and Exercise Training in Brown Adipose Tissue and Skeletal Muscle
Abstract
1. Introduction
2. Materials and Methods
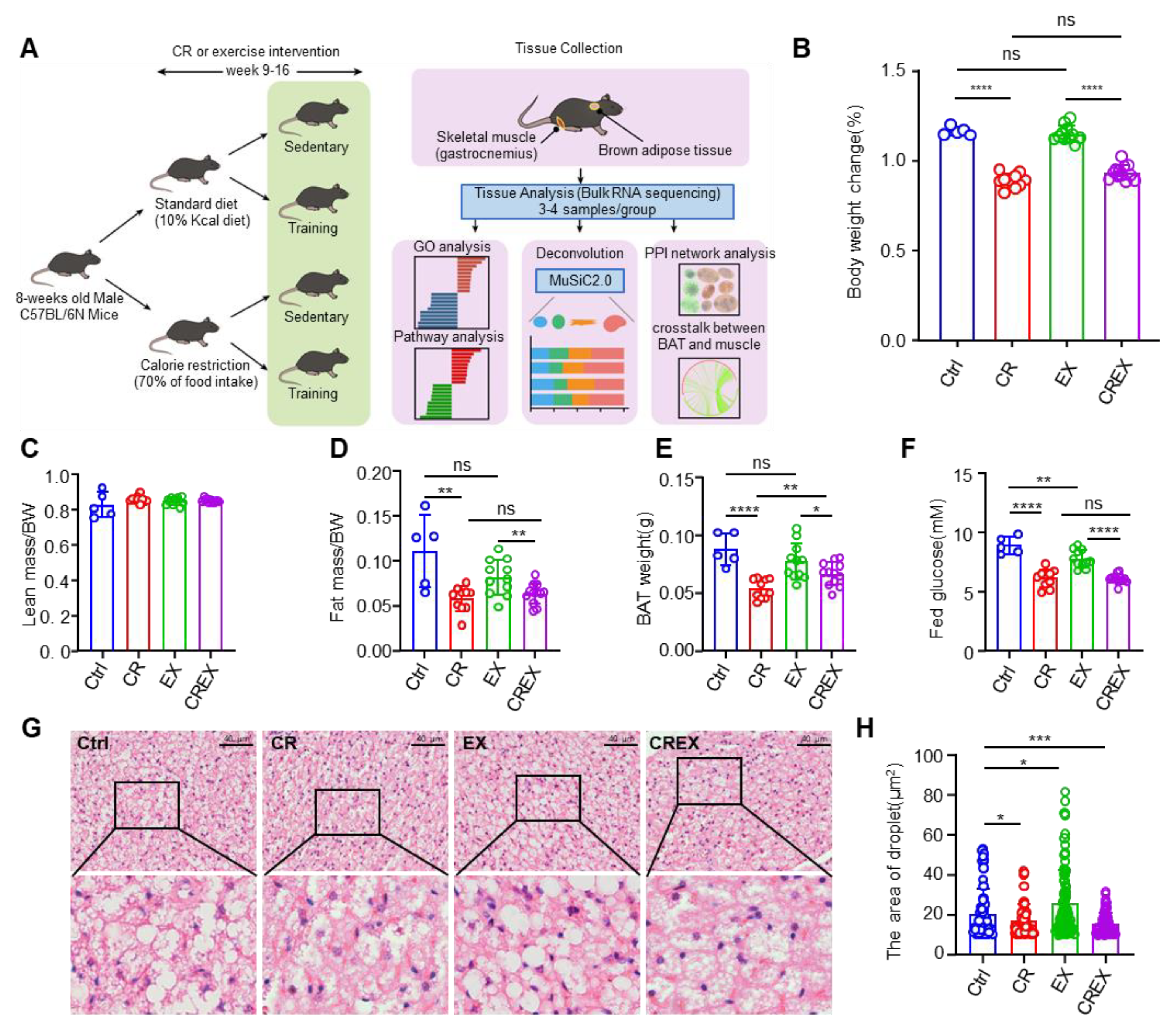
2.1. Experiment Model and Subject Details
2.2. CR
2.3. Chronic Treadmill Exercise Training
2.4. Body Composition Measurement
2.5. H&E Staining
2.6. Bulk mRNA Sequencing
2.7. Sequence Alignment and Gene Expression Analysis
2.8. Adipokine and Myokine Analysis
2.9. Deconvolution
2.10. BAT and Skeletal Muscle Tissue Crosstalk
2.11. Quantification and Statistical Analysis
3. Results
3.1. Phenotypic Response to CR and/or EX and Profiling of Two Metabolic Tissues
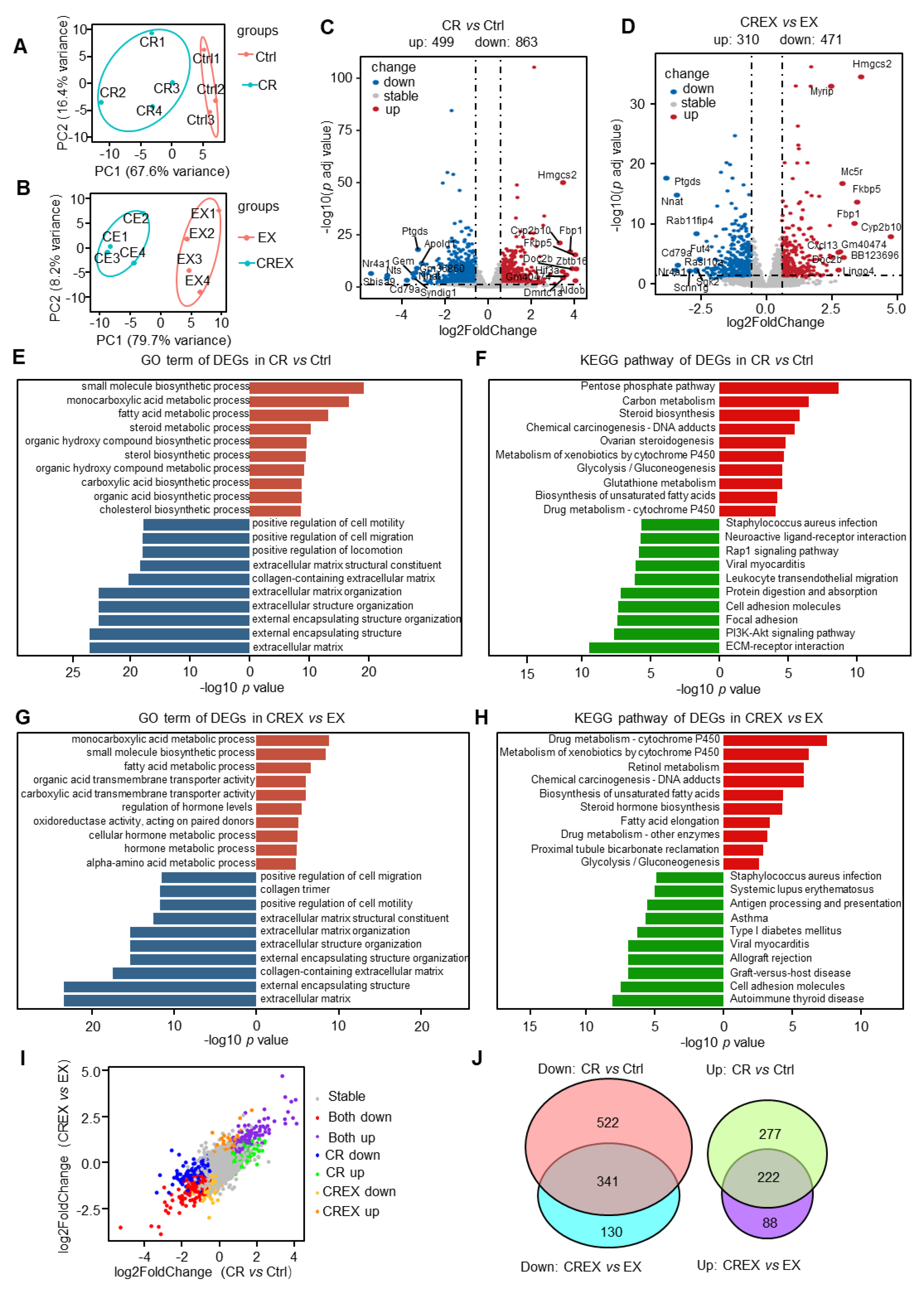
3.2. BAT-Level Gene and Pathway Alterations upon CR with or without EX
3.3. BAT-Level Gene Alteration upon EX with or without CR
3.4. Analysis of DEGs Encoding Adipokines upon CR with or without EX
3.5. Skeletal Muscle-Level Gene and Pathway Alterations upon CR with or without EX
3.6. Skeletal Muscle-Level Gene and Pathway Alterations upon EX with or without CR
3.7. Analysis of DEGs Encoding Myokines upon CR with or without EX
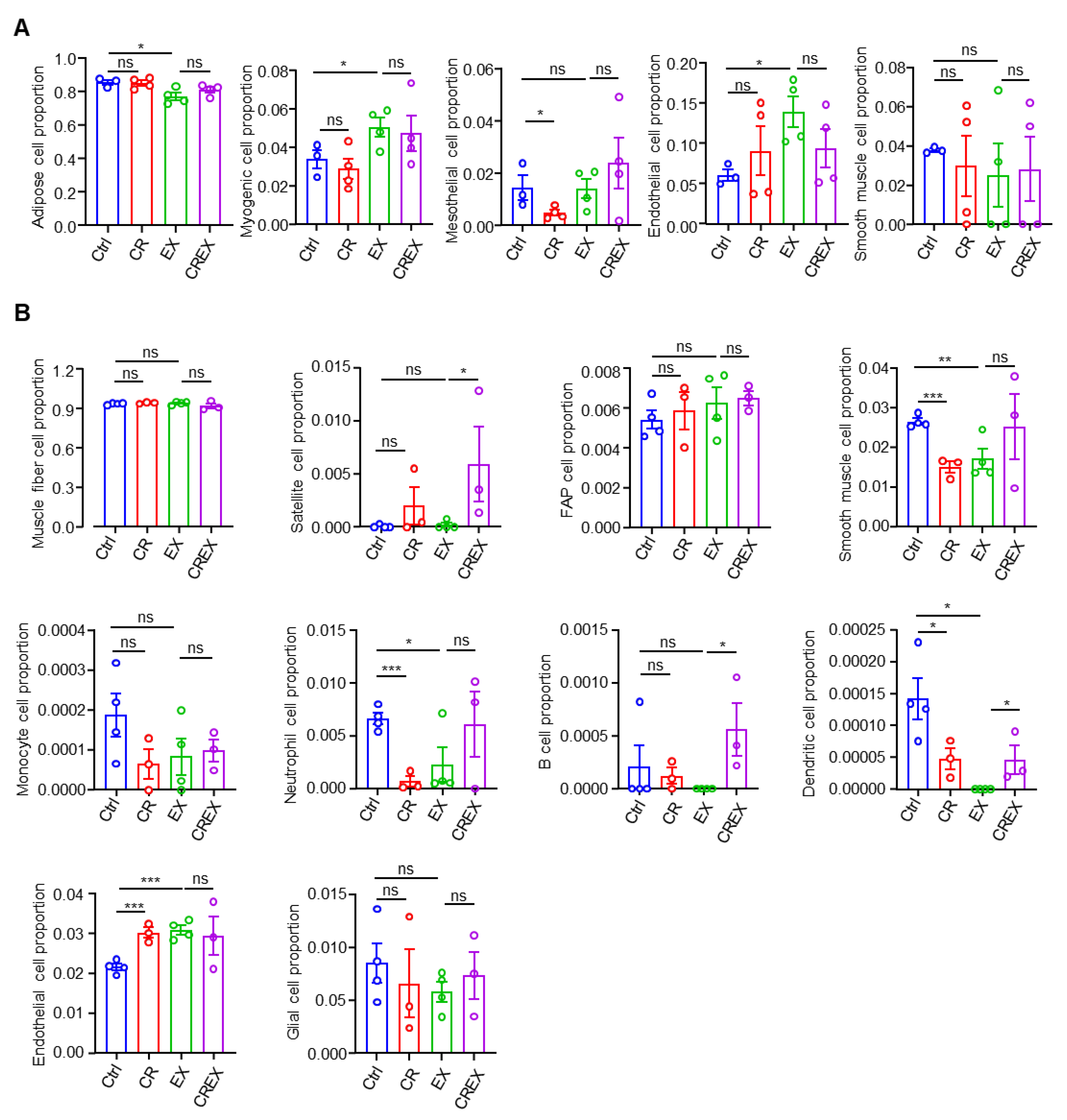
3.8. Cell Proportion Alterations resulting from CR and/or EX across the Two Tissues
3.9. The Crosstalk between BAT and Muscle upon CR with or without EX
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kong, X.; Yao, T.; Zhou, P.; Kazak, L.; Tenen, D.; Lyubetskaya, A.; Dawes, B.A.; Tsai, L.; Kahn, B.B.; Spiegelman, B.M.; et al. Brown Adipose Tissue Controls Skeletal Muscle Function via the Secretion of Myostatin. Cell Metab. 2018, 28, 631–643.e3. [Google Scholar] [CrossRef] [PubMed]
- Timmons, J.A.; Wennmalm, K.; Larsson, O.; Walden, T.B.; Lassmann, T.; Petrovic, N.; Hamilton, D.L.; Gimeno, R.E.; Wahlestedt, C.; Baar, K.; et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc. Natl. Acad. Sci. USA 2007, 104, 4401–4406. [Google Scholar] [CrossRef] [PubMed]
- Bal, N.C.; Maurya, S.K.; Pani, S.; Sethy, C.; Banerjee, A.; Das, S.; Patnaik, S.; Kundu, C.N. Mild cold induced thermogenesis: Are BAT and skeletal muscle synergistic partners? Biosci. Rep. 2017, 37, BSR20171087. [Google Scholar] [CrossRef] [PubMed]
- Aydin, J.; Shabalina, I.G.; Place, N.; Reiken, S.; Zhang, S.J.; Bellinger, A.M.; Nedergaard, J.; Cannon, B.; Marks, A.R.; Bruton, J.D.; et al. Nonshivering thermogenesis protects against defective calcium handling in muscle. FASEB J. 2008, 22, 3919–3924. [Google Scholar] [CrossRef] [PubMed]
- Dawkins, M.J.; Scopes, J.W. Non-shivering thermogenesis and brown adipose tissue in the human new-born infant. Nature 1965, 206, 201–202. [Google Scholar] [CrossRef]
- Enerback, S.; Jacobsson, A.; Simpson, E.M.; Guerra, C.; Yamashita, H.; Harper, M.E.; Kozak, L.P. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 1997, 387, 90–94. [Google Scholar] [CrossRef]
- Kong, X.; Banks, A.; Liu, T.; Kazak, L.; Rao, R.R.; Cohen, P.; Wang, X.; Yu, S.; Lo, J.C.; Tseng, Y.H.; et al. IRF4 is a key thermogenic transcriptional partner of PGC-1alpha. Cell 2014, 158, 69–83. [Google Scholar] [CrossRef]
- Colman, R.J.; Beasley, T.M.; Kemnitz, J.W.; Johnson, S.C.; Weindruch, R.; Anderson, R.M. Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat. Commun. 2014, 5, 3557. [Google Scholar] [CrossRef]
- Wei, M.; Fabrizio, P.; Hu, J.; Ge, H.; Cheng, C.; Li, L.; Longo, V.D. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008, 4, e13. [Google Scholar] [CrossRef]
- Bordone, L.; Guarente, L. Calorie restriction, SIRT1 and metabolism: Understanding longevity. Nat. Rev. Mol. Cell Biol. 2005, 6, 298–305. [Google Scholar] [CrossRef]
- Cerletti, M.; Jang, Y.C.; Finley, L.W.; Haigis, M.C.; Wagers, A.J. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell 2012, 10, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.G.; Zhong, L.; Xu, Z.J.; Tia, L.Y.; Ding, X.D.; Li, M.S.; Wang, G.L. Effects of low-calorie diet on steatohepatitis in rats with obesity and hyperlipidemia. World J. Gastroenterol. 2003, 9, 2045–2049. [Google Scholar] [CrossRef] [PubMed]
- Park, C.Y.; Park, S.; Kim, M.S.; Kim, H.K.; Han, S.N. Effects of mild calorie restriction on lipid metabolism and inflammation in liver and adipose tissue. Biochem. Biophys. Res. Commun. 2017, 490, 636–642. [Google Scholar] [CrossRef]
- Fabbiano, S.; Suarez-Zamorano, N.; Rigo, D.; Veyrat-Durebex, C.; Stevanovic Dokic, A.; Colin, D.J.; Trajkovski, M. Caloric Restriction Leads to Browning of White Adipose Tissue through Type 2 Immune Signaling. Cell Metab. 2016, 24, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Palou, M.; Priego, T.; Romero, M.; Szostaczuk, N.; Konieczna, J.; Cabrer, C.; Remesar, X.; Palou, A.; Pico, C. Moderate calorie restriction during gestation programs offspring for lower BAT thermogenic capacity driven by thyroid and sympathetic signaling. Int. J. Obes. 2015, 39, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Bonadonna, R.C.; Del Prato, S.; Saccomani, M.P.; Bonora, E.; Gulli, G.; Ferrannini, E.; Bier, D.; Cobelli, C.; DeFronzo, R.A. Transmembrane glucose transport in skeletal muscle of patients with non-insulin-dependent diabetes. J. Clin. Investig. 1993, 92, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Goodyear, L.J.; Kahn, B.B. Exercise, glucose transport, and insulin sensitivity. Annu. Rev. Med. 1998, 49, 235–261. [Google Scholar] [CrossRef]
- Dewal, R.S.; Stanford, K.I. Effects of exercise on brown and beige adipocytes. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 71–78. [Google Scholar] [CrossRef]
- Stanford, K.I.; Middelbeek, R.J.; Goodyear, L.J. Exercise Effects on White Adipose Tissue: Beiging and Metabolic Adaptations. Diabetes 2015, 64, 2361–2368. [Google Scholar] [CrossRef]
- Bostrom, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Bostrom, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef]
- Rao, R.R.; Long, J.Z.; White, J.P.; Svensson, K.J.; Lou, J.; Lokurkar, I.; Jedrychowski, M.P.; Ruas, J.L.; Wrann, C.D.; Lo, J.C.; et al. Meteorin-like is a hormone that regulates immune-adipose interactions to increase beige fat thermogenesis. Cell 2014, 157, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.D.; Bostrom, P.; O’Sullivan, J.F.; Schinzel, R.T.; Lewis, G.D.; Dejam, A.; Lee, Y.K.; Palma, M.J.; Calhoun, S.; Georgiadi, A.; et al. beta-Aminoisobutyric acid induces browning of white fat and hepatic beta-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab. 2014, 19, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, K.; Yoshida, T.; Wakabayashi, Y.; Nishioka, H.; Kondo, M. Effects of exercise training on brown adipose tissue thermogenesis in ovariectomized obese rats. Endocrinol. Jpn. 1989, 36, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Stanford, K.I.; Goodyear, L.J. Exercise regulation of adipose tissue. Adipocyte 2016, 5, 153–162. [Google Scholar] [CrossRef]
- Ignacio, D.L.; Fortunato, R.S.; Neto, R.A.; da Silva Silvestre, D.H.; Nigro, M.; Frankenfeld, T.G.; Werneck-de-Castro, J.P.; Carvalho, D.P. Blunted response of pituitary type 1 and brown adipose tissue type 2 deiodinases to swimming training in ovariectomized rats. Horm. Metab. Res. 2012, 44, 797–803. [Google Scholar] [CrossRef]
- Wu, M.V.; Bikopoulos, G.; Hung, S.; Ceddia, R.B. Thermogenic capacity is antagonistically regulated in classical brown and white subcutaneous fat depots by high fat diet and endurance training in rats: Impact on whole-body energy expenditure. J. Biol. Chem. 2014, 289, 34129–34140. [Google Scholar] [CrossRef]
- Vosselman, M.J.; Hoeks, J.; Brans, B.; Pallubinsky, H.; Nascimento, E.B.; van der Lans, A.A.; Broeders, E.P.; Mottaghy, F.M.; Schrauwen, P.; van Marken Lichtenbelt, W.D. Low brown adipose tissue activity in endurance-trained compared with lean sedentary men. Int. J. Obes. 2015, 39, 1696–1702. [Google Scholar] [CrossRef]
- Motiani, P.; Virtanen, K.A.; Motiani, K.K.; Eskelinen, J.J.; Middelbeek, R.J.; Goodyear, L.J.; Savolainen, A.M.; Kemppainen, J.; Jensen, J.; Din, M.U.; et al. Decreased insulin-stimulated brown adipose tissue glucose uptake after short-term exercise training in healthy middle-aged men. Diabetes Obes. Metab. 2017, 19, 1379–1388. [Google Scholar] [CrossRef]
- Hargreaves, M.; Spriet, L.L. Skeletal muscle energy metabolism during exercise. Nat. Metab. 2020, 2, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, B.K.; Febbraio, M.A. Muscles, exercise and obesity: Skeletal muscle as a secretory organ. Nat. Rev. Endocrinol. 2012, 8, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Aldiss, P.; Betts, J.; Sale, C.; Pope, M.; Budge, H.; Symonds, M.E. Exercise-induced ‘browning’ of adipose tissues. Metabolism 2018, 81, 63–70. [Google Scholar] [CrossRef]
- Green, C.L.; Mitchell, S.E.; Derous, D.; Wang, Y.; Chen, L.; Han, J.J.; Promislow, D.E.L.; Lusseau, D.; Douglas, A.; Speakman, J.R. The Effects of Graded Levels of Calorie Restriction: XIV. Global Metabolomics Screen Reveals Brown Adipose Tissue Changes in Amino Acids, Catecholamines, and Antioxidants After Short-Term Restriction in C57BL/6 Mice. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 218–229. [Google Scholar] [CrossRef]
- Yao, T.; Yan, H.; Zhu, X.; Zhang, Q.; Kong, X.; Guo, S.; Feng, Y.; Wang, H.; Hua, Y.; Zhang, J.; et al. Obese Skeletal Muscle-Expressed Interferon Regulatory Factor 4 Transcriptionally Regulates Mitochondrial Branched-Chain Aminotransferase Reprogramming Metabolome. Diabetes 2022, 71, 2256–2271. [Google Scholar] [CrossRef] [PubMed]
- Villarroya, F.; Cereijo, R.; Villarroya, J.; Giralt, M. Brown adipose tissue as a secretory organ. Nat. Rev. Endocrinol. 2017, 13, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cushman, S.W.; Pannell, L.K.; Hess, S. Quantitative proteomic analysis of the secretory proteins from rat adipose cells using a 2D liquid chromatography-MS/MS approach. J. Proteome Res. 2005, 4, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Mariman, E.; Keijer, J.; Bouwman, F.; Noben, J.P.; Robben, J.; Renes, J. Profiling of the secreted proteins during 3T3-L1 adipocyte differentiation leads to the identification of novel adipokines. Cell. Mol. Life Sci. 2004, 61, 2405–2417. [Google Scholar] [CrossRef] [PubMed]
- Lehr, S.; Hartwig, S.; Lamers, D.; Famulla, S.; Muller, S.; Hanisch, F.G.; Cuvelier, C.; Ruige, J.; Eckardt, K.; Ouwens, D.M.; et al. Identification and validation of novel adipokines released from primary human adipocytes. Mol. Cell. Proteom. 2012, 11, M111.010504. [Google Scholar] [CrossRef]
- Hartwig, S.; Raschke, S.; Knebel, B.; Scheler, M.; Irmler, M.; Passlack, W.; Muller, S.; Hanisch, F.G.; Franz, T.; Li, X.; et al. Secretome profiling of primary human skeletal muscle cells. Biochim. Biophys. Acta 2014, 1844, 1011–1017. [Google Scholar] [CrossRef]
- Henningsen, J.; Rigbolt, K.T.; Blagoev, B.; Pedersen, B.K.; Kratchmarova, I. Dynamics of the skeletal muscle secretome during myoblast differentiation. Mol. Cell. Proteom. 2010, 9, 2482–2496. [Google Scholar] [CrossRef] [PubMed]
- Bortoluzzi, S.; Scannapieco, P.; Cestaro, A.; Danieli, G.A.; Schiaffino, S. Computational reconstruction of the human skeletal muscle secretome. Proteins 2006, 62, 776–792. [Google Scholar] [CrossRef]
- Le Bihan, M.C.; Bigot, A.; Jensen, S.S.; Dennis, J.L.; Rogowska-Wrzesinska, A.; Laine, J.; Gache, V.; Furling, D.; Jensen, O.N.; Voit, T.; et al. In-depth analysis of the secretome identifies three major independent secretory pathways in differentiating human myoblasts. J. Proteom. 2012, 77, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, S.; Jokinen, R.; Rissanen, A.; Pietiläinen, K.H. White adipose tissue mitochondrial metabolism in health and in obesity. Obes. Rev. 2020, 21, e12958. [Google Scholar] [CrossRef] [PubMed]
- Herz, C.T.; Kiefer, F.W. Adipose tissue browning in mice and humans. J. Endocrinol. 2019, 241, R97–R109. [Google Scholar] [CrossRef]
- Yudasaka, M.; Okamatsu-Ogura, Y.; Tanaka, T.; Saeki, K.; Kataura, H. Cold-induced Conversion of Connective Tissue Skeleton in Brown Adipose Tissues. Acta Histochem. Cytochem. 2021, 54, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Heeren, J.; Scheja, L. Brown adipose tissue and lipid metabolism. Curr. Opin. Lipidol. 2018, 29, 180–185. [Google Scholar] [CrossRef]
- Okita, N.; Hayashida, Y.; Kojima, Y.; Fukushima, M.; Yuguchi, K.; Mikami, K.; Yamauchi, A.; Watanabe, K.; Noguchi, M.; Nakamura, M.; et al. Differential responses of white adipose tissue and brown adipose tissue to caloric restriction in rats. Mech. Ageing Dev. 2012, 133, 255–266. [Google Scholar] [CrossRef]
- Ham, D.J.; Borsch, A.; Chojnowska, K.; Lin, S.; Leuchtmann, A.B.; Ham, A.S.; Thurkauf, M.; Delezie, J.; Furrer, R.; Burri, D.; et al. Distinct and additive effects of calorie restriction and rapamycin in aging skeletal muscle. Nat. Commun. 2022, 13, 2025. [Google Scholar] [CrossRef]
- Gordon, P.M.; Liu, D.; Sartor, M.A.; IglayReger, H.B.; Pistilli, E.E.; Gutmann, L.; Nader, G.A.; Hoffman, E.P. Resistance exercise training influences skeletal muscle immune activation: A microarray analysis. J. Appl. Physiol. 2012, 112, 443–453. [Google Scholar] [CrossRef]
- Lecker, S.H.; Jagoe, R.T.; Gilbert, A.; Gomes, M.; Baracos, V.; Bailey, J.; Price, S.R.; Mitch, W.E.; Goldberg, A.L. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004, 18, 39–51. [Google Scholar] [CrossRef]
- Magkos, F.; Hjorth, M.F.; Astrup, A. Diet and exercise in the prevention and treatment of type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2020, 16, 545–555. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Y.; Cui, Z.; Lu, X.; Gong, H.; Liu, X.; Wang, H.; Cheng, H.; Gao, H.; Shi, X.; Li, Y.; et al. Transcriptomics Dissection of Calorie Restriction and Exercise Training in Brown Adipose Tissue and Skeletal Muscle. Nutrients 2023, 15, 1047. https://doi.org/10.3390/nu15041047
Feng Y, Cui Z, Lu X, Gong H, Liu X, Wang H, Cheng H, Gao H, Shi X, Li Y, et al. Transcriptomics Dissection of Calorie Restriction and Exercise Training in Brown Adipose Tissue and Skeletal Muscle. Nutrients. 2023; 15(4):1047. https://doi.org/10.3390/nu15041047
Chicago/Turabian StyleFeng, Yonghao, Zhicheng Cui, Xiaodan Lu, Hongyu Gong, Xiaoyu Liu, Hui Wang, Haoyu Cheng, Huanqing Gao, Xiaohong Shi, Yiming Li, and et al. 2023. "Transcriptomics Dissection of Calorie Restriction and Exercise Training in Brown Adipose Tissue and Skeletal Muscle" Nutrients 15, no. 4: 1047. https://doi.org/10.3390/nu15041047
APA StyleFeng, Y., Cui, Z., Lu, X., Gong, H., Liu, X., Wang, H., Cheng, H., Gao, H., Shi, X., Li, Y., Ye, H., Zhang, Q., & Kong, X. (2023). Transcriptomics Dissection of Calorie Restriction and Exercise Training in Brown Adipose Tissue and Skeletal Muscle. Nutrients, 15(4), 1047. https://doi.org/10.3390/nu15041047







